Abstract
Metabolic reprogramming is one of the hallmarks of tumors. Alterations of cellular metabolism not only contribute to tumor development, but also mediate the resistance of tumor cells to antitumor drugs. The metabolic response of tumor cells to various chemotherapy drugs can be analyzed by metabolomics. Although cancer cells have experienced metabolic reprogramming, the metabolism of drug resistant cancer cells has been further modified. Metabolic adaptations of drug resistant cells to chemotherapeutics involve redox, lipid metabolism, bioenergetics, glycolysis, polyamine synthesis and so on. The proposed metabolic mechanisms of drug resistance include the increase of glucose and glutamine demand, active pathways of glutaminolysis and glycolysis, promotion of NADPH from the pentose phosphate pathway, adaptive mitochondrial reprogramming, activation of fatty acid oxidation, and up-regulation of ornithine decarboxylase for polyamine production. Several genes are associated with metabolic reprogramming and drug resistance. Intervening regulatory points described above or targeting key genes in several important metabolic pathways may restore cell sensitivity to chemotherapy. This paper reviews the metabolic changes of tumor cells during the development of chemoresistance and discusses the potential of reversing chemoresistance by metabolic regulation.
1. Introduction
Chemotherapy and targeted therapy are common methods of tumor treatment, but tumor cells can develop resistance to drugs, leading to the failure of treatment. The development of tumor drug resistance involves general mechanisms and drug-specific or pathway-specific mechanisms (Figure 1) [1,2,3]. Pathway-specific mechanisms require the restoration of the tumor-driving signaling pathway. General mechanisms, common to many drugs, usually involve several aspects. First, the acquisition of resistance is often related to the failure of drug delivery. Multidrug resistance is mediated by the activation of ATP-binding cassette (ABC) transporters. The multidrug resistance protein 1 (MDR1) or P-glycoprotein is one of the members of ABC transporter family. The up-regulation of these proteins, especially MDR1, contributes to the acquired resistance to cytotoxic and targeted drugs. Drug escape can be rendered by cancer stem cells (CSCs). CSCs are a small population of tumor initiating cells that exist in tumors and are able to survive therapeutic intervention. The ability of CSCs resistance to chemotherapy attributes to their low proliferation rates, up-regulation of ABC transporter activity and DNA repair capacity, and down-regulation of apoptosis. Epithelial-mesenchymal transition (EMT) also contributes to the development of drug resistance. EMT is a transdifferentiation process in which epithelial cells lose their essential distinguishing characteristics and acquire instead mesenchymal features. Cells that undergo EMT can produce a CSC-like phenotype, leading to the resistance to tumor therapy. As most cancer drugs eradicate tumor cells by inducing apoptosis, the down-regulation of cell death pathways is another mechanism of drug resistance.
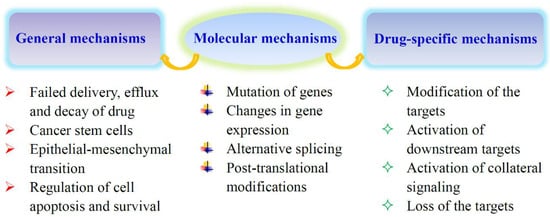
Figure 1.
Mechanisms of acquired tumor drug resistance. Some of the reasons for acquired drug resistance are common to many drugs, and others may be drug-specific or pathway-specific. They are all related to the expression and function of specific genes. The pathway-specific mechanisms usually involve the restoration of the tumor-driving signaling pathway.
Metabolic reprogramming, metabolic alterations occurring in tumor cells, is one of the hallmarks of tumors [4]. Enhancement of cell proliferation in tumors requires metabolic reprogramming, which leads to the changes of metabolic pathways to generate enough ATP and intermediates for macromolecular biosynthesis under stress to meet the needs of rapid cell division. It is believed that metabolic reprogramming of tumor cells is the consequence of metabolic adaptation, which involves mutations in oncogenes, consequently dysregulating the expression and activity of some metabolic enzymes and the flux of metabolic pathways, and finally affecting whole cellular metabolism. Changes in cellular metabolism not only promote tumor development [5,6,7,8,9], but also contribute to cell resistance to chemotherapeutics [10,11,12,13]. These changes may result from the metabolic adaptation of tumor cells to pharmacological stress, which include adaptations in oxidative stress, lipid metabolism, and bioenergetics; dependence on glucose and glycolysis; and an increase in polyamine production, etc. [14,15,16,17]. Although the underlying mechanism of metabolic adaptation for drug resistant cells has not been well understood, emerging evidences suggest that expression of some metabolic genes or some signal pathways in the drug resistant cells are deregulated [18,19,20]. This leads to an increase in the uptake of glucose and glutamine by drug resistant cells [21,22]; activation of pathways such as glutaminolysis, glycolysis, and fatty acid oxidation [18,23,24]; and promotion of antioxidant production [22,25] as well as metabolic reprogramming in mitochondria [20].
The study on the effect of cell metabolism on tumor survival and growth revealed that tumor cells are more sensitive to nutrient deprivation and more dependent on some metabolites. Fasting or fasting-mimicking diets have been used as an auxiliary means to improve the outcome of cancer treatment [26]. A specific dietary manipulation will affect the metabolism of tumor cells, thus affecting the treatment of tumors [27]. In addition to in vivo evidence described above, in vitro experimental results show that targeting cell metabolism is a strategy to improve tumor therapy [28]. Preliminary studies in vitro have shown that regulating the level of metabolites or targeting some genes in certain metabolic pathways can make cancer cells re-sensitize to a chemotherapeutic agent [19,23,29,30,31]. This paper reviews the metabolic changes of tumor cells during the development of chemoresistance and discusses the possibility of reversing chemoresistance by metabolic regulation.
2. Metabolic Reprogramming Contributes to Chemoresistance
The metabolic response of drug resistant cancer cells is an open question. Metabolomics has been used to analyze the metabolic changes of drug resistant tumor cells in response to various chemotherapies. Available data imply that resistant cancer cells undergo metabolic adaptation in several aspects.
2.1. Oxidative Stress Adaptation
Antioxidant capacity and redox homeostasis are essential for cell survival and growth. Cisplatin is a kind of platinum compound commonly used in cancer treatment. The antitumor mechanism of platinum involves DNA damage-induced cell apoptosis and reactive oxygen species (ROS)-mediated oxidative stress [32,33,34]. Cisplatin resistant ovarian cancer cells have an active glutathione (GSH) synthesis pathway and high level of intracellular GSH (Table 1), suggesting that an increase of cellular GSH is a beneficial mechanism to cope with cisplatin induced oxidative stress [21,32]. For example, cisplatin resistant lung cancer cells increase ROS production through metabolic reprogramming and become more dependent on oxidative metabolism rather than glycolysis [35,36,37]. High levels of ROS and other metabolic alterations promote EMT [36]. These cells take up more glutamine and are highly sensitive to glutamine deprivation. Glutamine is converted into glutamate to synthesize GSH, consequently reducing the cellular ROS level. Blocking glutamate flux selectively kills cisplatin resistant cells [35]. Glutamate and glycine from GSH are mainly glutamine-derived, but cysteine is not [25]. The thiol component of GSH is cysteine. Cysteine uptake and lower level of cysteine endogenous synthesis are mandatory for GSH bioavailability. The thiols from GSH as well as cysteine–glycine and cysteine can confiscate platinum molecules, avoiding them to reach cell components and form protein and DNA adducts [25]. The cysteine is essential for cancer proliferation and survival, and critical for metabolic rewiring of cancer [38,39,40,41,42]. Cysteine impacts the hypoxic adaptation of cancer cells, thus contributing to hypoxia-driven platinum-based chemotherapeutic agents’ resistance. Therefore, regulating cysteine bioavailability may serve as strategy to reverse resistance both to hypoxia and carboplatin [39,40]. Cysteine is also a valuable source of pyruvate for cancer cells replacing in part glucose; cysteine catabolism generates about 20% of intracellular pyruvate [41].

Table 1.
Main metabolic changes of chemoresistant tumor cells.
In addition to cisplatin, other antitumor agents also induce the changes of cellular GSH level. Paclitaxel (Taxol) targets cellular tubulin, which leads to defects in mitotic spindle assembly, chromosome segregation, and cell division. In triple negative breast cancer cells treated with paclitaxel, the levels of glutamine, glutamate, and GSH increase [43]. Sorafenib is a tyrosine kinase inhibitor. GSH synthesis in sorafenib resistant leukemia cells was enhanced, indicating that the resistant cells had specific metabolic and redox adaptations [17]. Erlotinib, an epidermal growth factor receptor (EGFR) inhibitor, is used for the therapy of mutated EGFR-driven cancers. The secondary mutation of T790M EGFR induces the acquisition of drug resistance in tumors. However, the metabolomic profiles of erlotinib sensitive and resistant cells showed that GSH levels are significantly down-regulated in T790M EGFR cells. The T790M mutation inhibits NFE2-related factor 2 (NRF2) activity, which down-regulates the expression of GSH-synthesizing enzymes [44]. Supplement of GSH in resistant cells re-sensitized them to erlotinib. In contrast, reducing GSH levels in sensitive cells made them resistant to erlotinib. Increasing intra-tumoral GSH levels re-sensitized resistant tumors to erlotinib in mice. Because the drug resistance of tumors is usually related to the increase of GSH level, the reason the decrease of GSH level is related to the resistance of EGFR tyrosine kinase inhibitor deserves further study.
Glucose was channeled to the pentose phosphate pathway (PPP) and serine synthesis pathway in bortezomib resistant cells, which increased antioxidant capacity of bortezomib resistant cells [49]. Bortezomib is a proteasome inhibitor. Proteasome inhibition causes endoplasmic reticulum stress and activation of the unfolded protein response, eventually leading to apoptosis through multiple pathways, including overproduction of ROS. The PPP can produce reduced form of nicotinamide adenine dinucleotide phosphate (NADPH), consequently maintaining intracellular GSH level and redox balance. High activity of serine synthesis pathway is attributed to the up-regulation of 3-phosphoglycerate dehydrogenase (PHGDH), the rate-limiting enzyme of serine synthesis. PHGDH decreased ROS through increasing GSH synthesis, thereby promoting cell growth and bortezomib resistance [50].
2.2. Adaptation of Lipid Metabolism
Increasing evidence suggests that lipid metabolism is reprogrammed when tumor cells respond to chemotherapy [51]. Early studies have shown that compared with cisplatin sensitive ovarian cancer cells, cisplatin resistant counterparts have higher basal content of intracellular mobile lipids originating from the hydrolyzed acyl chains in triacylglycerides (Table 1) [21,52,53]. Similar reprogramming in lipid metabolism was conformed in cisplatin resistant bladder cancer through a comparative lipidomic profiling [54]. The metabolomic analysis of lung cancer cells and their cisplatin resistant derivative showed that there were 40 differential metabolites, mainly involving phospholipids, fatty acids, amino acids, and metabolites related to energy metabolism [55]. The analysis of five metabolite groups showed that the level of short-chain acylcarnitines and selected lysophosphatidylcholines increased and the level of acyl-alkyl-phosphatidylcholines and one sphingolipid decreased in the erlotinib resistant human pancreatic cancer cells compared with the erlotinib sensitive parental cells. The results suggested that choline phospholipid and acetyl-CoA-associated metabolism was changed after the acquisition of erlotinib resistance in pancreatic cancer [16]. The content of choline was higher in docetaxel resistant BRCA1-mutated mouse mammary tumor tissues compared with the docetaxel sensitive controls [47]. Temozolomide is a chemotherapeutic drug that serves as a DNA alkylator. Temozolomide treatment showed up-regulation of choline and phosphorylcholine in temozolomide sensitive glioblastoma multiforme (GBM) cells compared to temozolomide resistant GBM cells (Table 1) [48]. With some exceptions, the increase of phospholipid appears to be related to cell resistance.
2.3. Bioenergetic Adaptation
Altered energetics is considered a hallmark of cancer. Bioenergetic adaptation is also a mechanism for cancer cells to cope with drug challenges. The cisplatin sensitive ovarian cancer cells have active glycolysis. However, their cisplatin resistant counterparts can enhance the glycolytic flux upon the treatment with ATP synthase inhibitor or gain greater oxidative phosphorylation (OXPHOS) compensation in the presence of glycolysis inhibition. The results indicated that resistant ovarian cancer cells are able to switch between OXPHOS and glycolysis [56]. Resistant chronic myelogenous leukemia (CML) cells elevated creatine production and creatine conversion to phosphocreatine [46]. The increase of phosphocreatine levels may provide an alternative energy reserve, allowing cells to escape imatinib-induced cell death [57].
2.4. Dependence on Glucose and Glycolysis
Tumor cells prefer to fuel glucose to the aerobic glycolysis pathway. Sorafenib is a tyrosine kinase inhibitor. Metabolism in sorafenib resistant leukemia cells was reprogrammed to increase glucose demand and decrease the flux of glucose into the PPP [17]. Imatinib is a breakpoint cluster region gene-Abelson murine leukemia viral oncogene homolog (BCR-ABL) tyrosine kinase inhibitor, and the metabolic changes of CML cells resistant to imatinib are contradictory. An early study found that compared with imatinib sensitive cells, the imatinib resistant BCR-ABL-positive cells increased glucose uptake, glycolysis, and lactate production in response to the treatment of imatinib [45]. However, Dewar et al. revealed that resistant CML cells reduced glucose consumption and lactate production [46]. The reasons for the inconsistent results are not clear. Flavopiridol is a pan-cyclin dependent kinase inhibitor inducing cancer cell apoptosis. Flavopiridol resistant human prostate cancer cells showed enhanced glycolysis and were less sensitive to the apoptosis induced by cisplatin and docetaxel [58]. Adriamycin (doxorubicin) is a topoisomerase II inhibitor. Compared with the sensitive cells, the central metabolism of adriamycin resistant leukemia cells has changed, which is characterized by increased dependence on glucose, decreased dependence on exogenous glutamine, and decreased pantothenic acid uptake and fatty acid β oxidation rate [59].
2.5. Synthesis of Polyamine
Polyamines are ubiquitous in living cells and are essential for eukaryotic cell growth [60]. Polyamine synthesis was reported as one of most significant metabolic changes in platinum resistant ovarian cancer cells [32]. Metabolomics analysis of the supernatant of tumor associated macrophages (TAMs) in colorectal cancer demonstrated that 5-Fluorouracil (5-FU) could stimulate the secretion of putrescine that induced the resistance of tumor cells to 5-FU-mediated apoptosis. Inhibition or down-regulation of ornithine decarboxylase, a key enzyme in the production of putrescine, reduced putrescine levels, thus preventing the TAMs-mediated 5-FU resistance and enhancing the 5-FU-induced growth inhibition of cancer cells [15].
Current evidence supports the existence of metabolic reprogramming in drug resistant cancer cells. It has been found that drug sensitive and resistant cancer cells respond differently to certain drugs [48,61]. The metabolic pattern of adriamycin sensitive cells is gradually similar to that of adriamycin resistant cells, which further supports a metabolic shift in the development of chemoresistance [61]. The metabolic changes induced by chemotherapy may be a dynamic response [62,63]. A short-term treatment of gastric cancer cells with 5-FU can reduce cellular proline and increase glutamate, which may be associated with the up-regulation of proline dehydrogenase (PRODH) that promotes production of glutamate from proline [63]. But the proline and glutamate levels are less affected in 5-FU resistant cells, where PRODH expression was not up-regulated following 5-FU treatment. PRODH catalyzes the first step of proline degradation, inducing superoxide and conversion of proline to glutamate. The inhibition of nucleotide synthesis by 5-FU and the resultant genetic stress induces PRODH activity following mitochondrial superoxide generation, contributing to drug resistance. In general, metabolic adaptation of resistant cancer cells may involve many aspects, including redox, lipid metabolism, bioenergetics, glycolysis, and synthesis of polyamine. The diversified antitumor mechanisms of different chemotherapeutic agents may lead to different resistance-related metabolic alterations. However, some drugs may induce similar metabolic change. The cells resistant to adriamycin and the cells resistant to cisplatin shared some similar differential metabolites compared with their drug sensitive counterparts [2]. In addition, the treatment of cells with different drug combinations may lead to different metabolic alterations. For example, the treatment of human breast cancer cells with a combination of cisplatin and tamoxifen significantly reduces cellular phosphocholine levels, while cisplatin combined with adriamycin can increase lactate levels [11]. Although prediction of such metabolic changes is still difficult, some studies have tried to identify the plasma markers from cancer patients for predicting drug resistance [64].
3. The Potential Mechanisms of Metabolic Alterations in Resistant Cells
We know that several metabolic enzymes, such as glycolytic enzymes, are targets for tumor therapy [65,66]. Many studies have explored the underlying mechanism of metabolic reprogramming in cancer cells, but the mechanism of metabolic adaptation in drug resistant cells is still unclear. Several potential mechanisms are proposed to explain metabolic reprogramming and drug resistance (Figure 2, Table 2).
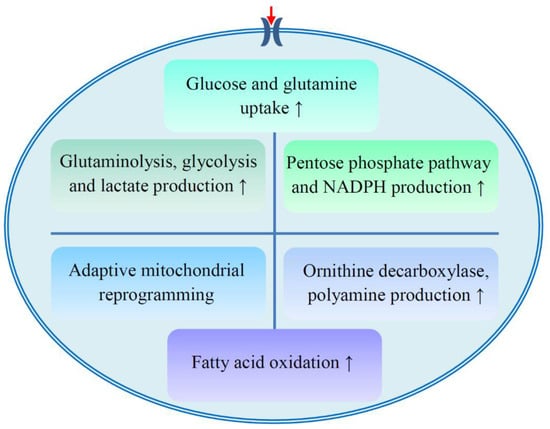
Figure 2.
Proposed metabolic mechanisms of drug resistance. The development of drug resistance involves several aspects of metabolism, including the enhancement of glucose and glutamine uptake; active glutaminolysis, glycolysis, pentose phosphate pathway, and fatty acid oxidation; polyamine production; and adaptive mitochondrial reprogramming. NADPH, reduced form of nicotinamide adenine dinucleotide phosphate.

Table 2.
Potential metabolic mechanisms of drug resistance.
First, the uptake of glucose and glutamine in resistant cells increases [21,22,24,67,68]. The ibrutinib resistant cells relied heavily on exogenous glutamine [24], and cisplatin resistant ovarian cancer cells increased glucose uptake and consumption [21,22]. Mucin1, a glycoprotein, was associated with chemoresistance and cancer aggression. Over-expression of mucin1 promoted glucose and glutamine uptake in breast cancer [68]. The expression of glucose transporter protein 1 (GLUT1) is up-regulated in oral cancer cells under hypoxia. Silencing of GLUT1 resulted in increased rates of cisplatin-induced apoptosis under hypoxia [67].
However, fatty acid oxidation is important for the resistance of certain cells (Figure 2, Table 2). Upon treatment of ibrutinib plus etomoxir, the carnitine palmitoyltransferase I (CPT-1) inhibitor suppressing β-oxidation of fatty acids in mitochondria, GLUT1 expression and glucose uptake in resistant chronic lymphocytic leukemia (CLL) cells were decreased compared to sensitive cells, suggesting the activation of fatty acid oxidation might be important for the sustenance of ibrutinib resistance in CLL cells [24].
Resistant cells exhibit active glutaminolysis and glycolysis pathways and increase lactate production (Figure 2, Table 2) [18,23,30,69,70]. The catabolism of glutamine in ibrutinib resistant cells was up-regulated [24]. Glutaminolysis was associated with the resistance of ovarian cancer cells to paclitaxel or cisplatin [23]. The expression of lactate dehydrogenase A (LDHA) in paclitaxel resistant cells increased [30]. Glycolytic enzymes, enolase and glyceraldehyde-3-phosphate dehydrogenase (GAPDH), are the resistance-related proteins of carboplatin and paclitaxel [18]. Enolase was also up-regulated in cisplatin resistant gastric cancer cells [70]. Melanoma cell adhesion molecule (MCAM), a cell surface receptor, was identified as being significantly up-regulated in chemoresistant lung cancer cells and patient-derived xenografts. Metabolomic profiling revealed that MCAM regulated lactate production and chemosensitivity of resistant cells through PI3K/AKT/SOX2 pathways [69].
Resistant cells channeled glucose to PPP and promoted the production of NADPH (Figure 2, Table 2) [19,22,24]. Glucose-6-phosphate dehydrogenase (G6PDH) and 6-phosphogluconate dehydrogenase (6PGD) are two important enzymes in PPP. The expression of G6PDH in cisplatin resistant cells was up-regulated [22]. 6PGD activity was related to sensitivity of hepatocellular carcinoma (HCC) to paclitaxel, adriamycin, and cisplatin [19]. PPP-mediated production of NADPH is beneficial for tumor cells to cope with chemotherapy-induced oxidative stress. Hepatocyte nuclear factor 1β (HNF1β) regulates γ-glutamylcysteine ligase expression and GSH production in ovarian cancer cells. HNF1β down-regulation sensitizes cells to carboplatin [25].
Resistant cells may have adaptive mitochondrial reprogramming (Figure 2, Table 2) [20,71,72,73]. PI3K therapy (targeting PI3K) induces global metabolic reprogramming in tumors and promotes the recruitment of Akt2 to mitochondria, which facilitates mitochondrial energy metabolism and reduces tumor cell death, conferring resistance to PI3K therapy [20]. Adenylate kinase 4 (AK4) is one of the key enzymes that catalyze the high-energy phosphoryl transfer reaction in mitochondria. AK4 knockdown increased the levels of fumarate and malate in tricarboxylic acid (TCA) cycle intermediates and enhanced the sensitivity of tumor cells to cisplatin and hypoxia. This may be due to its regulation of mitochondrial activity, including the increase of mitochondrial number and the up-regulation of gene expression of key enzymes in the TCA cycle, succinate dehydrogenase A, and oxoglutarate dehydrogenase L [71]. Tumor necrosis factor-associated protein 1 (TRAP1) facilitated a metabolic shift toward OXPHOS in ovarian cancer, which is related to cisplatin resistance [72].
Production of polyamines also contributes to chemoresistance (Figure 2, Table 2). Metabolic pathways of polyamines as well as amino acids and fatty acids were significantly changed in erlotinib resistant pancreatic cancer cells. Putrescine, the product catalyzed by ornithine decarboxylase (ODC), contributed to the acquisition of erlotinib resistance. ODC inhibition was able to restore erlotinib sensitivity in erlotinib resistant cells, which could be rescued by exogenous putrescine [31].
The mechanisms proposed may partly explain the metabolic adaptation during the development of drug resistance. One or several mechanisms may be used by cells to cope with a drug. Little is known about the correlation between the antitumor mechanism of a drug and its principle of metabolic adaptation, which is worth further study.
4. Potential Reversal Effect of Metabolic Regulation on Chemoresistant Tumor Cells
Since the development of chemoresistance is associated with metabolic reprogramming, it should be possible to restore the chemosensitivity through metabolic regulation (Figure 3). Some researchers have tried to do this and have achieved initial success. Metabolic regulation on resistant tumor cells may restore the cell sensitivity to chemotherapy [30].
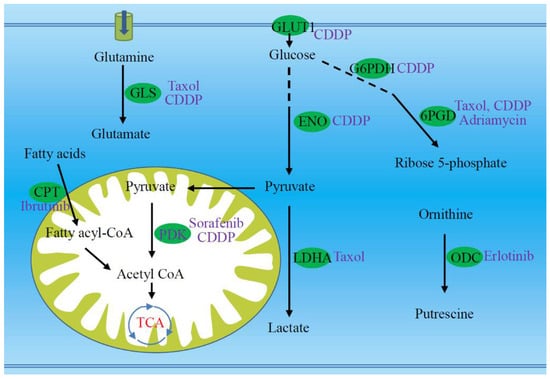
Figure 3.
Targeting several key enzymes or proteins (green ellipses) are potential to re-sensitize the resistant tumor cells to chemotherapeutic agents (purple font). Targets marked with green ellipses involve multiple metabolic pathways, including glutaminolysis, transport and glycolysis of glucose, pentose phosphate pathway, fatty acid oxidation, pyruvate dehydrogenation, and production of polyamine. The drugs that can be metabolically regulated are highlighted with purple font. 6PGD, 6-phosphogluconate dehydrogenase; CDDP, cisplatin; CoA, coenzyme A; CPT, carnitine palmitoyltransferase; ENO, enolase; G6PDH, glucose-6-phosphate dehydrogenase; GLS, glutaminase; GLUT1, glucose transporter protein 1; LDHA, lactate dehydrogenase A; ODC, ornithine decarboxylase; PDK, pyruvate dehydrogenase kinase.
First, metabolic regulation on glutamine in resistant cancer cells re-sensitizes their sensitivity to chemotherapeutics. It has been well known that cancer cells increase glutamine uptake, and even develop a reliance on glutamine. Cancer cells usually up-regulate the expression of glutaminase that catalyzes the first step of glutamine degradation (glutaminolysis). The treatment of paclitaxel or cisplatin resistant ovarian cancer cells with a glutaminase inhibitor increased their sensitivity to chemotherapy through inhibiting cell proliferation, regardless of their glutamine dependence status [23]. The expression of glutaminase 1 (GLS1) was enhanced in metastatic glutamine-dependent ovarian cancer cells and targeting GLS1 using siRNA sensitized the cancer cells to cisplatin (Figure 3). Argininosuccinate synthetase 1 (ASS1) is a chemosensitivity marker for arginine and glutamine starvation therapy. Up-regulation of ASS1 expression increased the resistance to glutamine starvation therapy. The supplement of fumarate suppressed ASS1 expression, which sensitized tumor cells to arginine and glutamine deprivation therapy [29].
Down-regulation of the glycolysis pathway in resistant cells may restore the cell sensitivity to chemotherapy. Cisplatin resistant gastric cancer cells highly depend on glycolysis through metabolic reprogramming. Disturbing cellular glycolysis via glucose starvation or 2-deoxyglucose treatment significantly reversed drug resistance. Enolase 1 is a glycolytic enzyme, and its up-regulation promotes the resistance of gastric cancer cells to cisplatin. Knockdown of enolase 1 reduced glycolysis and reversed drug resistance [70]. In paclitaxel resistant breast cancer cells, the expression and activity of LDHA increased compared with its parental cells. Silencing LDHA expression or inhibiting its activity re-sensitized the resistant cells to paclitaxel [30]. Pyruvate dehydrogenase kinase 4 (PDK4) is an inhibitor of mitochondrial pyruvate dehydrogenase, and inhibiting PDK4 reversed the resistance of HCC stem cells to sorafenib or cisplatin (Figure 3) [73].
Down-regulation of antioxidant capacity of resistant cells also contributes to the re-sensitivity of cells to drugs. Inhibition of GSH production by buthionine sulphoxamine (BSO) sensitizes ovarian clear cell carcinoma cells to carboplatin [25]. The dehydrogenation of glucose-6-phosphate and 6-phosphogluconate is two critical steps in the oxidative phase of PPP, in which NADPH is produced. The cisplatin resistant cells up-regulate expression and enzymatic activity of G6PDH and are more sensitive to G6PDH inhibition, suggesting that PPP is a potential target to overcome cisplatin resistance. Targeting G6PDH sensitized cisplatin resistant cells (Figure 3) [22]. 6PGD was found to be important for HCC growth and survival. 6PGD inhibition sensitized HCC to chemotherapy of paclitaxel, adriamycin, and cisplatin via AMP-activated protein kinase (AMPK)-dependent NADPH metabolic suppression and oxidative stress, suggesting that 6PGD is a promising therapeutic target to overcome chemoresistance [19]. In addition, serine starvation increased the sensitivity of multiple myeloma cells to bortezomib, suggesting the importance of serine metabolism in the cell response to bortezomib [49].
Fatty acid oxidation is another regulatory point. Carnitine palmitoyltransferase catalyzes the transport of long chain fatty acids into mitochondria for oxidation. Inhibition of carnitine palmitoyltransferase re-sensitized resistant leukemia cells to the tyrosine kinase inhibitor ibrutinib, suggesting that slowing down fatty acid oxidation may overcome ibrutinib resistance [24].
Of course, the regulation of drug degradation and excretion will affect the drug resistance of cells. Glucuronidation of drugs by uridine diphosphate-glucuronosyltransferases (UGT1As) contributes to the resistance of cancer cells to antitumor drugs. Selective UGT1As inhibitors specifically restored sensitivity of resistant cancer cells [74].
Several natural compounds have been shown to reverse chemoresistance [75,76,77]. Ursolic acid (UA) is a natural compound with antitumor activity. When combined with adriamycin, it can reverse the multidrug resistance of breast cancer cells, which is attributed to the changes of energy metabolism and amino acid metabolism induced by UA [76]. Treatment with UA reduces the level of intracellular metabolites, including lactate, pyruvate, glucose, α-ketoglutarate, and several amino acids. β-elemene, extracted from Curcuma zedoaria Roscoe, is potential to reverse the resistance to erlotinib [78]. Melittin, a main peptide with a potential for anticancer activity in bee venom, has a synergistic effect with cisplatin on ovarian cancer cells in several key metabolic pathways, such as TCA cycle, OXPHOS, purine and pyrimidine metabolism, and the arginine/proline pathway [77].
5. Conclusions
Drug resistance is the determinant of chemotherapy failure. The metabolic adaptability of cancer cells has been linked to their resistance to antitumor drugs. Resistant cells adapt to oxidative stress and maintain intracellular redox homeostasis by increasing the production of GSH and NADPH. Resistant cells can also increase fatty acid oxidation and phospholipid biosynthesis to obtain sufficient energy supply and maintain the integrity of cell membrane. Bioenergetic adaptation provides an alternative energy for resistant cells to survive drug intervention. Some resistant cells are highly dependent on glucose and glutamine. Glycolysis and glutaminolysis provide energy and intermediates for cell proliferation. Polyamines have essential roles in cell proliferation [79]. TAMs in tumors can secrete putrescine to induce drug resistance of tumor cells. According to different chemotherapeutic agents, resistant cells may adopt different adaptive mechanisms. Metabolic adaptation can be attributed to the mutation or deregulation of some genes, which lead to increased glucose and glutamine uptake, enhanced glutaminolysis and glycolysis pathways, boosted pentose phosphate pathway and antioxidant capacity, adaptive mitochondrial reprogramming, etc. This also provides an option to re-sensitize resistant cells to chemotherapy by regulating cell metabolism. There are some successful examples for the reversal of chemoresistance, which aim to target some key genes in the metabolic pathway or regulate the supply of metabolites.
There are two issues to be addressed here. First, many data discussed above are obtained by two-dimensional cell culture of established cell lines, with little regard for the untransformed state and the role of cell–cell interactions that modulate gene expression and enzyme activity. This has led to some observations that are true in cell culture, but do not always hold up in more complex models. Therefore, it is necessary to adopt different research models, such as ganoids and animal models, and combine with clinical data. Since metabolism does not operate in isolation from gene expression, mutational events, and the subsequent protein activities, more use of other ‘omics’ technologies such as transcriptomics and proteomics will help to understand the potential mechanism of metabolic reprogramming in response to chemotherapy. It also helps to assess whether the altered metabolism observed is a driver or a bystander. Another concern is the diverse metabolic responses to chemotherapeutics, which are affected by many factors. Glutathione levels increased in response to low dose curcumin in breast cancer cells, but decreased at high dose, indicating a biphasic response [80]. Triple-negative and hormone receptor-positive breast cancer cells had different metabolic responses to paclitaxel [43]. Comparable tumors, such as those from the same source, may respond differently to certain chemotherapy [11]. Better understanding of drug resistance-related metabolic reprogramming may help to develop new therapeutic strategies [57,59].
Author Contributions
X.C. wrote the manuscript; S.C. and D.Y. supervised the project; S.C. and D.Y. reviewed and edited the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This study was funded by the National Natural Science Foundation of China (No. 81873711 and No. 31670788) and Open Fund of Guangdong Key Laboratory of Pharmaceutical Functional Genes (No. 2014B030301028 and No. 2017B030314021).
Conflicts of Interest
The authors confirm that there are no conflicts of interest.
References
- Aleksakhina, S.N.; Kashyap, A.; Imyanitov, E.N. Mechanisms of acquired tumor drug resistance. Biochim. Biophys. Acta Rev. Cancer 2019, 1872, 188310. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Zhuang, X.; Zong, L.; Liu, S.; Liu, Z.; Song, F. Investigations on the cell metabolomics basis of multidrug resistance from tumor cells by ultra-performance liquid chromatography-mass spectrometry. Anal. Bioanal. Chem. 2016, 408, 5843–5854. [Google Scholar] [CrossRef] [PubMed]
- Guo, Q.; Cao, H.; Qi, X.; Li, H.; Ye, P.; Wang, Z.; Wang, D.; Sun, M. Research Progress in Reversal of Tumor Multi-Drug Resistance via Natural Products. Anticancer Agents Med. Chem. 2017, 17, 1466–1476. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef]
- Faubert, B.; Solmonson, A.; DeBerardinis, R.J. Metabolic reprogramming and cancer progression. Science 2020, 368, eaaw5473. [Google Scholar] [CrossRef]
- Qiao, S.; Koh, S.B.; Vivekanandan, V.; Salunke, D.; Patra, K.C.; Zaganjor, E.; Ross, K.; Mizukami, Y.; Jeanfavre, S.; Chen, A.; et al. REDD1 loss reprograms lipid metabolism to drive progression of RAS mutant tumors. Genes Dev. 2020, 34, 751–756. [Google Scholar] [CrossRef]
- Lee, N.C.W.; Carella, M.A.; Papa, S.; Bubici, C. High Expression of Glycolytic Genes in Cirrhosis Correlates With the Risk of Developing Liver Cancer. Front. Cell Dev. Biol. 2018, 6, 138. [Google Scholar] [CrossRef] [PubMed]
- Vernucci, E.; Abrego, J.; Gunda, V.; Shukla, S.K.; Dasgupta, A.; Rai, V.; Chaika, N.; Buettner, K.; Illies, A.; Yu, F.; et al. Metabolic Alterations in Pancreatic Cancer Progression. Cancers 2019, 12, 2. [Google Scholar] [CrossRef]
- Boroughs, L.K.; DeBerardinis, R.J. Metabolic pathways promoting cancer cell survival and growth. Nat. Cell Biol. 2015, 17, 351–359. [Google Scholar] [CrossRef]
- Cardoso, M.R.; Santos, J.C.; Ribeiro, M.L.; Talarico, M.C.R.; Viana, L.R.; Derchain, S.F.M. A Metabolomic Approach to Predict Breast Cancer Behavior and Chemotherapy Response. Int. J. Mol. Sci. 2018, 19, 617. [Google Scholar] [CrossRef]
- Maria, R.M.; Altei, W.F.; Selistre-de-Araujo, H.S.; Colnago, L.A. Impact of chemotherapy on metabolic reprogramming: Characterization of the metabolic profile of breast cancer MDA-MB-231 cells using H-1 HR-MAS NMR spectroscopy. J. Pharm. Biomed. 2017, 146, 324–328. [Google Scholar] [CrossRef]
- Rahman, M.; Hasan, M.R. Cancer Metabolism and Drug Resistance. Metabolites 2015, 5, 571–600. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.; Cao, M.; Liu, J.; Yang, Q.; Miao, X.; Go, V.L.W.; Lee, P.W.N.; Xiao, G.G. Metabolic Regulation in Mitochondria and Drug Resistance. Adv. Exp. Med. Biol. 2017, 1038, 149–171. [Google Scholar] [CrossRef]
- Zhang, C.; Ma, Q.; Shi, Y.; Li, X.; Wang, M.; Wang, J.; Ge, J.; Chen, Z.; Wang, Z.; Jiang, H. A novel 5-fluorouracil-resistant human esophageal squamous cell carcinoma cell line Eca-109/5-FU with significant drug resistance-related characteristics. Oncol. Rep. 2017, 37, 2942–2954. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Chen, Y.; Hao, L.; Hou, A.; Chen, X.; Li, Y.; Wang, R.; Luo, P.; Ruan, Z.; Ou, J.; et al. Macrophages induce resistance to 5-fluorouracil chemotherapy in colorectal cancer through the release of putrescine. Cancer Lett. 2016, 381, 305–313. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Jang, W.J.; Choi, B.; Joo, S.H.; Jeong, C.H. Comparative metabolomic analysis of HPAC cells following the acquisition of erlotinib resistance. Oncol. Lett. 2017, 13, 3437–3444. [Google Scholar] [CrossRef]
- You, X.; Jiang, W.; Lu, W.; Zhang, H.; Yu, T.; Tian, J.; Wen, S.; Garcia-Manero, G.; Huang, P.; Hu, Y. Metabolic reprogramming and redox adaptation in sorafenib-resistant leukemia cells: Detected by untargeted metabolomics and stable isotope tracing analysis. Cancer Commun. 2019, 39, 17. [Google Scholar] [CrossRef] [PubMed]
- Cruz, I.N.; Coley, H.M.; Kramer, H.B.; Madhuri, T.K.; Safuwan, N.A.; Angelino, A.R.; Yang, M. Proteomics Analysis of Ovarian Cancer Cell Lines and Tissues Reveals Drug Resistance-Associated Proteins. Cancer Genom. Proteom. 2017, 14, 35–51. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Wu, D.; Bao, L.; Yin, T.; Lei, D.; Yu, J.; Tong, X. 6PGD inhibition sensitizes hepatocellular carcinoma to chemotherapy via AMPK activation and metabolic reprogramming. Biomed. Pharmacother. 2019, 111, 1353–1358. [Google Scholar] [CrossRef]
- Ghosh, J.C.; Siegelin, M.D.; Vaira, V.; Faversani, A.; Tavecchio, M.; Chae, Y.C.; Lisanti, S.; Rampini, P.; Giroda, M.; Caino, M.C.; et al. Adaptive mitochondrial reprogramming and resistance to PI3K therapy. J. Natl. Cancer Inst. 2015, 107, dju502. [Google Scholar] [CrossRef] [PubMed]
- Montopoli, M.; Bellanda, M.; Lonardoni, F.; Ragazzi, E.; Dorigo, P.; Froldi, G.; Mammi, S.; Caparrotta, L. “Metabolic Reprogramming” in Ovarian Cancer Cells Resistant to Cisplatin. Curr. Cancer Drug Targets 2011, 11, 226–235. [Google Scholar] [CrossRef] [PubMed]
- Catanzaro, D.; Gaude, E.; Orso, G.; Giordano, C.; Guzzo, G.; Rasola, A.; Ragazzi, E.; Caparrotta, L.; Frezza, C.; Montopoli, M. Inhibition of glucose-6-phosphate dehydrogenase sensitizes cisplatin-resistant cells to death. Oncotarget 2015, 6, 30102–30114. [Google Scholar] [CrossRef] [PubMed]
- Masamha, C.P.; LaFontaine, P. Molecular targeting of glutaminase sensitizes ovarian cancer cells to chemotherapy. J. Cell. Biochem. 2018, 119, 6136–6145. [Google Scholar] [CrossRef] [PubMed]
- Galicia-Vazquez, G.; Aloyz, R. Ibrutinib Resistance Is Reduced by an Inhibitor of Fatty Acid Oxidation in Primary CLL Lymphocytes. Front. Oncol. 2018, 8, 411. [Google Scholar] [CrossRef]
- Lopes-Coelho, F.; Gouveia-Fernandes, S.; Goncalves, L.G.; Nunes, C.; Faustino, I.; Silva, F.; Felix, A.; Pereira, S.A.; Serpa, J. HNF1beta drives glutathione (GSH) synthesis underlying intrinsic carboplatin resistance of ovarian clear cell carcinoma (OCCC). Tumour Biol. 2016, 37, 4813–4829. [Google Scholar] [CrossRef] [PubMed]
- Nencioni, A.; Caffa, I.; Cortellino, S.; Longo, V.D. Fasting and cancer: Molecular mechanisms and clinical application. Nat. Rev. Cancer 2018, 18, 707–719. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Sanderson, S.M.; Dai, Z.; Reid, M.A.; Cooper, D.E.; Lu, M.; Richie, J.P., Jr.; Ciccarella, A.; Calcagnotto, A.; Mikhael, P.G.; et al. Dietary methionine influences therapy in mouse cancer models and alters human metabolism. Nature 2019, 572, 397–401. [Google Scholar] [CrossRef]
- Zhao, Y.; Butler, E.B.; Tan, M. Targeting cellular metabolism to improve cancer therapeutics. Cell Death Dis. 2013, 4, e532. [Google Scholar] [CrossRef]
- Long, Y.; Tsai, W.B.; Wang, D.J.; Hawke, D.H.; Savaraj, N.; Feun, L.G.; Hung, M.C.; Chen, H.H.W.; Kuo, M.T. Argininosuccinate synthetase 1 (ASS1) is a common metabolic marker of chemosensitivity for targeted arginine- and glutamine-starvation therapy. Cancer Lett. 2017, 388, 54–63. [Google Scholar] [CrossRef]
- Zhou, M.; Zhao, Y.H.; Ding, Y.; Liu, H.; Liu, Z.X.; Fodstad, O.; Riker, A.I.; Kamarajugadda, S.; Lu, J.R.; Owen, L.B.; et al. Warburg effect in chemosensitivity: Targeting lactate dehydrogenase-A re-sensitizes Taxol-Resistant cancer cells to Taxol. Mol. Cancer 2010, 9, 1–12. [Google Scholar] [CrossRef]
- Jang, W.J.; Choi, B.; Song, S.H.; Lee, N.; Kim, D.J.; Lee, S.; Jeong, C.H. Multi-Omics analysis reveals that ornithine decarboxylase contributes to erlotinib resistance in pancreatic cancer cells. Oncotarget 2017, 8, 92727–92742. [Google Scholar] [CrossRef] [PubMed]
- Poisson, L.M.; Munkarah, A.; Madi, H.; Datta, I.; Hensley-Alford, S.; Tebbe, C.; Buekers, T.; Giri, S.; Rattan, R. A metabolomic approach to identifying platinum resistance in ovarian cancer. J. Ovarian Res. 2015, 8, 13. [Google Scholar] [CrossRef] [PubMed]
- Marullo, R.; Werner, E.; Degtyareva, N.; Moore, B.; Altavilla, G.; Ramalingam, S.S.; Doetsch, P.W. Cisplatin induces a mitochondrial-ROS response that contributes to cytotoxicity depending on mitochondrial redox status and bioenergetic functions. PLoS ONE 2013, 8, e81162. [Google Scholar] [CrossRef]
- Kim, S.J.; Kim, H.S.; Seo, Y.R. Understanding of ROS-Inducing Strategy in Anticancer Therapy. Oxidative Med. Cell Longev. 2019, 2019, 5381692. [Google Scholar] [CrossRef] [PubMed]
- Wangpaichitr, M.; Wu, C.; Li, Y.Y.; Nguyen, D.J.M.; Kandemir, H.; Shah, S.; Chen, S.; Feun, L.G.; Prince, J.S.; Kuo, M.T.; et al. Exploiting ROS and metabolic differences to kill cisplatin resistant lung cancer. Oncotarget 2017, 8, 49275–49292. [Google Scholar] [CrossRef] [PubMed]
- Wangpaichitr, M.; Kandemir, H.; Li, Y.Y.; Wu, C.; Nguyen, D.; Feun, L.G.; Kuo, M.T.; Savaraj, N. Relationship of Metabolic Alterations and PD-L1 Expression in Cisplatin Resistant Lung Cancer. Cell Dev. Biol. 2017, 6, 183. [Google Scholar] [CrossRef] [PubMed]
- Cruz-Bermudez, A.; Laza-Briviesca, R.; Vicente-Blanco, R.J.; Garcia-Grande, A.; Coronado, M.J.; Laine-Menendez, S.; Palacios-Zambrano, S.; Moreno-Villa, M.R.; Ruiz-Valdepenas, A.M.; Lendinez, C.; et al. Cisplatin resistance involves a metabolic reprogramming through ROS and PGC-1alpha in NSCLC which can be overcome by OXPHOS inhibition. Free Radic. Biol. Med. 2019, 135, 167–181. [Google Scholar] [CrossRef]
- Kang, Y.P.; Torrente, L.; Falzone, A.; Elkins, C.M.; Liu, M.; Asara, J.M.; Dibble, C.C.; DeNicola, G.M. Cysteine dioxygenase 1 is a metabolic liability for non-small cell lung cancer. Elife 2019, 8, e45572. [Google Scholar] [CrossRef]
- Nunes, S.C.; Lopes-Coelho, F.; Gouveia-Fernandes, S.; Ramos, C.; Pereira, S.A.; Serpa, J. Cysteine boosters the evolutionary adaptation to CoCl2 mimicked hypoxia conditions, favouring carboplatin resistance in ovarian cancer. BMC Evol. Biol. 2018, 18, 97. [Google Scholar] [CrossRef]
- Nunes, S.C.; Ramos, C.; Lopes-Coelho, F.; Sequeira, C.O.; Silva, F.; Gouveia-Fernandes, S.; Rodrigues, A.; Guimaraes, A.; Silveira, M.; Abreu, S.; et al. Cysteine allows ovarian cancer cells to adapt to hypoxia and to escape from carboplatin cytotoxicity. Sci. Rep. 2018, 8, 1–17. [Google Scholar] [CrossRef]
- Yu, L.; Teoh, S.T.; Ensink, E.; Ogrodzinski, M.P.; Yang, C.; Vazquez, A.I.; Lunt, S.Y. Cysteine catabolism and the serine biosynthesis pathway support pyruvate production during pyruvate kinase knockdown in pancreatic cancer cells. Cancer Metab. 2019, 7, 1–13. [Google Scholar] [CrossRef]
- Combs, J.A.; DeNicola, G.M. The Non-Essential Amino Acid Cysteine Becomes Essential for Tumor Proliferation and Survival. Cancers 2019, 11, 678. [Google Scholar] [CrossRef]
- Stewart, D.A.; Winnike, J.H.; McRitchie, S.L.; Clark, R.F.; Pathmasiri, W.W.; Sumner, S.J. Metabolomics Analysis of Hormone-Responsive and Triple-Negative Breast Cancer Cell Responses to Paclitaxel Identify Key Metabolic Differences. J. Proteome Res. 2016, 15, 3225–3240. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Stokes, W.; Chater, E.; Roy, R.; de Bruin, E.; Hu, Y.; Liu, Z.; Smit, E.F.; Heynen, G.J.; Downward, J.; et al. Decreased glutathione biosynthesis contributes to EGFR T790M-driven erlotinib resistance in non-small cell lung cancer. Cell Discov. 2016, 2, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Kominsky, D.J.; Klawitter, J.; Brown, J.L.; Boros, L.G.; Melo, J.V.; Eckhardt, S.G.; Serkova, N.J. Abnormalities in glucose uptake and metabolism in imatinib-resistant human BCR-ABL-positive cells. Clin. Cancer Res. 2009, 15, 3442–3450. [Google Scholar] [CrossRef] [PubMed]
- Dewar, B.J.; Keshari, K.; Jeffries, R.; Dzeja, P.; Graves, L.M.; Macdonald, J.M. Metabolic assessment of a novel chronic myelogenous leukemic cell line and an imatinib resistant subline by H-1 NMR spectroscopy. Metabolomics 2010, 6, 439–450. [Google Scholar] [CrossRef]
- van Asten, J.J.A.; Vettukattil, R.; Buckle, T.; Rottenberg, S.; van Leeuwen, F.; Bathen, T.F.; Heerschap, A. Increased levels of choline metabolites are an early marker of docetaxel treatment response in BRCA1-mutated mouse mammary tumors: An assessment by ex vivo proton magnetic resonance spectroscopy. J. Transl. Med. 2015, 13, 114. [Google Scholar] [CrossRef]
- St-Coeur, P.D.; Poitras, J.J.; Cuperlovic-Culf, M.; Touaibia, M.; Morin, P. Investigating a signature of temozolomide resistance in GBM cell lines using metabolomics. J. Neuro-Oncol. 2015, 125, 91–102. [Google Scholar] [CrossRef]
- Zaal, E.A.; Wu, W.; Jansen, G.; Zweegman, S.; Cloos, J.; Berkers, C.R. Bortezomib resistance in multiple myeloma is associated with increased serine synthesis. Cancer Metab. 2017, 5, 7. [Google Scholar] [CrossRef]
- Wu, X.; Xia, J.; Zhang, J.; Zhu, Y.; Wu, Y.; Guo, J.; Chen, S.; Lei, Q.; Meng, B.; Kuang, C.; et al. Phosphoglycerate dehydrogenase promotes proliferation and bortezomib resistance through increasing reduced glutathione synthesis in multiple myeloma. Br. J. Haematol. 2020, 190, 52–66. [Google Scholar] [CrossRef]
- Cao, Y. Adipocyte and lipid metabolism in cancer drug resistance. J. Clin. Invest. 2019, 129, 3006–3017. [Google Scholar] [CrossRef] [PubMed]
- Musacchio, T.; Toniutti, M.; Kautz, R.; Torchilin, V.P. 1H NMR detection of mobile lipids as a marker for apoptosis: The case of anticancer drug-loaded liposomes and polymeric micelles. Mol. Pharm. 2009, 6, 1876–1882. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Luciani, A.M.; Grande, S.; Palma, A.; Rosi, A.; Giovannini, C.; Sapora, O.; Viti, V.; Guidoni, L. Characterization of 1H NMR detectable mobile lipids in cells from human adenocarcinomas. FEBS J. 2009, 276, 1333–1346. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.Y.; Yeon, A.; Shahid, M.; Cho, E.; Sairam, V.; Figlin, R.; Kim, K.H.; Kim, J. Reprogrammed lipid metabolism in bladder cancer with cisplatin resistance. Oncotarget 2018, 9, 13231–13243. [Google Scholar] [CrossRef]
- Hong, W.; Zhao, Y.; Cao, L.; Cao, D.; Zhao, Z.; Jin, J. Metabolomics Study on the Differences of Endogenous Small Molecule between A549/DDP and A549 Cells Based on High Solution UPLC-TOF-MS. Zhongguo Fei Ai Za Zhi 2018, 21, 571–577. [Google Scholar] [CrossRef]
- Dar, S.; Chhina, J.; Mert, I.; Chitale, D.; Buekers, T.; Kaur, H.; Giri, S.; Munkarah, A.; Rattan, R. Bioenergetic Adaptations in Chemoresistant Ovarian Cancer Cells. Sci. Rep. 2017, 7, 1–17. [Google Scholar] [CrossRef]
- Puchades-Carrasco, L.; Pineda-Lucena, A. Metabolomics Applications in Precision Medicine: An Oncological Perspective. Curr. Top. Med. Chem. 2017, 17, 2740–2751. [Google Scholar] [CrossRef]
- Li, X.R.; Lu, J.; Kan, Q.C.; Li, X.L.; Fan, Q.; Li, Y.Q.; Huang, R.X.; Slipicevic, A.; Dong, H.P.; Eide, L.; et al. Metabolic reprogramming is associated with flavopiridol resistance in prostate cancer DU145 cells. Sci. Rep. 2017, 7, 1–20. [Google Scholar] [CrossRef]
- Staubert, C.; Bhuiyan, H.; Lindahl, A.; Broom, O.J.; Zhu, Y.; Islam, S.; Linnarsson, S.; Lehtio, J.; Nordstrom, A. Rewired metabolism in drug-resistant leukemia cells: A metabolic switch hallmarked by reduced dependence on exogenous glutamine. J. Biol. Chem. 2015, 290, 8348–8359. [Google Scholar] [CrossRef] [PubMed]
- Nowotarski, S.L.; Feith, D.J.; Shantz, L.M. Skin Carcinogenesis Studies Using Mouse Models with Altered Polyamines. Cancer Growth Metastasis 2015, 8, 17–27. [Google Scholar] [CrossRef]
- Cao, B.; Li, M.J.; Zha, W.B.; Zhao, Q.J.; Gu, R.R.; Liu, L.S.; Shi, J.; Zhou, J.; Zhou, F.; Wu, X.L.; et al. Metabolomic approach to evaluating adriamycin pharmacodynamics and resistance in breast cancer cells. Metabolomics 2013, 9, 960–973. [Google Scholar] [CrossRef] [PubMed]
- Klawitter, J.; Anderson, N.; Klawitter, J.; Christians, U.; Leibfritz, D.; Eckhardt, S.G.; Serkova, N.J. Time-Dependent effects of imatinib in human leukaemia cells: A kinetic NMR-profiling study. Br. J. Cancer 2009, 100, 923–931. [Google Scholar] [CrossRef]
- Sasada, S.; Miyata, Y.; Tsutani, Y.; Tsuyama, N.; Masujima, T.; Hihara, J.; Okada, M. Metabolomic analysis of dynamic response and drug resistance of gastric cancer cells to 5-fluorouracil. Oncol. Rep. 2013, 29, 925–931. [Google Scholar] [CrossRef]
- Wu, W.; Wang, Q.; Yin, F.; Yang, Z.; Zhang, W.; Gabra, H.; Li, L. Identification of proteomic and metabolic signatures associated with chemoresistance of human epithelial ovarian cancer. Int. J. Oncol. 2016, 49, 1651–1665. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.; Chen, X.; Wang, L.; Chen, S. The sweet trap in tumors: Aerobic glycolysis and potential targets for therapy. Oncotarget 2016, 7, 38908–38926. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.; Chen, X.; Sun, X.Q.; Wang, L.T.; Chen, S.W. The Glycolytic Switch in Tumors: How Many Players Are Involved? J. Cancer 2017, 8, 3430–3440. [Google Scholar] [CrossRef]
- Shimanishi, M.; Ogi, K.; Sogabe, Y.; Kaneko, T.; Dehari, H.; Miyazaki, A.; Hiratsuka, H. Silencing of GLUT-1 inhibits sensitization of oral cancer cells to cisplatin during hypoxia. J. Oral Pathol. Med. 2013, 42, 382–388. [Google Scholar] [CrossRef] [PubMed]
- Goode, G.; Gunda, V.; Chaika, N.V.; Purohit, V.; Yu, F.; Singh, P.K. MUC1 facilitates metabolomic reprogramming in triple-negative breast cancer. PLoS ONE 2017, 12, e0179098. [Google Scholar] [CrossRef]
- Tripathi, S.C.; Fahrmann, J.F.; Celiktas, M.; Aguilar, M.; Marini, K.D.; Jolly, M.K.; Katayama, H.; Wang, H.; Murage, E.N.; Dennison, J.B.; et al. MCAM Mediates Chemoresistance in Small-Cell Lung Cancer via the PI3K/AKT/SOX2 Signaling Pathway. Cancer Res. 2017, 77, 4414–4425. [Google Scholar] [CrossRef]
- Qian, X.; Xu, W.; Xu, J.; Shi, Q.; Li, J.; Weng, Y.; Jiang, Z.; Feng, L.; Wang, X.; Zhou, J.; et al. Enolase 1 stimulates glycolysis to promote chemoresistance in gastric cancer. Oncotarget 2017, 8, 47691–47708. [Google Scholar] [CrossRef]
- Fujisawa, K.; Terai, S.; Takami, T.; Yamamoto, N.; Yamasaki, T.; Matsumoto, T.; Yamaguchi, K.; Owada, Y.; Nishina, H.; Noma, T.; et al. Modulation of anti-cancer drug sensitivity through the regulation of mitochondrial activity by adenylate kinase 4. J. Exp. Clin. Cancer Res. 2016, 35, 48. [Google Scholar] [CrossRef]
- Amoroso, M.R.; Matassa, D.S.; Agliarulo, I.; Avolio, R.; Maddalena, F.; Condelli, V.; Landriscina, M.; Esposito, F. Stress-Adaptive Response in Ovarian Cancer Drug Resistance: Role of TRAP1 in Oxidative Metabolism-Driven Inflammation. Adv. Protein Chem. Struct. Biol. 2017, 108, 163–198. [Google Scholar] [CrossRef] [PubMed]
- Fekir, K.; Dubois-Pot-Schneider, H.; Desert, R.; Daniel, Y.; Glaise, D.; Rauch, C.; Morel, F.; Fromenty, B.; Musso, O.; Cabillic, F.; et al. Retrodifferentiation of Human Tumor Hepatocytes to Stem Cells Leads to Metabolic Reprogramming and Chemoresistance. Cancer Res. 2019, 79, 1869–1883. [Google Scholar] [CrossRef] [PubMed]
- Osborne, M.J.; de Oliveira, L.C.; Volpon, L.; Zahreddine, H.A.; Borden, K.L.B. Overcoming Drug Resistance through the Development of Selective Inhibitors of UDP-Glucuronosyltransferase Enzymes. J. Mol. Biol. 2019, 431, 258–272. [Google Scholar] [CrossRef]
- Zhou, J.X.; Wink, M. Reversal of Multidrug Resistance in Human Colon Cancer and Human Leukemia Cells by Three Plant Extracts and Their Major Secondary Metabolites. Medicines 2018, 5, 123. [Google Scholar] [CrossRef] [PubMed]
- Zong, L.; Cheng, G.; Liu, S.; Pi, Z.; Liu, Z.; Song, F. Reversal of multidrug resistance in breast cancer cells by a combination of ursolic acid with doxorubicin. J. Pharm. Biomed. Anal. 2019, 165, 268–275. [Google Scholar] [CrossRef]
- Alonezi, S.; Tusiimire, J.; Wallace, J.; Dufton, M.J.; Parkinson, J.A.; Young, L.C.; Clements, C.J.; Park, J.K.; Jeon, J.W.; Ferro, V.A.; et al. Metabolomic Profiling of the Synergistic Effects of Melittin in Combination with Cisplatin on Ovarian Cancer Cells. Metabolites 2017, 7, 14. [Google Scholar] [CrossRef]
- Lin, L.; Li, L.; Chen, X.; Zeng, B.; Lin, T. Preliminary evaluation of the potential role of beta-elemene in reversing erlotinib-resistant human NSCLC A549/ER cells. Oncol. Lett. 2018, 16, 3380–3388. [Google Scholar] [CrossRef]
- Capella Roca, B.; Lao, N.; Barron, N.; Doolan, P.; Clynes, M. An arginase-based system for selection of transfected CHO cells without the use of toxic chemicals. J. Biol. Chem. 2019, 294, 18756–18768. [Google Scholar] [CrossRef]
- Bayet-Robert, M.; Morvan, D. Metabolomics reveals metabolic targets and biphasic responses in breast cancer cells treated by curcumin alone and in association with docetaxel. PLoS ONE 2013, 8, e57971. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).