Polygenic Risk Scores for Personalized Cardiovascular Pharmacogenomics―A Scoping Review
Abstract
:1. Introduction
2. Methods
2.1. Data Source and Search Strategy
2.2. Inclusion Criteria
2.3. Main Outcomes
2.4. Data Extraction
3. Results
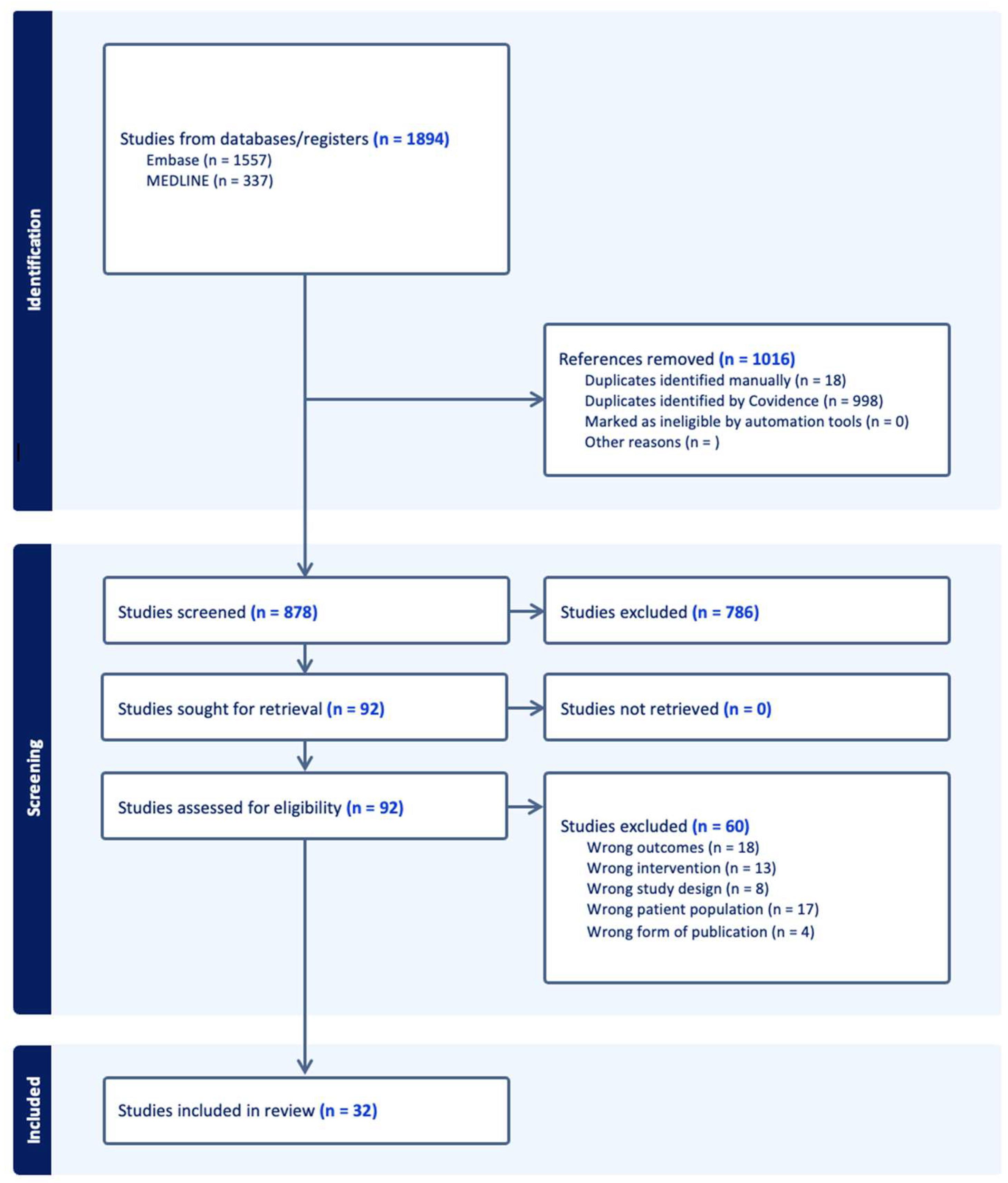
3.1. Study Selection
3.2. Characteristics of Included Studies
3.3. Lipid-Lowering Medications
3.4. Antihypertensives (Calcium Channel Blockers (CCBs), Beta-Blockers (BBs), Thiazide Diuretics)
3.5. Rate-Control Medications
3.6. Antiarrhythmics
3.7. Antiplatelets
3.8. Acetaminophen and Ibuprofen
4. Discussion
4.1. Nature Versus Nurture: A Clinician’s Dilemma
4.2. PRS-PGx and Healthcare Costs in a Clinical Context
4.3. Challenges in Using PRS in Cardiovascular Pharmacogenomics
4.4. Strengths and Limitations
4.5. Future Directions
4.5.1. Short-Term Priorities (Within 2–5 Years)
4.5.2. Medium-Term Priorities (Within 5–10 Years)
4.5.3. Long-Term Priorities (Within 10–15 Years)
4.6. Potential Strategies to Streamline Implementation
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Cardiovascular Diseases (CVDs). Available online: https://www.who.int/news-room/fact-sheets/detail/cardiovascular-diseases-(cvds) (accessed on 31 October 2024).
- Khera, A.V.; Won, H.-H.; Peloso, G.M.; Lawson, K.S.; Bartz, T.M.; Deng, X.; van Leeuwen, E.M.; Natarajan, P.; Emdin, C.A.; Bick, A.G.; et al. Diagnostic Yield and Clinical Utility of Sequencing Familial Hypercholesterolemia Genes in Patients With Severe Hypercholesterolemia. J. Am. Coll. Cardiol. 2016, 67, 2578–2589. [Google Scholar] [CrossRef] [PubMed]
- Khera, A.V.; Emdin, C.A.; Drake, I.; Natarajan, P.; Bick, A.G.; Cook, N.R.; Chasman, D.I.; Baber, U.; Mehran, R.; Rader, D.J.; et al. Genetic Risk, Adherence to a Healthy Lifestyle, and Coronary Disease. N. Engl. J. Med. 2016, 375, 2349–2358. [Google Scholar]
- Kathiresan, S.; Srivastava, D. Genetics of Human Cardiovascular Disease. Cell 2012, 148, 1242–1257. [Google Scholar] [CrossRef] [PubMed]
- Kirk, B.W.; Feinsod, M.; Favis, R.; Kliman, R.M.; Barany, F. Single nucleotide polymorphism seeking long term association with complex disease. Nucleic Acids Res. 2002, 30, 3295–3311. [Google Scholar] [CrossRef]
- Humphries, S.E. Common Variants for Cardiovascular Disease. Circulation 2017, 135, 2102–2105. [Google Scholar] [CrossRef]
- Sitinjak, B.D.P.; Murdaya, N.; Rachman, T.A.; Zakiyah, N.; Barliana, M.I. The Potential of Single Nucleotide Polymorphisms (SNPs) as Biomarkers and Their Association with the Increased Risk of Coronary Heart Disease: A Systematic Review. Vasc. Health Risk Manag. 2023, 19, 289–301. [Google Scholar] [CrossRef]
- Zhu, C.; Baumgarten, N.; Wu, M.; Wang, Y.; Das, A.P.; Kaur, J.; Ardakani, F.B.; Duong, T.T.; Pham, M.D.; Duda, M.; et al. CVD-associated SNPs with regulatory potential reveal novel non-coding disease genes. Hum. Genom. 2023, 17, 1–17. [Google Scholar] [CrossRef]
- Lewis, C.M.; Vassos, E. Polygenic risk scores: From research tools to clinical instruments. Genome Med. 2020, 12, 44. [Google Scholar] [CrossRef]
- Collister, J.A.; Liu, X.; Clifton, L. Calculating Polygenic Risk Scores (PRS) in UK Biobank: A Practical Guide for Epidemiologists. Front. Genet. 2022, 13, 818574. [Google Scholar]
- Samani, N.J.; Beeston, E.; Greengrass, C.; Riveros-McKay, F.; Debiec, R.; Lawday, D.; Wang, Q.; Budgeon, C.A.; Braund, P.S.; Bramley, R.; et al. Polygenic risk score adds to a clinical risk score in the prediction of cardiovascular disease in a clinical setting. Eur. Heart J. 2024, 45, 3152–3160. [Google Scholar] [CrossRef]
- Marston, N.A.; Pirruccello, J.P.; Melloni, G.E.M.; Koyama, S.; Kamanu, F.K.; Weng, L.-C.; Roselli, C.; Kamatani, Y.; Komuro, I.; Aragam, K.G.; et al. Predictive Utility of a Coronary Artery Disease Polygenic Risk Score in Primary Prevention. JAMA Cardiol. 2023, 8, 130–137. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Pennells, L.; Kaptoge, S.; Nelson, C.P.; Ritchie, S.C.; Abraham, G.; Arnold, M.; Bell, S.; Bolton, T.; Burgess, S.; et al. Polygenic risk scores in cardiovascular risk prediction: A cohort study and modelling analyses. PLoS Med. 2021, 18, e1003498. [Google Scholar] [CrossRef]
- Relling, M.V.; Evans, W.E. Pharmacogenomics in the clinic. Nature 2015, 526, 343–350. [Google Scholar] [PubMed]
- O’shea, J.; Ledwidge, M.; Gallagher, J.; Keenan, C.; Ryan, C. Pharmacogenetic interventions to improve outcomes in patients with multimorbidity or prescribed polypharmacy: A systematic review. Pharmacogenom. J. 2022, 22, 89–99. [Google Scholar] [CrossRef]
- Igo, R.P.; Kinzy, T.G.; Cooke Bailey, J.N. Genetic Risk Scores. Curr. Protoc. Hum. Genet. 2019, 104, e95. [Google Scholar] [PubMed]
- Oni-Orisan, A.; Haldar, T.; Cayabyab, M.A.; Ranatunga, D.K.; Hoffmann, T.J.; Iribarren, C.; Krauss, R.M.; Risch, N. Polygenic Risk Score and Statin Relative Risk Reduction for Primary Prevention of Myocardial Infarction in a Real-World Population. Clin. Pharmacol. Ther. 2022, 112, 1070–1078. [Google Scholar] [CrossRef]
- Mega, J.L.; Stitziel, N.O.; Smith, J.G.; Chasman, D.I.; Caulfield, M.J.; Devlin, J.J.; Nordio, F.; Hyde, C.L.; Cannon, C.P.; Sacks, F.M.; et al. Genetic risk, coronary heart disease events, and the clinical benefit of statin therapy: An analysis of primary and secondary prevention trials. Lancet 2015, 385, 2264–2271. [Google Scholar] [CrossRef]
- Natarajan, P.; Young, R.; Stitziel, N.O.; Padmanabhan, S.; Baber, U.; Mehran, R.; Sartori, S.; Fuster, V.; Reilly, D.F.; Butterworth, A.; et al. Polygenic risk score identifies subgroup with higher burden of atherosclerosis and greater relative benefit from statin therapy in the primary prevention setting. Circulation 2017, 135, 2091–2101. [Google Scholar]
- Jarmul, J.; Pletcher, M.J.; Lich, K.H.; Wheeler, S.B.; Weinberger, M.; Avery, C.L.; Jonas, D.E.; Earnshaw, S.; Pignone, M. Cardiovascular Genetic Risk Testing for Targeting Statin Therapy in the Primary Prevention of Atherosclerotic Cardiovascular Disease. Circ. Cardiovasc. Qual. Outcomes 2018, 11, e004171. [Google Scholar] [CrossRef]
- Leusink, M.; der Zee, A.H.M.-V.; Ding, B.; Drenos, F.; van Iperen, E.P.; Warren, H.R.; Caulfield, M.J.; Cupples, L.A.; Cushman, M.; Hingorani, A.D.; et al. A Genetic Risk Score is Associated with Statin-Induced Low-Density Lipoprotein Cholesterol Lowering. Pharmacogenomics 2016, 17, 583–591. [Google Scholar] [CrossRef]
- Hamrefors, V.; Orho-Melander, M.; Krauss, R.M.; Hedblad, B.; Almgren, P.; Berglund, G.; Melander, O. A gene score of nine LDL and HDL regulating genes is associated with fluvastatin-induced cholesterol changes in women. J. Lipid Res. 2010, 51, 625–634. [Google Scholar] [CrossRef] [PubMed]
- Marston, N.A.; Kamanu, F.K.; Nordio, F.; Gurmu, Y.; Roselli, C.; Sever, P.S.; Pedersen, T.R.; Keech, A.C.; Wang, H.; Lira Pineda, A.; et al. Predicting Benefit From Evolocumab Therapy in Patients With Atherosclerotic Disease Using a Genetic Risk Score Results from the FOURIER Trial. Circulation 2020, 141, 616–623. [Google Scholar]
- D’Erasmo, L.; Minicocci, I.; Di Costanzo, A.; Pigna, G.; Commodari, D.; Ceci, F.; Montali, A.; Brancato, F.; Stanca, I.; Nicolucci, A.; et al. Clinical implications of monogenic versus polygenic hypercholesterolemia: Long-term response to treatment, coronary atherosclerosis burden, and cardiovascular events. J. Am. Heart Assoc. 2021, 10, 18932. [Google Scholar]
- Kiflen, M.; Le, A.; Mao, S.; Lali, R.; Narula, S.; Xie, F.; Paré, G. Cost-Effectiveness of Polygenic Risk Scores to Guide Statin Therapy for Cardiovascular Disease Prevention. Circ. Genom. Precis. Med. 2022, 15, 386–395. [Google Scholar] [CrossRef]
- Damask, A.; Steg, P.G.; Schwartz, G.G.; Szarek, M.; Hagström, E.; Badimon, L.; Chapman, M.J.; Boileau, C.; Tsimikas, S.; Ginsberg, H.N.; et al. Patients With High Genome-Wide Polygenic Risk Scores for Coronary Artery Disease May Receive Greater Clinical Benefit From Alirocumab Treatment in the ODYSSEY OUTCOMES Trial. Circulation 2020, 141, 624–636. [Google Scholar] [CrossRef] [PubMed]
- Mayerhofer, E.; Malik, R.; Parodi, L.; Burgess, S.; Harloff, A.; Dichgans, M.; Rosand, J.; Anderson, C.D.; Georgakis, M.K. Genetically predicted on-statin LDL response is associated with higher intracerebral haemorrhage risk. Brain 2022, 145, 2677–2686. [Google Scholar] [CrossRef]
- Pechlivanis, S.; Jung, D.; Moebus, S.; Lehmann, N.; Mahabadi, A.A.; Hoffmann, P.; Erbel, R.; Nöthen, M.M.; Bachmann, H.S. Pharmacogenetic association of diabetes-associated genetic risk score with rapid progression of coronary artery calcification following treatment with HMG-CoA-reductase inhibitors —Results of the Heinz Nixdorf Recall Study. Naunyn Schmiedebergs Arch. Pharmacol. 2021, 394, 1713–1725. [Google Scholar]
- Zhai, Y.; Chen, H.; Che, B.; Liu, Y.; Peng, Y.; Chen, J.; Xu, T.; He, J.; Zhang, Y.; Zhong, C. Efficacy of Immediate Antihypertensive Treatment in Patients With Acute Ischemic Stroke With Different Blood Pressure Genetic Variants. Hypertension 2024, 81, 658–667. [Google Scholar] [CrossRef]
- Marques-Vidal, P.; Chekanova, V.; Vaucher, J. Association between genetic risk of high SBP and hypertension control: The CoLausjPsyColaus study. J. Hypertens 2022, 40, 1388–1393. [Google Scholar]
- Network, O.B.O.E.; El Rouby, N.; McDonough, C.W.; Gong, Y.; McClure, L.A.; Mitchell, B.D.; Horenstein, R.B.; Talbert, R.L.; Crawford, D.C.; Gitzendanner, M.A.; et al. Genome-wide association analysis of common genetic variants of resistant hypertension. Pharmacogenom. J. 2018, 19, 295–304. [Google Scholar] [CrossRef]
- McDonough, C.W.; Gong, Y.; Padmanabhan, S.; Burkley, B.; Langaee, T.Y.; Melander, O.; Pepine, C.J.; Dominiczak, A.F.; Cooper-DeHoff, R.M.; Johnson, J.A. Pharmacogenomic association of nonsynonymous SNPs in SIGLEC12, A1BG, and the selectin region and cardiovascular outcomes. Hypertension 2013, 62, 48–54. [Google Scholar] [CrossRef] [PubMed]
- Gong, Y.; McDonough, C.W.; Wang, Z.; Hou, W.; Cooper-DeHoff, R.M.; Langaee, T.Y.; Beitelshees, A.L.; Chapman, A.B.; Gums, J.G.; Bailey, K.R.; et al. Hypertension susceptibility loci and blood pressure response to antihypertensives: Results from the phar-macogenomic evaluation of antihypertensive responses study. Circ. Cardiovasc. Genet. 2012, 5, 686–691. [Google Scholar]
- Lynch, A.I.; Eckfeldt, J.H.; Davis, B.R.; Ford, C.E.; Boerwinkle, E.; Leiendecker-Foster, C.; Arnett, D.K. Gene panels to help identify subgroups at high and low risk of coronary heart disease among those randomized to antihypertensive treatment: The GenHAT study. Pharmacogenet Genom. 2012, 22, 355–366. [Google Scholar] [CrossRef]
- Sánez Tähtisalo, H.; Ruotsalainen, S.; Mars, N.; Porthan, K.; Oikarinen, L.; Virolainen, J.; Fyhrquist, F.; Ripatti, S.; Kontula, K.K.; Hiltunen, T.P. Human essential hypertension: No significant association of polygenic risk scores with antihyperten-sive drug responses. Sci. Rep. 2020, 10, 11940. [Google Scholar]
- Türkmen, D.; Bowden, J.; Masoli, J.A.H.; Delgado, J.; Kuo, C.-L.; Melzer, D.; Pilling, L.C. Polygenic scores for cardiovascular risk factors improve estimation of clinical outcomes in CCB treatment compared to pharmacogenetic variants alone. Pharmacogenom. J. 2024, 24, 1–7. [Google Scholar] [CrossRef]
- Åberg, F.; Kantojärvi, K.; Männistö, V.; But, A.; Salomaa, V.; Niiranen, T.; Färkkilä, M.; Luukkonen, P.; Männistö, S.; Lundqvist, A.; et al. Association between arterial hypertension and liver outcomes using polygenic risk scores: A population-based study. Sci. Rep. 2022, 12, 15581. [Google Scholar]
- Acosta, J.N.; Both, C.P.; Demarais, Z.S.; Conlon, C.J.; Leasure, A.C.; Torres-Lopez, V.M.; de Havenon, A.; Petersen, N.H.; Gill, T.M.; Sansing, L.H.; et al. Polygenic Susceptibility to Hypertension and Blood Pressure Control in Stroke Survivors. Neurology 2023, 100, e1587–e1597. [Google Scholar] [CrossRef]
- Maroteau, C.; Siddiqui, M.K.; Veluchamy, A.; Carr, F.; White, M.; Cassidy, A.J.; Baranova, E.V.; Rasmussen, E.R.; Eriksson, N.; Bloch, K.M.; et al. Exome Sequencing Reveals Common and Rare Variants in F5 Associated With ACE Inhibitor and Angio-tensin Receptor Blocker–Induced Angioedema. Clin. Pharmacol. Ther. 2020, 108, 1195–1202. [Google Scholar]
- Narang, R.K.; Gamble, G.; Phipps-Green, A.J.; Topless, R.; Cadzow, M.; Stamp, L.K.; Merriman, T.R.; Dalbeth, N. Do Serum Urate–associated Genetic Variants Influence Gout Risk in People Taking Diuretics? Analysis of the UK Biobank. J. Rheumatol. 2020, 47, 1704–1711. [Google Scholar] [CrossRef] [PubMed]
- Barrett, T.W.; Self, W.H.; Darbar, D.; Jenkins, C.A.; Wasserman, B.S.; Kassim, N.A.; Casner, M.; Shoemaker, M.B. Association of atrial fibrillation risk alleles and response to acute rate control therapy. Am. J. Emerg. Med. 2016, 34, 735–740. [Google Scholar] [CrossRef]
- Luzum, J.A.; Campos-Staffico, A.M.; Li, J.; She, R.; Gui, H.; Peterson, E.L.; Liu, B.; Sabbah, H.N.; Donahue, M.P.; Kraus, W.E.; et al. Genome-Wide Association Study of Beta-Blocker Survival Benefit in Black and White Patients with Heart Failure with Reduced Ejection Fraction. Genes 2023, 14, 2019. [Google Scholar] [CrossRef] [PubMed]
- Lanfear, D.E.; Luzum, J.A.; She, R.; Gui, H.; Donahue, M.P.; O’connor, C.M.; Adams, K.F.; Wijk, S.S.-V.; Zeld, N.; Maeder, M.T.; et al. Polygenic Score for β-Blocker Survival Benefit in European Ancestry Patients With Reduced Ejection Fraction Heart Failure. Circ. Heart Fail. 2020, 13, e007012. [Google Scholar] [CrossRef]
- Strauss, D.G.; Vicente, J.; Johannesen, L.; Blinova, K.; Mason, J.W.; Weeke, P.; Behr, E.R.; Roden, D.M.; Woosley, R.; Kosova, G.; et al. Common Genetic Variant Risk Score Is Associated With Drug-Induced QT Prolongation and Torsade de Pointes Risk. Circulation 2017, 135, 1300–1310. [Google Scholar] [CrossRef]
- Tadros, R.; Tan, H.L.; ESCAPE-NET Investigators; El Mathari, S.; Kors, J.A.; Postema, P.G.; Lahrouchi, N.; Beekman, L.; Radivojkov-Blagojevic, M.; Amin, A.S.; et al. Predicting cardiac electrical response to sodium-channel blockade and Brugada syndrome using polygenic risk scores. Eur. Heart J. 2019, 40, 3097–3107. [Google Scholar] [CrossRef]
- Lewis, J.P.; Backman, J.D.; Reny, J.-L.; Bergmeijer, T.O.; Mitchell, B.D.; Ritchie, M.D.; Déry, J.-P.; E Pakyz, R.; Gong, L.; Ryan, K.; et al. Pharmacogenomic polygenic response score predicts ischaemic events and cardiovascular mortality in clopidogrel-treated patients. Eur. Heart J.-Cardiovasc. Pharmacother. 2020, 6, 203–210. [Google Scholar] [CrossRef] [PubMed]
- Lacaze, P.; Bakshi, A.; Riaz, M.; Polekhina, G.; Owen, A.; Bhatia, H.S.; Natarajan, P.; Wolfe, R.; Beilin, L.; Nicholls, S.J.; et al. Aspirin for Primary Prevention of Cardiovascular Events in Relation to Lipoprotein(a) Genotypes. J. Am. Coll. Cardiol. 2022, 80, 1287–1298. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhou, C.; Yang, S.; Zhang, Y.; Ye, Z.; He, P.; Liu, M.; Wu, Q.; Qin, X. Association of regular use of ibuprofen and paracetamol, genetic susceptibility, and new-onset dementia in the older population. Gen Hosp Psychiatry 2023, 84, 226–233. [Google Scholar]
- Phulka, J.S.; Ashraf, M.; Bajwa, B.K.; Pare GLaksman, Z. Current State and Future of Polygenic Risk Scores in Cardi-ometabolic Disease: A Scoping Review. Circ. Genom. Precis Med. 2023, 16, 286–313. [Google Scholar] [CrossRef]
- Peng, L.; Zhong, X. Epigenetic regulation of drug metabolism and transport. Acta Pharm. Sin. B 2015, 5, 106–112. [Google Scholar] [CrossRef]
- de La Harpe, R.; Thorball, C.W.; Redin, C.; Fournier, S.; Müller, O.; Strambo, D.; Michel, P.; Vollenweider, P.; Marques-Vidal, P.; Fellay, J.; et al. Combining European and U.S. risk prediction models with polygenic risk scores to refine cardiovascular prevention: The CoLaus|PsyCoLaus Study. Eur. J. Prev. Cardiol. 2023, 30, 561–571. [Google Scholar] [CrossRef]
- O’sullivan, J.W.; Shcherbina, A.; Justesen, J.M.; Turakhia, M.; Perez, M.; Wand, H.; Tcheandjieu, C.; Clarke, S.L.; Rivas, M.A.; Ashley, E.A. Combining Clinical and Polygenic Risk Improves Stroke Prediction Among Individuals With Atrial Fibrillation. Circ. Genom. Precis. Med. 2021, 14, 339–347. [Google Scholar] [CrossRef]
- Zhu, Y.; Swanson, K.M.; Rojas, R.L.; Wang, Z.; Sauver, J.L.; Visscher, S.L.; Prokop, L.J.; Bielinski, S.J.; Wang, L.; Weinshilboum, R.; et al. Systematic review of the evidence on the cost-effectiveness of pharmacogenomics-guided treatment for cardio-vascular diseases. Genet. Med. 2019, 22, 475. [Google Scholar]
- National Health Expenditure Trends|CIHI. Available online: https://www.cihi.ca/en/national-health-expenditure-trends (accessed on 11 January 2025).
- Smith, J.L.; Tcheandjieu, C.; Dikilitas, O.; Iyer, K.; Miyazawa, K.; Hilliard, A.; Lynch, J.; Rotter, J.I.; Chen, Y.D.; Sheu, W.H.; et al. Multi-Ancestry Polygenic Risk Score for Coronary Heart Disease Based on an Ancestrally Diverse Ge-nome-Wide Association Study and Population-Specific Optimization. Circ. Genom. Precis. Med. 2024, 17, e004272. [Google Scholar] [CrossRef] [PubMed]
- Moreno-Grau, S.; Vernekar, M.; Lopez-Pineda, A.; Mas-Montserrat, D.; Barrabés, M.; Quinto-Cortés, C.D.; Moatamed, B.; Lee, M.T.M.; Yu, Z.; Numakura, K.; et al. Polygenic risk score portability for common diseases across genetically diverse populations. Hum. Genom. 2024, 18, 1–12. [Google Scholar] [CrossRef]
- UK-Canadian AI Initiative to Create Equitable Multi-Ethnic Polygenic Risk Scores That Improve Clinical Care-HDR UK. Available online: https://www.hdruk.ac.uk/projects/uk-canadian-ai-initiative-to-create-equitable-multi-ethnic-polygenic-risk-scores-that-improve-clinical-care/ (accessed on 10 January 2025).
- Slunecka, J.L.; van der Zee, M.D.; Beck, J.J.; Johnson, B.N.; Finnicum, C.T.; Pool, R.; Hottenga, J.-J.; de Geus, E.J.C.; Ehli, E.A. Implementation and implications for polygenic risk scores in healthcare. Hum. Genom. 2021, 15, 1–18. [Google Scholar] [CrossRef]
- Ayatollahi, H.; Hosseini, S.F.; Hemmat, M. Integrating Genetic Data into Electronic Health Records: Medical Geneticists’ Perspectives. Healt Informatics Res. 2019, 25, 289–296. [Google Scholar] [CrossRef]
- Torkamani, A.; Wineinger, N.E.; Topol, E.J. The personal and clinical utility of polygenic risk scores. Nat. Rev. Genet. 2018, 19, 581–590. [Google Scholar] [CrossRef] [PubMed]
- Lapointe, J.; Buron, A.-C.; Mbuya-Bienge, C.; Dorval, M.; Pashayan, N.; Brooks, J.D.; Walker, M.J.; Chiquette, J.; Eloy, L.; Blackmore, K.; et al. Polygenic risk scores and risk-stratified breast cancer screening: Familiarity and perspectives of health care professionals. Genet. Med. 2022, 24, 2380–2388. [Google Scholar] [CrossRef]
- Brar, S.; Townsend, J.; Phulka, J.; Halperin, L.; Liew, J.; Parker, J.; Brunham, L.R.; Laksman, Z. Knowledge, attitudes and demand toward cardiovascular polygenic risk testing in clinical practice: Cross-sectional survey of patients. Eur. J. Hum. Genet. 2024, 1–7. [Google Scholar] [CrossRef]
- Cai, X.; Li, H.; Cao, X.; Ma, X.; Zhu, W.; Xu, L.; Yang, S.; Yu, R.; Huang, P. Integrating transcriptomic and polygenic risk scores to enhance predictive accuracy for ischemic stroke subtypes. Hum. Genet. 2024, 144, 43–54. [Google Scholar] [CrossRef]
- Liang, Y.; Pividori, M.; Manichaikul, A.; Palmer, A.A.; Cox, N.J.; Wheeler, H.E.; Im, H.K. Polygenic transcriptome risk scores (PTRS) can improve portability of polygenic risk scores across ancestries. Genome Biol. 2022, 23, 1–18. [Google Scholar] [CrossRef]
- Zhai, S.; Zhang, H.; Mehrotra, D.V.; Shen, J. Pharmacogenomics polygenic risk score for drug response prediction using PRS-PGx methods. Nat. Commun. 2022, 13, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Kadlec, H.; Hollander, M.J.; Clelland, C.; Kallstrom, L. Family physicians enhance end-of-life care: Evaluation of a new continuing medical education learning module in British Columbia. BMC Med. Educ. 2015, 15, 119. [Google Scholar] [CrossRef] [PubMed]
- Martin, M.A.; Kroetz, D.L. Abacavir Pharmacogenetics—From Initial Reports to Standard of Care. Pharmacother. J. Hum. Pharmacol. Drug Ther. 2013, 33, 765–775. [Google Scholar] [CrossRef]
- Abramowitz, S.A.; Boulier, K.; Keat, K.; Cardone, K.M.; Shivakumar, M.; DePaolo, J.; Judy, R.; Bermudez, F.; Mimouni, N.; Neylan, C.; et al. Evaluating Performance and Agreement of Coronary Heart Disease Polygenic Risk Scores. JAMA 2025, 333, 60–70. [Google Scholar] [CrossRef]
| Authors (y) | Refs. | Participant Race/Ancestry | Medication Class |
|---|---|---|---|
| Oni-Orisan A. et al., 2022 | [17] | White, Black, Latinx, and East Asian | Lipid-Lowering |
| Mega et al., 2015 | [18] | White | Lipid-Lowering |
| Natarajan et al., 2017 | [19] | White | Lipid-Lowering |
| Jarmul et al., 2018 | [20] | N/A | Lipid-Lowering |
| Leusink et al., 2016 | [21] | White | Lipid-Lowering |
| Hamrefors et al., 2010 | [22] | N/A | Lipid-Lowering |
| Marston et al., 2020 | [23] | White | Lipid-Lowering |
| Erasmo et al., 2021 | [24] | Italian | Lipid-Lowering |
| Kiflen et al., 2022 | [25] | White | Lipid-Lowering |
| Damask et al., 2020 | [26] | White, Asian, Black, and Others | Lipid-Lowering |
| Mayerhofer et al., 2022 | [27] | White | Lipid-Lowering |
| Pechlivanis et al., 2021 | [28] | White | Lipid-Lowering |
| Zhai et al., 2024 | [29] | Chinese | Antihypertensives |
| Vidal et al., 2022 | [30] | Not Mentioned Explicitly—Participants from Lausanne, Switzerland | Antihypertensives |
| Rouby et al., 2019 | [31] | White and Hispanic | Antihypertensives |
| McDonough et al., 2013 | [32] | White and Hispanic | Antihypertensives |
| Gong et al., 2012 | [33] | White and Black | Antihypertensives |
| Lynch et al., 2012 | [34] | White, Black, American Indians/Alaskan Natives, Asians/Pacific Islanders, and Others | Antihypertensives |
| Tähtisalo et al., 2020 | [35] | Finnish | Antihypertensives |
| Türkmen et al., 2024 | [36] | White | Antihypertensives |
| Åberg et al., 2022 | [37] | Finnish | Antihypertensives |
| Acosta et al., 2023 | [38] | White | Antihypertensives |
| Maroteau et al., 2020 | [39] | White and Black | Antihypertensives |
| Narang et al., 2020 | [40] | White | Antihypertensives |
| Barrett et al., 2016 | [41] | White | Rate-Control Agents |
| Luzum et al., 2023 | [42] | White and Black | Rate-Control Agents |
| Lanfear et al., 2020 | [43] | White | Rate-Control Agents |
| Strauss et al., 2017 | [44] | White, Black, and Asian | Antiarrhythmics |
| Tadros et al., 2019 | [45] | White | Antiarrhythmics |
| Lewis et al., 2020 | [46] | White | Antiplatelets |
| Lacaze et al., 2022 | [47] | White | Antiplatelets |
| Zhang et al., 2023 | [48] | White | Anti-Inflammatory |
| Authors (y) | Discovery GWAS (n) | Validation/Test Dataset (n) | PRS | SNPs in PRS | HR/OR (95% CI) | Clinical Factors Included in Model | Main Findings |
|---|---|---|---|---|---|---|---|
| Oni-Orisan A. et al., 2022 [1] | N/A | 32,736 | CHD PRS | 164 | Statin effectiveness in high PRS, HR = 0.41 (0.31–0.53) | Age, sex, HTN, diabetes, smoking, LDLc levels, ASCVD risk score | Statin effectiveness was highest in the high-PRS group, intermediate in the intermediate group, and lowest in the low PRS group, while ASCVD risk and statin LDLc lowering were similar across all PRS groups. |
| Mega et al., 2015 [2] | N/A | 48,421 | CHD PRS | 27 | Statin effectiveness in high PRS, HR = 0.52 (0.37–0.71) | Age, sex, diabetes status, smoking, race (if applicable), Fx of CHD, HDLc LDLc, HTN | PRS identified individuals at higher risk for incident and recurrent CHD events, with those having a high PRS experiencing the greatest benefit from statin therapy. For primary prevention, high-PRS individuals had a threefold-lower NNT compared to low-risk groups. |
| Natarajan et al., 2017 [3] | N/A | WOSCOPS (4910), CARDIA (1154), BioImage (4392) | CHD PRS | 57 | Statin effectiveness in high PRS, HR = 0.56 (0.40–0.78) | Age, sex, DM, smoking, baseline LDLc, baseline HDLc, SBP, antihypertensive medication status, Fx of MI, or stroke | Statin therapy led to greater RR reduction in those at high genetic risk for CHD. High genetic risk was associated with greater burden of subclinical atherosclerosis. |
| Jarmul et al., 2018 [4] | N/A | 10,000 | Cardiovascular PRS (cPRS) | 27 | N/A | Age, sex, ASCVD risk factors (SBP, total cholesterol, HDLc, smoking, antihypertensive medication use) | Testing a cPRS is usually not cost-effective for guiding statin therapy in primary ASCVD prevention among low- to intermediate-risk patients. However, in specific cases, cPRS testing may be cost-effective under certain assumptions. |
| Leusink et al., 2016 [5] | 1991 | 5314 | PRS for statin response | 50,000 (discovery), 3 (risk score) | N/A | Age, sex, baseline LDLc, population stratification | No new SNPs were linked to statin-induced LDLc reduction. The PRS had a minor but significant effect, with each allele reducing LDLc response by 2%. Since statins effectively lower LDLc regardless of genotype, genetic testing is unlikely to significantly influence statin therapy decisions. |
| Hamrefors et al., 2010 [6] | N/A | 395 | LDL + HDL PRS | 9 | N/A | Age, BMI reduction, baseline blood glucose, % of BMI change | PRS was associated with fluvastatin-induced changes in LDL and HDL in women but not in men. Higher PRS correlated with smaller LDL reductions and greater HDL increases. Additionally, higher LDL + HDL and HDL-specific scores were linked to larger HDL increases in women. |
| Marston et al., 2020 [7] | N/A | 14,298 | CHD PRS | 27 | Evolocumab effectiveness in high PRS, HR = 0.75 (0.60–0.94) | Age, sex, HTN, DM, smoking, eGFR, ancestry (using the first 5 PCs) | Individuals with intermediate and high PRS had 1.32- and 1.66-fold-increased hazard for major coronary events, respectively. Patients with high PRS had the greatest benefit from evolocumab, reducing their event rates to levels similar to patients with low genetic risk. |
| Erasmo et al., 2021 [8] | N/A | 370 | LDLc PRS | 6 | N/A | Age, gender, smoking, LDLc levels, and LLT intensity | Monogenic FH patients showed higher baseline LDLc and poorer LLT response compared to polygenic FH and undefined groups. |
| Kiflen et al., 2022 [9] | NA | 96,116 | CAD PRS | N/A | N/A | FRS for intermediate CVD risk classification (age, sex, LDLc, non-HDLc, ApoB, additional CVD risk factors) | The most cost-effective strategy was prescribing statins to intermediate-risk individuals with PRS in the top 70%, excluding those in the bottom 1%. |
| Damask et al., 2020 [10] | 184,305 | 11,953 | CAD PRS | 6,579,025 | Alirocumab effectiveness in high PRS, HR = 0.63 (0.46–0.86) | Ancestry, age, sex, baseline LDLc, lipoprotein(a), Fx of premature CHD, medical characteristics before the index ACS (MI, Percutaneous Coronary Intervention, CABG, and CHF) | Patients with high PRS for CAD had a greater incidence of MACE. Alirocumab treatment resulted in greater absolute and RR of MACE in high-PRS patients compared to lower-PRS patients. |
| Mayerhofer et al., 2022 [11] | 40,914 | 225,195 | On-Statin LDL PRS | 35 | ICH risk in high-statin-response group: HR = 1.16 (1.05–1.28) Statin effectiveness in high-response group: MI, HR = 0.98 (0.96–0.99); PAD, HR = 0.93 (0.87–0.99) | Age, sex, BMI, smoking, history of diabetes, SBP, cumulative statin dose exposure, use of anticoagulation and antiplatelet drugs, PCs 1–10, race, kinship, and genotyping assay | A genetically predicted greater LDL reduction from statins was linked to lower LDL levels and reduced risks of MI and PAD. However, among statin users, a higher predicted statin response was also associated with an increased risk of ICH. |
| Pechlivanis et al., 2021 [12] | N/A | 3157 | DM PRS | 100 | Interaction between DM PRS and statin use on CAC, RR = 1.08 (95% CI: 0.83; 1.41) | Age, sex, baseline CAC, statin intake | There was no significant association between the weighted DM PRS and rapid CAC progression, and no interaction between the PRS and statin use was observed for CAC progression. |
| Authors (y) | Discovery GWAS (n) | Validation/Test Dataset (n) | PRS | SNPs in PRS | HR/OR (95% CI) | Clinical Factors Included in Model | AUC/C-Statistic: PRS Alone | AUC/C-Statistic: PRS + Clinical Risk Factors | Main Findings |
|---|---|---|---|---|---|---|---|---|---|
| Zhai et al., 2024 [13] | N/A | 2590 | BP PRS | 5 | Antihypertensive effectiveness in high PRS, OR = 0.91 (0.69–1.22) | Age, sex, baseline SBP, BMI, diabetes, smoking, stroke subtype, baseline National Institutes of Health Stroke Scale score, and time from onset to randomization | N/A | N/A | Early antihypertensive treatment had neutral effects on clinical outcomes in acute ischemic stroke patients across 5 BP-related genetic variants, with no outcome differences between treatment and control groups based on genotype subgroup. |
| Vidal et al., 2022 [14] | N/A | 1097 (baseline), 1126 (1st follow-up), 1020 (2nd follow-up), 809 (3rd follow-up) | SBP PRS | 362 | N/A | Age, sex, marital status, smoking, education, and BMI | N/A | N/A | There was no link between the PRS and hypertension control, but higher PRSs were associated with increased SBP levels in untreated individuals. |
| Rouby et al., 2019 [15] | 1194 | SPS3: 585 eMERGE: 2417 | RHTN PRS | 3 | Variants associated with RHTN despite antihypertensive therapy: rs11749255: OR = 1.60 (1.3–1.9) rs6487504: OR = 1.81 (1.5–2.3) rs324498: OR = 1.62 (1.3–2.0) | Age, sex, BMI, smoking, diabetes, HF, MI, and PVD | N/A | N/A | Associations with RHTN were identified and replicated in the MSX2, IFLTD1, and PTPRD regions. A PRS based on these SNPs was linked to an increased risk of RHTN and was significant in both discovery and replication cohorts. |
| McDonough et al., 2013 [16] | 1345 | 4196 | CVD PRS | 3 | Risk of adverse cardiovascular outcomes: high PRS, OR = 1.31 (1.08–1.59) (favouring BB); low PRS: OR = 0.60 (95% CI: 0.42–0.86) (favouring CCB). | Age, sex, MI, HF, DM, and PCs for ancestry | N/A | N/A | SIGLEC12 rs16982743 and rs893184 significantly interacted with treatment strategies for adverse CV outcomes. A PRS including these SNPs and F5 rs4525 was associated with different CV outcomes based on antihypertensive treatment. Patients with a low PRS benefited more from CCBs, while those with a high PRS had better outcomes with BBs. |
| Gong et al., 2012 [17] | N/A | 768 | BP PRS | 37 | N/A | Baseline BP, age, sex, and PCs for ancestry | N/A | N/A | No individual SNPs reached genome-wide significance (6 had p < 0.05 and 3 had p < 0.01). However, PRS for atenolol and hydrochlorothiazide BP-lowering alleles were significantly associated with BP response. |
| Lynch et al., 2012 [26] | 39,114 | N/A | CHD PRS | 78 candidates Chlorthalidone: 5 AMP: 5 Lisinopril: 6 Doxazosin: 6 | N/A | Sex, age, race, type 2 diabetes, smoking, LVH, total cholesterol, HDLc, SBP, and DBP | N/A | Chlorthalidone Group: AUC(RF): 0.6529 AUC(RF+PRS): 0.6601 Amlodipine Group: AUC(RF): 0.6429 AUC(RF+PRS): 0.6548 Lisinopril Group: AUC(RF): 0.6584 AUC(RF+PRS): 0.6693 Doxazosin Group: AUC(RF): 0.6516 AUC(RF+PRS):0.6705 | Identified treatment-specific PRS that modestly enhanced CHD outcome predictions in hypertensive patients randomized to different antihypertensive drugs. PRS provided a small but statistically significant improvement in CHD prediction within each treatment group. |
| Tähtisalo et al., 2020 [20] | N/A | GENRES cohort: (205 AMP, 207 BPL, 206 HTZ 203 LSN) LIFE cohort: 401 Finnish patients on monotherapy for LSN or ATL | HTN PRS | 793 for Top_PRS, over 1 million for GW_PRS | N/A | Sex, age, BMI, smoking, antihypertensive medication, daily urinary sodium excretion, and serum creatinine | HTZ AUC SBP response (PRS) = 0.64 AUC DBP response (PRS) = 0.63 | N/A | No significant associations were found between PRSs and antihypertensive drug responses after Bonferroni correction. However, higher PRSs were weakly linked to reduced responsiveness to diuretics. Additionally, the GW PRS for SBP correlated with ECG-estimated QRS area and was significantly higher in individuals with drug-resistant HTN compared to those with controlled HTN. |
| Türkmen et al., 2024 [25] | N/A | 32,360 | SBP PRS DBP PRS Body fat mass PRS Waist/hip PRS Lean mass PRS Serum calcium PRS eGFR PRS Lipoprotein(a) PRS Urinary sodium PRS Liver fibrosis PRS | Varies by trait (e.g., SBP = 240 S, DBP = 297). Refer to Table 1 | HF risk despite CCB treatment, HR = 1.14 (1.09–1.19) | Sex, age, and genetic PCs 1–10 | N/A | N/A | Genetically predicted body fat mass, lean mass, and lipoprotein(a) were linked to adverse outcomes in hypertensive patients on CCBs. Genetic predisposition to HF also increased the risk of incident HF in these patients. Individual pharmacogenetic effects were modest, but combining high PRS significantly elevated risk. |
| Åberg et al., 2022 [27] | 757,601 | 33,770 | SBP PRS, DBP PRS | SBP PRS (1,072,098) DBP PRS (1,073,588) | Antihypertensive medication and adverse liver outcomes, HR = 0.55 (0.31–0.97) | Sex and age | N/A | N/A | In the highest quintile of the SBP PRS, new initiation of antihypertensive medication was associated with reduced rates of liver-related outcomes. |
| Acosta et al., 2023 [23] | 5940 | 1750 | SBP PRS, DBP PRS | 732 | RHTN: Highest quintile SBP PRS, OR = 2.28 (1.61–3.28); Highest quintile DBP PRS, OR = 2.27 (1.62–3.22) | Age, sex, and vascular risk factors | N/A | Uncontrolled BP C-index (RF): 0.61 C-index (RF+PRS): 0.62 Resistant BP C-index (RF): 0.75 C-index (RF+PRS): 0.76 | A higher polygenic susceptibility to hypertension is associated with worse BP control in stroke survivors. |
| Maroteau et al., 2020 [24] | 1066 | 652 | F5 Variants PRS | Multiple (11 common and rare nonsynonymous variants in the F5 locus) | ARB-AE/ACE-AE odds with at-least one F5 variant, OR = 2.21 (1.49–3.27) | Sex, age, centre, sequencing batch, PCs 1–10, and covariates associated with intolerance, such as sex and age | N/A | N/A | F5 rs6025 was significantly associated with ACEi-AE and ARB-AE. A combined PRS showed that individuals with at least one variant had significantly higher odds of ACEi-AE or ARB-AE. |
| Narang et al., 2020 [29] | N/A | 359,876 | Gout PRS | 10 | High PRS for gout odds: Non-users, OR = 2.63 (2.49–2.79). Loop, OR = 2.04 (1.65–2.53). Thiazide, OR = 2.70 (2.26–3.23). Thiazide-like, OR = 2.11 (1.37–3.25) | Age, sex, BMI, HTN, renal failure, and HF | N/A | N/A | Serum urate-associated variants strongly increase gout risk in diuretic users, similar to those not taking diuretics. No nonadditive gene–diuretic interactions were observed. |
| Authors (y) | Discovery GWAS (n) | Validation/Test Dataset (n) | PRS | SNPs in PRS | HR/OR (95% CI) | Clinical Factors Included in Model | Main Findings |
|---|---|---|---|---|---|---|---|
| Barrett et al., 2016 [33] | N/A | 142 | AVN conduction PRS, resting heart rate PRS, and AF susceptibility PRS | 24: AVN conduction (4), resting heart rate (12), or AF susceptibility (8) | N/A | Age, sex, baseline ventricular rate, total weight-based diltiazem dose (mg/kg) received in the first 4 h of treatment, and AF susceptibility PRS | Genetic variants related to AVN conduction, resting heart rate, or AF susceptibility did not significantly predict successful rate-control with IV diltiazem for acute AF. |
| Luzum et al., 2023 [32] | 928 | 867 | BB survival benefit PRS | 229: Black patients 18: White patients | rs16844448 × BB survival interaction in Black patients Discovery HR = 73.7 (15.4–353.5) Validation HR = 55.1 (3.4–865.8) | Age, sex, ischemic etiology, AF, stroke, diabetes, BMI, SBP, heart rate, NT pro-BNP, serum creatinine, MAGGIC risk score, BB exposure | Discovery GWAS identified potential genetic variants associated with BB survival benefit in HFrEF patients, but none was validated in an independent dataset. However, rs16844448 in LRP1B showed a suggestive association in Black patients. |
| Lanfear et al., 2020 [40] | 248 | 1188 | BB survival benefit in HFrEF PRP | 44 | BB exposure: high PRS, HR = 0.84 (0.53–1.3) low PRS, HR = 0.19 (0.04–0.51) | MAGGIC score (without BB), BB propensity score, AF, EF, cardiovascular death, age, sex, creatinine, ischemic etiology, stroke, COPD, PVD, HTN | PRP showed that BBs significantly improved survival in patients with low PRP scores but not in those with high scores. The PRP effectively distinguished patients who received substantial survival benefits from BB from those who did not. |
| Authors (y) | Discovery GWAS (n) | Validation/Test Dataset (n) | PRS | SNPs in PRS | HR/OR (95% CI) | Clinical Factors Included in Model | AUC/C-Statistic: PRS Alone | AUC/C-Statistic: PRS + Clinical Risk Factors | Main Findings |
|---|---|---|---|---|---|---|---|---|---|
| Strauss et al., 2017 [39] | N/A | 22 healthy subjects in drug response study, 216 TdP cases, and 771 controls in Torsade de Pointes study | Genetic QT score | 61 | N/A | Age, sex, BMI, baseline QTc, electrolyte levels | N/A | N/A | The genetic QT score was associated with drug-induced QTc prolongation and significantly predicted TdP risk, accounting for 12% of TdP risk variability. |
| Tadros et al., 2019 [41] | N/A | 1368 | PRSPR, PRSQRS, PRSBrS | PR interval GWAS: 44 QRS GWAS: 26 BrS GWAS: 3 | PRSBrS-based odds for ajmaline-induced BrS diagnosis, OR = 1.17 (1.14–1.21) | Age, sex, baseline PR, baseline QRS, presence of Type II or III BrS ECG, family history of BrS | C-statistic (PRSBrS): 0.68 (0.65–0.71) | C-statistic (PRSBrS + RF): 0.741 (0.710–0.773) | Higher PRSPR, baseline PR, and female sex were linked to a pronounced PR slope, while PRSQRS and age were associated with QRS slope. PRSBrS, baseline QRS duration, presence of Type II or III BrS ECG at baseline, and a family history of BrS independently predicted the occurrence of a Type I BrS ECG. |
| Authors (y) | Discovery GWAS (n) | Validation/Test Dataset (n) | PRS | SNPs in PRS | HR/OR (95% CI) | Clinical Factors Included in Model | Main Findings |
|---|---|---|---|---|---|---|---|
| Lewis et al., 2020 [42] | N/A | 3391 | ADP-stimulated platelet reactivity PgxRS | 31 candidates,6 included in PgxRS | Age, sex, site, BMI, smoking, diabetes, proton pump inhibitor use | 8 or more risk alleles odds: CVE OR = 1.78 (1.14–2.76) Cardiovascular death OR = 4.39 (1.35–14.27) | Multiple polymorphisms influence clopidogrel response. A PgxRS predicts cardiovascular events and death in patients with more alleles linked to higher platelet reactivity. Clopidogrel users with eight or more risk alleles have significantly increased odds of such events and mortality. |
| Lacaze et al., 2022 [43] | N/A | 12,815 | LPa PRS | 43 | Age, sex, smoking, alcohol, BMI, previous regular aspirin use, HTN, diabetes, CKD, NSAID use | MACE risk: rs3798220-C carriers (Aspirin group), HR = 0.54 (0.17–1.70) High LPa-PRS (aspirin group), HR = 1.41 (0.90–2.23) | Aspirin may reduce MACE risk in individuals with elevated LPa genotypes, particularly rs3798220-C carriers and those in the highest PRS quintile. However, there was no significant interaction between aspirin use and the highest LPa-PRS quintile for MACE. While aspirin increased clinically significant bleeding overall, it did not significantly raise bleeding risk among rs3798220-C carriers or those in the top LPa-PRS quintile. |
| Authors (y) | Discovery GWAS (n) | Validation/Test Dataset (n) | PRS | SNPs in PRS | HR/OR (95% CI) | Clinical Factors Included in Model | Main Findings |
|---|---|---|---|---|---|---|---|
| Zhang et al., 2023 [44] | N/A | 212,968 | Alzheimer’s disease PRS | 25 | Paracetamol All-Cause Dementia risk, HR = 1.18 (1.10–1.26) Ibuprofen All-Cause Dementia risk, HR = 1.06 (0.97–1.16) | Age, sex, race, BMI, socioeconomic deprivation, smoking, alcohol, income, education, physical activity, healthy diet scores, albumin, C-reactive protein, dementia family history, HTN, diabetes, CVD, self-reported joint pain, self-reported arthritis, aspirin use, APOE ε4 dosage, and PRS of Alzheimer’s disease | Regular paracetamol use was linked to a higher risk of new-onset dementia, whereas ibuprofen was not. PRS did not significantly affect the association between either drug and the incidence of all-cause dementia. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Published by MDPI on behalf of the Österreichische Pharmazeutische Gesellschaft. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dwivedi, A.; Phulka, J.S.; Namdarimoghaddam, P.; Laksman, Z. Polygenic Risk Scores for Personalized Cardiovascular Pharmacogenomics―A Scoping Review. Sci. Pharm. 2025, 93, 18. https://doi.org/10.3390/scipharm93020018
Dwivedi A, Phulka JS, Namdarimoghaddam P, Laksman Z. Polygenic Risk Scores for Personalized Cardiovascular Pharmacogenomics―A Scoping Review. Scientia Pharmaceutica. 2025; 93(2):18. https://doi.org/10.3390/scipharm93020018
Chicago/Turabian StyleDwivedi, Aaryan, Jobanjit S. Phulka, Peyman Namdarimoghaddam, and Zachary Laksman. 2025. "Polygenic Risk Scores for Personalized Cardiovascular Pharmacogenomics―A Scoping Review" Scientia Pharmaceutica 93, no. 2: 18. https://doi.org/10.3390/scipharm93020018
APA StyleDwivedi, A., Phulka, J. S., Namdarimoghaddam, P., & Laksman, Z. (2025). Polygenic Risk Scores for Personalized Cardiovascular Pharmacogenomics―A Scoping Review. Scientia Pharmaceutica, 93(2), 18. https://doi.org/10.3390/scipharm93020018






