Diagnostic Potential of the Plasma Lipidome in Infectious Disease: Application to Acute SARS-CoV-2 Infection
Abstract
:1. Introduction
2. Methods
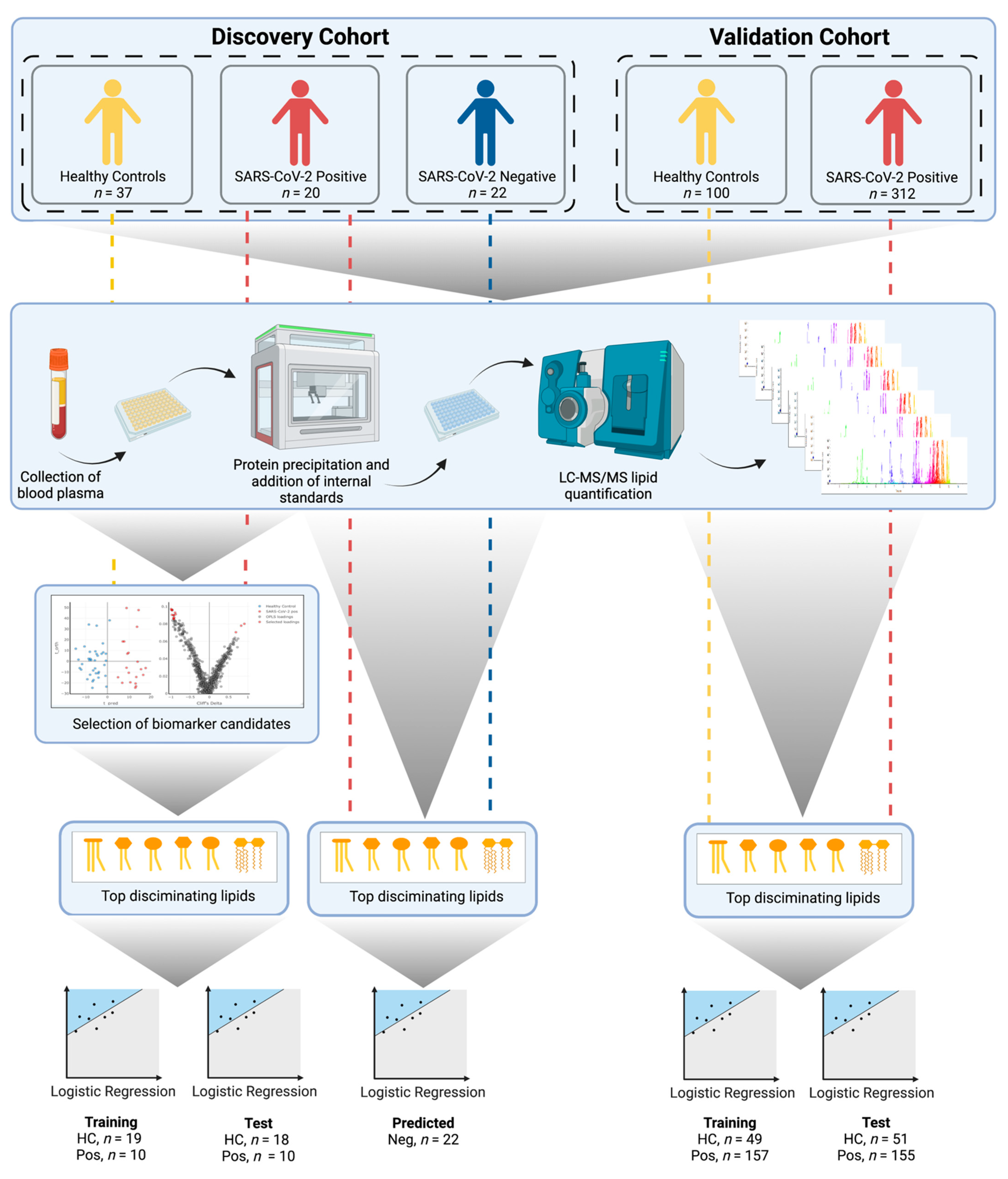
2.1. Discovery Cohort Patients and Sample Collection (Australia)
2.2. Validation Cohort Patients and Sample Collection (Spain)
2.3. LC–MS/MS Lipid Analysis
2.4. Data Pre-Processing and Quality Control
2.5. Statistics
3. Results
3.1. Lipid Analysis
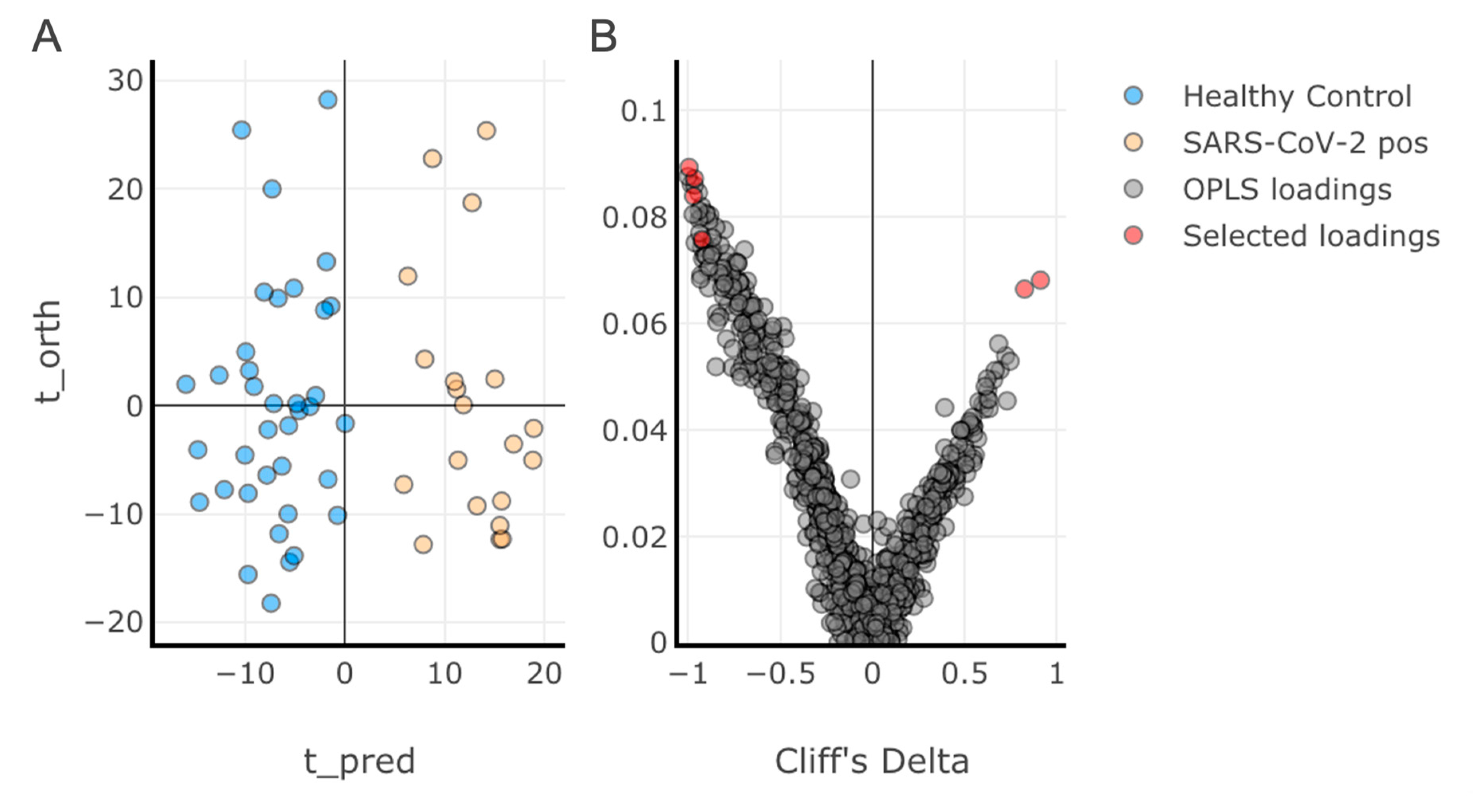
3.2. Multivariate Analysis of the Discovery Cohort and Identification of Individual Lipid Species as Biomarker Candidates
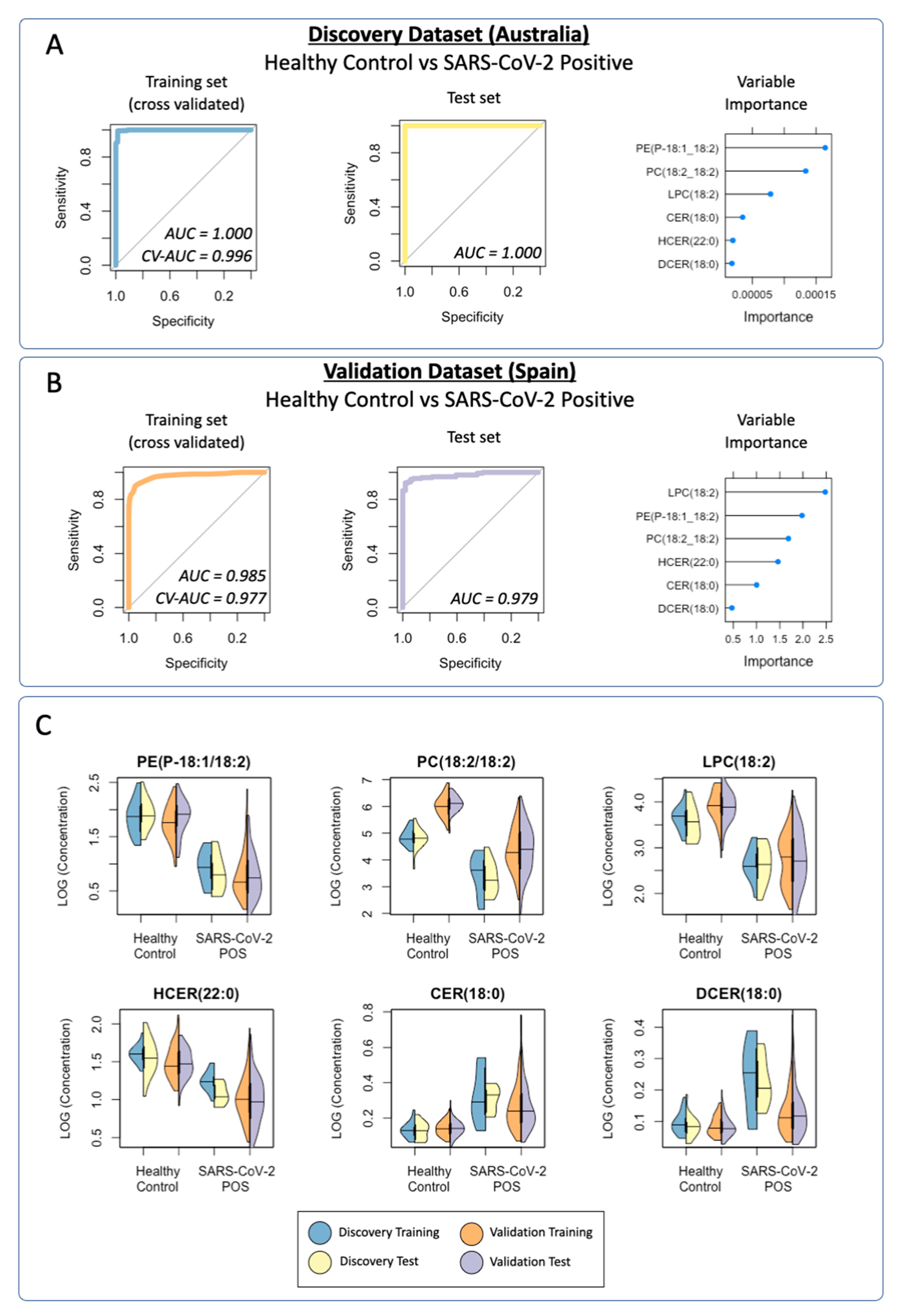
3.3. Diagnostic Performance of the Lipid Panel in the Discovery and Validation Cohorts
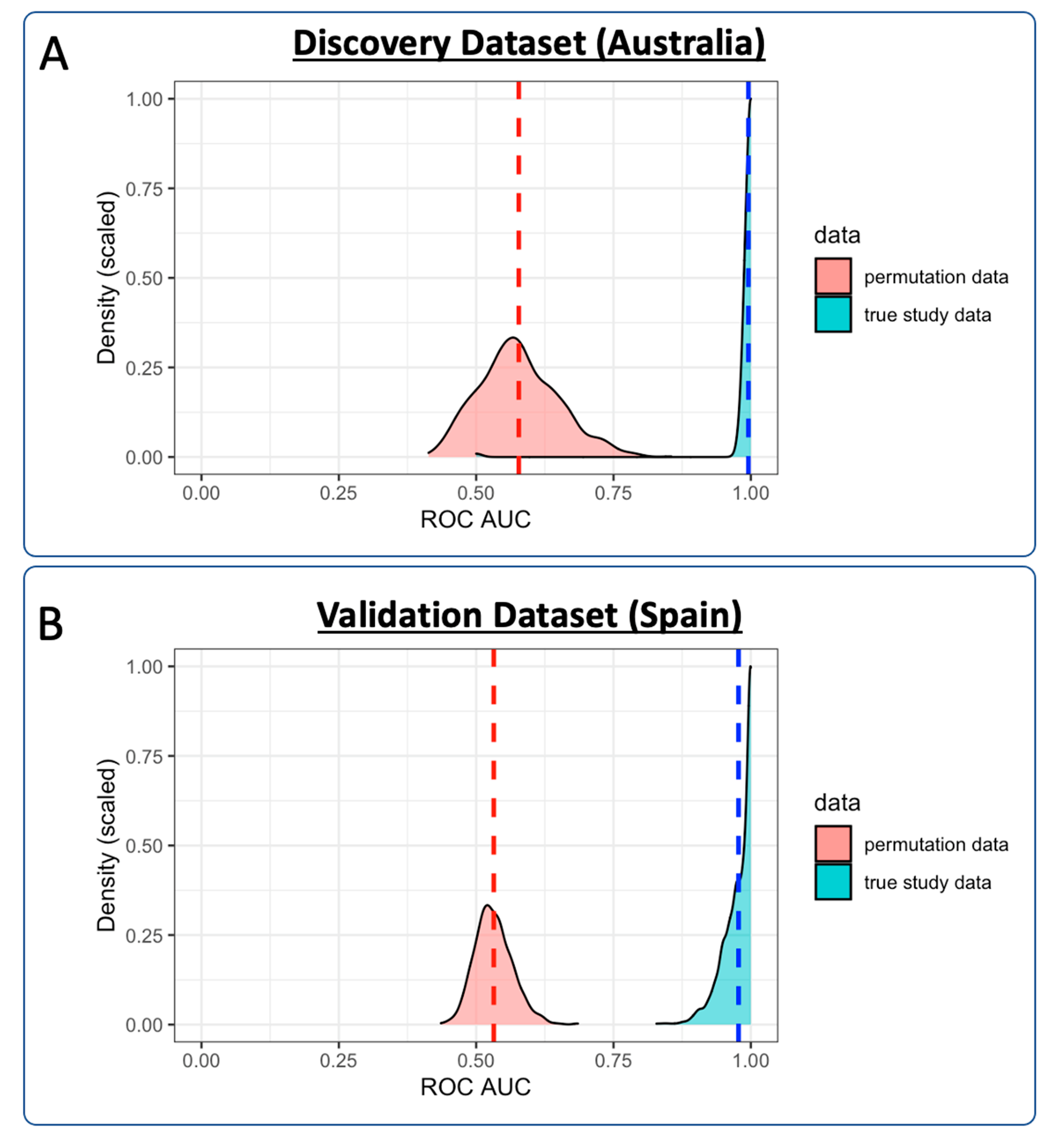
3.3.1. Discovery Cohort Training Model: SARS-CoV-2-Positive vs. Healthy Controls
3.3.2. Prediction of the Discovery Cohort Test Data Using the Cross-Validated Training Model: SARS-CoV-2-Positive vs. Healthy Control
3.3.3. Prediction of the Discovery Cohort Test Data Using the Cross-Validated Training Model: SARS-CoV-2-Negative
3.3.4. Validation Cohort Training Model: SARS-CoV-2-Positive vs. Healthy Controls
3.3.5. Prediction of the Validation Cohort Test Data Using the Cross-Validated Training Model: SARS-CoV-2-Positive vs. Healthy Controls
3.3.6. Visualization of the Distribution of the Lipid Classification Panel in SARS-CoV-2 Infection
3.3.7. Effects of Age and Sex on Lipid Concentrations
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kucirka, L.M.; Lauer, S.A.; Laeyendecker, O.; Boon, D.; Lessler, J. Variation in False-Negative Rate of Reverse Transcriptase Polymerase Chain Reaction-Based SARS-CoV-2 Tests by Time Since Exposure. Ann. Intern. Med. 2020, 173, 262–267. [Google Scholar] [CrossRef]
- Ismail, I.T.; Showalter, M.R.; Fiehn, O. Inborn Errors of Metabolism in the Era of Untargeted Metabolomics and Lipidomics. Metabolites 2019, 9, 242. [Google Scholar] [CrossRef]
- Miller, M.J.; Kennedy, A.D.; Eckhart, A.D.; Burrage, L.C.; Wulff, J.E.; Miller, L.A.D.; Milburn, M.V.; Ryals, J.A.; Beaudet, A.L.; Sun, Q.; et al. Untargeted Metabolomic Analysis for the Clinical Screening of Inborn Errors of Metabolism. J. Inherit. Metab. Dis. 2015, 38, 1029–1039. [Google Scholar] [CrossRef]
- Piraud, M.; Ruet, S.; Boyer, S.; Acquaviva, C.; Clerc-Renaud, P.; Cheillan, D.; Vianey-Saban, C. Amino Acid Profiling for the Diagnosis of Inborn Errors of Metabolism. Methods Mol. Biol. 2011, 708, 25–53. [Google Scholar]
- Fernández-García, M.; Rojo, D.; Rey-Stolle, F.; García, A.; Barbas, C. Metabolomic-Based Methods in Diagnosis and Monitoring Infection Progression. Experientia Suppl. 2018, 109, 283–315. [Google Scholar]
- Kimhofer, T.; Lodge, S.; Whiley, L.; Gray, N.; Loo, R.L.; Lawler, N.G.; Nitschke, P.; Bong, S.-H.; Morrison, D.L.; Begum, S.; et al. Integrative Modeling of Quantitative Plasma Lipoprotein, Metabolic, and Amino Acid Data Reveals a Multiorgan Pathological Signature of SARS-CoV-2 Infection. J. Proteome Res. 2020, 19, 4442–4454. [Google Scholar] [CrossRef] [PubMed]
- Lawler, N.G.; Gray, N.; Kimhofer, T.; Boughton, B.; Gay, M.; Yang, R.; Morillon, A.-C.; Chin, S.-T.; Ryan, M.; Begum, S.; et al. Systemic Perturbations in Amine and Kynurenine Metabolism Associated with Acute SARS-CoV-2 Infection and Inflammatory Cytokine Responses. J. Proteome Res. 2021, 20, 2796–2811. [Google Scholar] [CrossRef] [PubMed]
- Blasco, H.; Bessy, C.; Plantier, L.; Lefevre, A.; Piver, E.; Bernard, L.; Marlet, J.; Stefic, K.; Benz-de Bretagne, I.; Cannet, P.; et al. The Specific Metabolome Profiling of Patients Infected by SARS-COV-2 Supports the Key Role of Tryptophan-Nicotinamide Pathway and Cytosine Metabolism. Sci. Rep. 2020, 10, 16824. [Google Scholar] [CrossRef]
- Overmyer, K.A.; Shishkova, E.; Miller, I.J.; Balnis, J.; Bernstein, M.N.; Peters-Clarke, T.M.; Meyer, J.G.; Quan, Q.; Muehlbauer, L.K.; Trujillo, E.A.; et al. Large-Scale Multi-Omic Analysis of COVID-19 Severity. Cell Syst. 2021, 12, 23–40.e7. [Google Scholar] [CrossRef] [PubMed]
- Thomas, T.; Stefanoni, D.; Reisz, J.A.; Nemkov, T.; Bertolone, L.; Francis, R.O.; Hudson, K.E.; Zimring, J.C.; Hansen, K.C.; Hod, E.A.; et al. COVID-19 Infection Alters Kynurenine and Fatty Acid Metabolism, Correlating with IL-6 Levels and Renal Status. JCI Insight 2020, 5. [Google Scholar] [CrossRef]
- Lodge, S.; Nitschke, P.; Kimhofer, T.; Coudert, J.D.; Begum, S.; Bong, S.-H.; Richards, T.; Edgar, D.; Raby, E.; Spraul, M.; et al. NMR Spectroscopic Windows on the Systemic Effects of SARS-CoV-2 Infection on Plasma Lipoproteins and Metabolites in Relation to Circulating Cytokines. J. Proteome Res. 2021, 20, 1382–1396. [Google Scholar] [CrossRef] [PubMed]
- Bruzzone, C.; Bizkarguenaga, M.; Gil-Redondo, R.; Diercks, T.; Arana, E.; García de Vicuña, A.; Seco, M.; Bosch, A.; Palazón, A.; San Juan, I.; et al. SARS-CoV-2 Infection Dysregulates the Metabolomic and Lipidomic Profiles of Serum. iScience 2020, 23, 101645. [Google Scholar] [CrossRef] [PubMed]
- Bley, H.; Schöbel, A.; Herker, E. Whole Lotta Lipids-from HCV RNA Replication to the Mature Viral Particle. Int. J. Mol. Sci. 2020, 21, 2888. [Google Scholar] [CrossRef] [PubMed]
- Eisfeld, A.J.; Halfmann, P.J.; Wendler, J.P.; Kyle, J.E.; Burnum-Johnson, K.E.; Peralta, Z.; Maemura, T.; Walters, K.B.; Watanabe, T.; Fukuyama, S.; et al. Multi-Platform ’Omics Analysis of Human Ebola Virus Disease Pathogenesis. Cell Host Microbe 2017, 22, 817–829.e8. [Google Scholar] [CrossRef] [PubMed]
- Kyle, J.E.; Burnum-Johnson, K.E.; Wendler, J.P.; Eisfeld, A.J.; Halfmann, P.J.; Watanabe, T.; Sahr, F.; Smith, R.D.; Kawaoka, Y.; Waters, K.M.; et al. Plasma Lipidome Reveals Critical Illness and Recovery from Human Ebola Virus Disease. Proc. Natl. Acad. Sci. USA 2019, 116, 3919–3928. [Google Scholar] [CrossRef] [PubMed]
- Funderburg, N.T.; Mehta, N.N. Lipid Abnormalities and Inflammation in HIV Inflection. Curr. HIV/AIDS Rep. 2016, 13, 218–225. [Google Scholar] [CrossRef] [PubMed]
- Negro, F. Abnormalities of Lipid Metabolism in Hepatitis C Virus Infection. Gut 2010, 59, 1279–1287. [Google Scholar] [CrossRef]
- Melo, C.F.O.R.; de Oliveira, D.N.; Lima, E.D.O.; Guerreiro, T.M.; Esteves, C.Z.; Beck, R.M.; Padilla, M.A.; Milanez, G.P.; Arns, C.W.; Proença-Modena, J.L.; et al. A Lipidomics Approach in the Characterization of Zika-Infected Mosquito Cells: Potential Targets for Breaking the Transmission Cycle. PLoS ONE 2016, 11, e0164377. [Google Scholar] [CrossRef]
- Tisoncik-Go, J.; Gasper, D.J.; Kyle, J.E.; Eisfeld, A.J.; Selinger, C.; Hatta, M.; Morrison, J.; Korth, M.J.; Zink, E.M.; Kim, Y.-M.; et al. Integrated Omics Analysis of Pathogenic Host Responses during Pandemic H1N1 Influenza Virus Infection: The Crucial Role of Lipid Metabolism. Cell Host Microbe 2016, 19, 254–266. [Google Scholar] [CrossRef]
- Wu, Q.; Zhou, L.; Sun, X.; Yan, Z.; Hu, C.; Wu, J.; Xu, L.; Li, X.; Liu, H.; Yin, P.; et al. Altered Lipid Metabolism in Recovered SARS Patients Twelve Years after Infection. Sci. Rep. 2017, 7, 9110. [Google Scholar] [CrossRef]
- Yan, B.; Chu, H.; Yang, D.; Sze, K.-H.; Lai, P.-M.; Yuan, S.; Shuai, H.; Wang, Y.; Kao, R.Y.-T.; Chan, J.F.-W.; et al. Characterization of the Lipidomic Profile of Human Coronavirus-Infected Cells: Implications for Lipid Metabolism Remodeling upon Coronavirus Replication. Viruses 2019, 11, 73. [Google Scholar] [CrossRef] [PubMed]
- Bai, Y.; Huang, W.; Li, Y.; Lai, C.; Huang, S.; Wang, G.; He, Y.; Hu, L.; Chen, C. Lipidomic Alteration of Plasma in Cured COVID-19 Patients Using Ultra High-Performance Liquid Chromatography with High-Resolution Mass Spectrometry. Biosci. Rep. 2021, 41. [Google Scholar] [CrossRef] [PubMed]
- Barberis, E.; Timo, S.; Amede, E.; Vanella, V.V.; Puricelli, C.; Cappellano, G.; Raineri, D.; Cittone, M.G.; Rizzi, E.; Pedrinelli, A.R.; et al. Large-Scale Plasma Analysis Revealed New Mechanisms and Molecules Associated with the Host Response to SARS-CoV-2. Int. J. Mol. Sci. 2020, 21, 8623. [Google Scholar] [CrossRef] [PubMed]
- Caterino, M.; Gelzo, M.; Sol, S.; Fedele, R.; Annunziata, A.; Calabrese, C.; Fiorentino, G.; D’Abbraccio, M.; Dell’Isola, C.; Fusco, F.M.; et al. Dysregulation of Lipid Metabolism and Pathological Inflammation in Patients with COVID-19. Sci. Rep. 2021, 11, 2941. [Google Scholar] [CrossRef]
- Wu, D.; Shu, T.; Yang, X.; Song, J.-X.; Zhang, M.; Yao, C.; Liu, W.; Huang, M.; Yu, Y.; Yang, Q.; et al. Plasma Metabolomic and Lipidomic Alterations Associated with COVID-19. Natl. Sci. Rev. 2020, 7, 1157–1168. [Google Scholar] [CrossRef]
- Ren, Z.; Wang, H.; Cui, G.; Lu, H.; Wang, L.; Luo, H.; Chen, X.; Ren, H.; Sun, R.; Liu, W.; et al. Alterations in the Human Oral and Gut Microbiomes and Lipidomics in COVID-19. Gut 2021, 70, 1253–1265. [Google Scholar] [CrossRef]
- Song, J.W.; Lam, S.M.; Fan, X.; Cao, W.J.; Wang, S.Y.; Tian, H.; Chua, G.H.; Zhang, C.; Meng, F.P.; Xu, Z.; et al. Omics-Driven Systems Interrogation of Metabolic Dysregulation in COVID-19 Pathogenesis. Cell Metab. 2020, 32, 188–202 e5. [Google Scholar] [CrossRef]
- Adams, K.J.; Pratt, B.; Bose, N.; Dubois, L.G.; St John-Williams, L.; Perrott, K.M.; Ky, K.; Kapahi, P.; Sharma, V.; MacCoss, M.J.; et al. Skyline for Small Molecules: A Unifying Software Package for Quantitative Metabolomics. J. Proteome Res. 2020, 19, 1447–1458. [Google Scholar] [CrossRef]
- Storey, J.D. The Positive False Discovery Rate: A Bayesian Interpretation and the Q-Value. Ann. Stat. 2003, 31, 2013–2035. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Bravo-Merodio, L.; Acharjee, A.; Hazeldine, J.; Bentley, C.; Foster, M.; Gkoutos, G.V.; Lord, J.M. Machine Learning for the Detection of Early Immunological Markers as Predictors of Multi-Organ Dysfunction. Sci. Data 2019, 6, 328. [Google Scholar] [CrossRef]
- Heinze, G.; Wallisch, C.; Dunkler, D. Variable Selection—A Review and Recommendations for the Practicing Statistician. Biom. J. 2018, 60, 431–449. [Google Scholar] [CrossRef] [PubMed]
- Maile, M.D.; Standiford, T.J.; Engoren, M.C.; Stringer, K.A.; Jewell, E.S.; Rajendiran, T.M.; Soni, T.; Burant, C.F. Associations of the Plasma Lipidome with Mortality in the Acute Respiratory Distress Syndrome: A Longitudinal Cohort Study. Respir. Res. 2018, 19, 60. [Google Scholar] [CrossRef] [PubMed]
- Queiroz, A.; Pinto, I.F.D.; Lima, M.; Giovanetti, M.; de Jesus, J.G.; Xavier, J.; Barreto, F.K.; Canuto, G.A.B.; do Amaral, H.R.; de Filippis, A.M.B.; et al. Lipidomic Analysis Reveals Serum Alteration of Plasmalogens in Patients Infected with ZIKA Virus. Front. Microbiol. 2019, 10, 753. [Google Scholar] [CrossRef] [PubMed]
- Shan, J.; Qian, W.; Shen, C.; Lin, L.; Xie, T.; Peng, L.; Xu, J.; Yang, R.; Ji, J.; Zhao, X. High-Resolution Lipidomics Reveals Dysregulation of Lipid Metabolism in Respiratory Syncytial Virus Pneumonia Mice. RSC Adv. 2018, 8, 29368–29377. [Google Scholar] [CrossRef]
- Hurley, B.P.; McCormick, B.A. Multiple Roles of Phospholipase A2 during Lung Infection and Inflammation. Infect. Immun. 2008, 76, 2259–2272. [Google Scholar] [CrossRef]
- Li, Y.-F.; Li, R.-S.; Samuel, S.B.; Cueto, R.; Li, X.-Y.; Wang, H.; Yang, X.-F. Lysophospholipids and Their G Protein-Coupled Receptors in Atherosclerosis. Front. Biosci. 2016, 21, 70–88. [Google Scholar] [CrossRef]
- Grossmayer, G.E.; Keppeler, H.; Boeltz, S.; Janko, C.; Rech, J.; Herrmann, M.; Lauber, K.; Muñoz, L.E. Elevated Serum Lysophosphatidylcholine in Patients with Systemic Lupus Erythematosus Impairs Phagocytosis of Necrotic Cells In Vitro. Front. Immunol. 2017, 8, 1876. [Google Scholar] [CrossRef]
- Liu, Y.; Sawalha, A.H.; Lu, Q. COVID-19 and Autoimmune Diseases. Curr. Opin. Rheumatol. 2021, 33, 155–162. [Google Scholar] [CrossRef]
- Zhao, W.; Wang, X.; Deik, A.A.; Hanna, D.B.; Wang, T.; Haberlen, S.A.; Shah, S.J.; Lazar, J.M.; Hodis, H.N.; Landay, A.L.; et al. Elevated Plasma Ceramides Are Associated with Antiretroviral Therapy Use and Progression of Carotid Artery Atherosclerosis in HIV Infection. Circulation 2019, 139, 2003–2011. [Google Scholar] [CrossRef]
- Drobnik, W.; Liebisch, G.; Audebert, F.-X.; Frohlich, D.; Gluck, T.; Vogel, P.; Rothe, G.; Schmitz, G. Plasma Ceramide and Lysophosphatidylcholine Inversely Correlate with Mortality in Sepsis Patients. J. Lipid Res. 2003, 44, 754–761. [Google Scholar] [CrossRef]
- Cogolludo, A.; Villamor, E.; Perez-Vizcaino, F.; Moreno, L. Ceramide and Regulation of Vascular Tone. Int. J. Mol. Sci. 2019, 20, 411. [Google Scholar] [CrossRef]
- Tippetts, T.S.; Holland, W.L.; Summers, S.A. The Ceramide Ratio: A Predictor of Cardiometabolic Risk. J. Lipid Res. 2018, 59, 1549–1550. [Google Scholar] [CrossRef] [PubMed]
- Delogu, G.; Famularo, G.; Amati, F.; Signore, L.; Antonucci, A.; Trinchieri, V.; Di Marzio, L.; Cifone, M.G. Ceramide Concentrations in Septic Patients: A Possible Marker of Multiple Organ Dysfunction Syndrome. Crit. Care Med. 1999, 27, 2413–2417. [Google Scholar] [CrossRef] [PubMed]
- Lopes-Pacheco, M.; Silva, P.L.; Cruz, F.F.; Battaglini, D.; Robba, C.; Pelosi, P.; Morales, M.M.; Caruso Neves, C.; Rocco, P.R.M. Pathogenesis of Multiple Organ Injury in COVID-19 and Potential Therapeutic Strategies. Front. Physiol. 2021, 12, 593223. [Google Scholar] [CrossRef] [PubMed]
- Peng, H.; Li, C.; Kadow, S.; Henry, B.D.; Steinmann, J.; Becker, K.A.; Riehle, A.; Beckmann, N.; Wilker, B.; Li, P.-L.; et al. Acid Sphingomyelinase Inhibition Protects Mice from Lung Edema and Lethal Staphylococcus Aureus Sepsis. J. Mol. Med. 2015, 93, 675–689. [Google Scholar] [CrossRef] [PubMed]
- Becker, K.A.; Riethmüller, J.; Lüth, A.; Döring, G.; Kleuser, B.; Gulbins, E. Acid Sphingomyelinase Inhibitors Normalize Pulmonary Ceramide and Inflammation in Cystic Fibrosis. Am. J. Respir. Cell Mol. Biol. 2010, 42, 716–724. [Google Scholar] [CrossRef] [PubMed]
- Hellerbrand, C. Inhibition of Monoacylglycerol Lipase for the Treatment of Liver Disease: Tempting but Still Playing with Fire. Gut 2019, 68, 382–384. [Google Scholar] [CrossRef]
- Mulvihill, M.M.; Nomura, D.K. Therapeutic Potential of Monoacylglycerol Lipase Inhibitors. Life Sci. 2013, 92, 492–497. [Google Scholar] [CrossRef] [PubMed]
| Healthy | SARS-CoV-2 Pos | Sex (F/M) | Age (Std Dev) | ||
|---|---|---|---|---|---|
| Discovery(Australia) | Training Set | 19 | 10 | 17/13 | 53.0 (18.4) |
| Test Set | 18 | 10 | 12/15 | 56.1 (18.1) | |
| All | 37 | 20 | 28/29 | 54.5 (18.2) | |
| Validation(Spain) | Training Set | 49 | 157 | 104/102 | 62.9 (20.0) |
| Test Set | 51 | 155 | 118/88 | 62.3 (20.3) | |
| All | 100 | 312 | 222/190 | 62.6 (20.1) | |
| Training Set | Test Set | Sex (F/M) | Age (Std Dev) | ||
| Discovery(Australia) | Healthy | 19 | 18 | 15/22 | 47.54 (16.9) |
| SARS-CoV-2 Pos | 10 | 10 | 14/6 | 67.32 (12.8) | |
| Healthy and SARS-CoV-2 Pos | 29 | 28 | 28/29 | 54.50 (18.2) | |
| SARS-CoV-2 Neg subcohort | 22 | 13/9 | 47.64 (15.05) | ||
| Validation(Spain) | Healthy | 49 | 51 | 50/50 | 42.87 (12.5) |
| SARS-CoV-2 Pos | 157 | 155 | 172/140 | 68.89 (17.9) | |
| Healthy and SARS-CoV-2 Pos | 206 | 206 | 222/190 | 54.50 (18.2) | |
| Discovery (Australia) | |||||||
|---|---|---|---|---|---|---|---|
| Lipid | Healthy Control Mean (sd) | SARS-CoV-2 Pos Mean (sd) | Mann–Whitney p | BH q Value | Cliff’s Delta | Cliff’s Delta 95%CI—Lower | Cliff’s Delta 95%CI—Upper |
| PE(P-18:1/18:2) | 5.91 (2.09) | 1.54 (0.84) | 6.61 × 10−15 | 3.97 × 10−14 | 0.99 | 0.97 | 1.00 |
| PC(18:2/18:2) | 131.92 (46.05) | 35.19 (22.5) | 6.16 × 10−13 | 1.23 × 10−12 | 0.96 | 0.87 | 0.99 |
| LPC(18:2) | 37.31 (12.8) | 13.9 (5.51) | 3.22 × 10−13 | 9.66 × 10−13 | 0.97 | 0.89 | 0.99 |
| HCER(22:0) | 3.94 (0.89) | 2.23 (0.51) | 3.03 × 10−11 | 4.54 × 10−11 | 0.92 | 0.77 | 0.98 |
| CER(18:0) | 0.14 (0.05) | 0.38 (0.17) | 8.77 × 10−11 | 1.05 × 10−10 | −0.91 | −0.97 | −0.72 |
| DCER(18:0) | 0.10 (0.04) | 0.27 (0.12) | 1.94 × 10−8 | 1.94 × 10−8 | −0.82 | −0.94 | −0.56 |
| Validation (Spain) | |||||||
| Lipid | Healthy Control Mean (sd) | SARS-CoV-2 Pos Mean (sd) | Mann–Whitney p | BH q Value | Cliff’s Delta | Cliff’s Delta 95%CI—Lower | Cliff’s Delta 95%CI—Upper |
| PE(P-18:1/18:2) | 5.4 (1.99) | 1.41 (1.26) | 1.74 × 10−43 | 5.22 × 10−43 | 0.92 | 0.88 | 0.95 |
| PC(18:2/18:2) | 451.11 (156.5) | 110.91 (100.96) | 8.21 × 10−45 | 4.93 × 10−44 | 0.93 | 0.9 | 0.96 |
| LPC(18:2) | 51.23 (17.35) | 17.19 (11.06) | 6.38 × 10−43 | 1.28 × 10−42 | 0.91 | 0.86 | 0.94 |
| HCER(22:0) | 3.51 (0.96) | 1.84 (0.92) | 3.70 × 10−35 | 5.55 × 10−35 | 0.82 | 0.76 | 0.87 |
| CER(18:0) | 0.15 (0.05) | 0.31 (0.18) | 1.17 × 10−22 | 1.40 × 10−22 | −0.65 | −0.73 | −0.56 |
| DCER(18:0) | 0.09 (0.04) | 0.14 (0.09) | 6.93 × 10−9 | 6.93 × 10−9 | −0.38 | −0.48 | −0.28 |
| Discovery Dataset (SARS-CoV-2-Positive vs. Healthy Controls) | ||||
|---|---|---|---|---|
| Training | Test | |||
| Healthy Control Actual | SARS-CoV-2 POS Actual | Healthy Control Actual | SARS-CoV-2 POS Actual | |
| Model healthy control predicted | 19 | 0 | 17 | 0 |
| Model SARS-CoV-2 POS predicted | 0 | 10 | 1 | 10 |
| Validation Dataset (SARS-CoV-2 Positive vs. Healthy Control) | ||||
| Training | Test | |||
| Healthy Control Actual | SARS-CoV-2 POS Actual | Healthy Control Actual | SARS-CoV-2 POS Actual | |
| Model healthy control predicted | 44 | 6 | 47 | 8 |
| Model SARS-CoV-2 POS predicted | 5 | 151 | 4 | 147 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gray, N.; Lawler, N.G.; Zeng, A.X.; Ryan, M.; Bong, S.H.; Boughton, B.A.; Bizkarguenaga, M.; Bruzzone, C.; Embade, N.; Wist, J.; et al. Diagnostic Potential of the Plasma Lipidome in Infectious Disease: Application to Acute SARS-CoV-2 Infection. Metabolites 2021, 11, 467. https://doi.org/10.3390/metabo11070467
Gray N, Lawler NG, Zeng AX, Ryan M, Bong SH, Boughton BA, Bizkarguenaga M, Bruzzone C, Embade N, Wist J, et al. Diagnostic Potential of the Plasma Lipidome in Infectious Disease: Application to Acute SARS-CoV-2 Infection. Metabolites. 2021; 11(7):467. https://doi.org/10.3390/metabo11070467
Chicago/Turabian StyleGray, Nicola, Nathan G. Lawler, Annie Xu Zeng, Monique Ryan, Sze How Bong, Berin A. Boughton, Maider Bizkarguenaga, Chiara Bruzzone, Nieves Embade, Julien Wist, and et al. 2021. "Diagnostic Potential of the Plasma Lipidome in Infectious Disease: Application to Acute SARS-CoV-2 Infection" Metabolites 11, no. 7: 467. https://doi.org/10.3390/metabo11070467
APA StyleGray, N., Lawler, N. G., Zeng, A. X., Ryan, M., Bong, S. H., Boughton, B. A., Bizkarguenaga, M., Bruzzone, C., Embade, N., Wist, J., Holmes, E., Millet, O., Nicholson, J. K., & Whiley, L. (2021). Diagnostic Potential of the Plasma Lipidome in Infectious Disease: Application to Acute SARS-CoV-2 Infection. Metabolites, 11(7), 467. https://doi.org/10.3390/metabo11070467








