Abstract
Acetaminophen (APAP) is a widely used analgesic and antipyretic, whereas benzo(a)pyrene (B[a]P) is a carcinogen with significant global health risks due to environmental exposure. While APAP is generally safe at therapeutic doses, co-exposure to B[a]P can exacerbate its toxicity. This study aimed to identify potential human target proteins for B[a]P and APAP through inverse molecular docking and molecular dynamics simulations. We performed inverse docking with B[a]P, APAP, and three APAP metabolites against 689 human proteins involved in various biological processes. Five proteins were selected based on high docking affinity and their involvement in multiple pathways. Molecular dynamics simulations revealed that B[a]P primarily interacted via hydrophobic and π-stacking interactions with proteins like LXR-β, HSP90α, HSP90β, and AKT1, while AM404 formed hydrogen bonds and hydrophobic interactions. The simulations confirmed that the complexes had high conformational stability, except for protein AKT1. These results provide insights into the potential impacts of B[a]P and AM404 on protein functions and their implications for understanding the toxic effects of combined exposure.
1. Introduction
Acetaminophen (APAP) is the most used over-the-counter analgesic and the preferred first-line option for patients with mild to moderate acute pain worldwide [1]. This drug is an active ingredient in more than 600 pharmaceuticals [2]. Acetaminophen is available without a medical prescription because it is considered safe when used in therapeutic doses [3]. Once in the human body, APAP is primarily metabolized by the liver through conjugation with sulfate and glucuronide. A small fraction of the drug (5–10%) is oxidized by several P450 cytochromes to N-acetyl-p-benzoquinone imine (NAPQI), a highly reactive metabolite responsible for the toxicity of acetaminophen. In the brain and the spinal cord, N-arachidonoylphenolamine (AM404) is produced through the conjugation of p-aminophenol with arachidonic acid [4]. Finally, NAPQI is eliminated through its conjugation with glutathione (GSH) [5,6] (Figure 1).
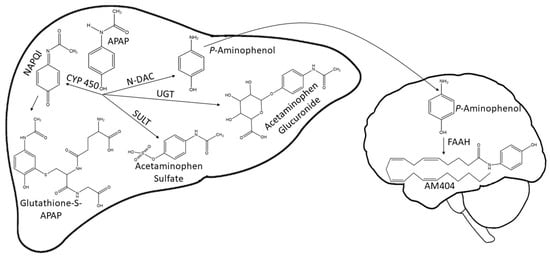
Figure 1.
APAP and metabolite structures generated in the liver and brain during APAP biotransformation. APAP is mainly metabolized in the liver by the enzymes UGT and SULT to form acetaminophen glucuronide and acetaminophen sulfate. A small amount of APAP is metabolized by CYP P450 into the highly reactive metabolite NAPQI. NAPQI is detoxified by conjugating with GSH. In the brain, p-aminophenol is conjugated to arachidonic acid by FAAH to yield AM404. APAP, acetaminophen; NAPQI, N-acetyl-p-benzoquinone imine; CYP 450, cytochrome P450; SULT, sulfotransferase; UGT, UDP-glucuronosyltransferase; GSH, glutathione; N-DAC, N- deacetylase; FAAH, fatty acid amide hydrolase.
The lack of knowledge about therapeutic doses, the inadvertent combination of multiple APAP-containing preparations, and the influence of endogenous and exogenous factors on susceptibility to acetaminophen poisoning can lead to glutathione depletion and subsequent liver damage [6,7]. Another emerging health concern involves the increasing release of polycyclic aromatic hydrocarbons, such as benzo(a)pyrene (B[a]P), into the environment [8]. This environmental exposure stems from various sources, including cigarette smoke, automobile emissions, and the combustion of domestic fuels, presenting a widespread risk to human health [9]. The association between B[a]P, categorized as a Group 1 carcinogen [10], and various types of cancers highlights the diverse health implications associated with its exposure. Additionally, B[a]P exposure is linked to immunosuppression, neurotoxicity, teratogenicity, and endocrine disruption, among other risks [11,12,13,14].
The widespread use of acetaminophen [15] and the increased release of B[a]P into the environment from different emission sources [9] create a scenario of potential co-exposure to both xenobiotics. This co-exposure could result in serious health conditions in humans, including liver damage, cancer, alterations in lipid metabolism, and neurotoxicity. Such outcomes are possible even when patients use therapeutic doses of acetaminophen. This is due to the biochemical processes involved in the toxicity of B[a]P, which lead to disturbances in antioxidant responses, including decreased glutathione levels [16,17]. Glutathione constitutes the primary detoxification mechanism for NAPQI [4], a metabolite produced during the metabolism of APAP via cytochrome P450 activation [4]. Additionally, the molecular mechanisms involved in the toxicity of B[a]P via the ligand-dependent nuclear receptor (AHR) have been reported [18]. Upon the binding of B[a]P to AHR, the formation of the AhR/ARNT heterodimer is induced, which then binds to xenobiotic-responsive elements (XREs), leading to the regulation of various genes such as cytochrome P450s (CYPs), intercellular cell adhesion molecule 1 (ICAM1), vascular cell adhesion molecule 1 (VCAM1), prostaglandin endoperoxide synthase 2 (PTGS2), and MYC (MYC Proto-Oncogene, BHLH Transcription Factor), among others [18,19]. Therefore, AHR activation may lead to the expression of genes responsible for the oxidation of acetaminophen to the reactive metabolite NAPQI via CYP450s, resulting in increased susceptibility to liver failure [20].
We recently reported that prior exposure to B[a]P increases susceptibility to APAP-induced liver injury in mice, even at therapeutic doses of APAP, resulting in changes in gene expression related to metabolism, redox balance, and cell proliferation [21]. Thus, the objective of this work is to identify possible human target proteins for benzo(a)pyrene and acetaminophen (and its metabolites) through inverse molecular docking and molecular dynamics simulations.
2. Materials and Methods
The identification of possible human target proteins for benzo(a)pyrene and acetaminophen (and its metabolites) was conducted using inverse molecular docking. Proteins were selected based on their affinity scores when docked with B[a]P and APAP or at least one of its metabolites and were required to be involved in at least three biological pathways, as determined through a protein–protein interaction (PPI) network analysis. After selecting these proteins, the primary interactions between the ligands and the protein binding sites were examined, followed by molecular validation using experimental data for the native ligands of the selected proteins.
2.1. Virtual Screening for Selection of Human Proteins
Inverse molecular docking was performed with a set of molecules, including B[a]P, APAP, and three of its metabolites, against 689 human proteins involved in pathways such as oxidative stress, apoptosis, inflammation, metabolism, cancer, cell proliferation, insulin signaling, coagulation, epigenetic changes, and angiogenesis, among others. These proteins were selected from the Automatic Docking Generation (ADGEN AV/SM) DataBank (An internally generated database created by the Environmental and Computational Chemistry Research Group). The protein structures were optimized by removing water molecules and co-crystallized ligands and then minimized using AMBER/Kollman charges with a gradient threshold of 0.005 kcal/mol-Å. Docking calculations were performed with the AutoDock Vina program [22]. Three docking runs were conducted for each ligand, and the averages of the best affinity scores (kcal/mol) for each protein–ligand pair were recorded.
The 100 proteins with the highest affinity for docking with both benzo(a)pyrene and acetaminophen (or its metabolites) were compiled and used to construct a protein–protein interaction network using the STRING tool (version 12.0) (https://string-db.org/, accessed on 17 April 2023) [23]. A high confidence level of 0.7 was set as a STRING parameter, and a stringent PPI enrichment p-value (<1.0 × 10−16) was applied, resulting in a total of 37 nodes. Each node represents a protein, with connections based on physical or functional interactions. Finally, among the proteins identified in the protein–protein interaction network, five were selected based on their involvement in at least three biological processes.
2.2. Molecular Docking with AutoDock Vina
2.2.1. Compounds’ Structures
The structures of compounds were constructed using the GaussView 5.08 [24] program, and geometry optimizations were carried out with the PM6 semi-empirical method in the Gaussian09 program [25]. Following optimization, the output file was converted to .pdb format using the Open Babel program (Version 2.3) [26] and to .pdbqt format using the the AutoDock Tools program (Version 4.2.6).
2.2.2. Preparation of Protein Structures
The proteins selected through the initial virtual screening process were downloaded from the Protein Data Bank (PDB) database (https://www.rcsb.org/, accessed on 18 August 2023) in PDB format and prepared using CHARMM-GUI [27] and the Chimera 1.17.3 program [28]. The 3D structures of the proteins were uploaded to CHARMM-GUI, which added the missing amino acids using the CHARMM force field at a pH of 7.0.
2.2.3. Docking Calculations
Five proteins were docked against B[a]P, APAP, Am404, NAPQI, and acetaminophen glucuronide conjugate according to the virtual screening process initially carried out.
Docking calculations were performed using the AutoDock Vina program [22] on the selected protein structures (1P8D, 3NMQ, 3O96, 4ACG, and 5J20). A grid box was defined at the geometric center of the co-crystallized ligand in each protein, with dimensions of 40 × 40 × 40 Å, covering the active site. A grid point spacing of 0.375 Å was employed. The x, y, and z coordinates of the center of the grid boxes on each protein were as follows: 62.037, 0.233, and 92.221 for 1P8D; 1.857, 10.362, and 27.023 for 3NMQ; 9.657, −7.831, and 10.604 for 3O96; 23.804, 20.383, and 9.494 for 4ACG; and 1.757, 33.414, and 23.389 for 5J20.
Three docking runs were performed for each ligand using the flexible side chains protocol based on the Iterated Local Search Global Optimizer Algorithm of AutoDock Vina. The pose with the best affinity (kcal/mol) was selected and saved. The maximum number of binding modes was set to 20, the energy range to 1.5, and exhaustiveness to 25. Finally, the average of the best affinity scores (in kcal/mol) was used to rank the complexes.
2.3. Protein–Ligand Interaction Prediction
The nature interactions present in the ligand–protein complex was identified using the LigandScout 4.5 program [29]. This was achieved through pharmacophore analysis to detect the types of ligand–residue interactions within the protein active site for each specific protein–ligand complex [30]. Additionally, the PLIP web server [31] was used to analyze the interactions in the protein–ligand complexes.
2.4. Validation of Docking Protocol
To validate the docking protocols, the native ligands were re-docked into the binding sites of the proteins using the AutoDock Vina program [22].
Each native ligand was extracted from the selected human proteins and saved in PDB file format using the Chimera 1.17.3 program [28].
Subsequently, each native ligand was docked into the active site of the respective protein using the same parameters described above for docking B[a]P, acetaminophen, Am404, NAPQI, and acetaminophen glucuronide conjugate on selected proteins. Three docking runs were performed for each native ligand, and the averages of the best affinity scores (kcal/mol) of each protein–ligand pair were calculated. The re-docked complexes were then superimposed onto the reference co-crystallized complexes, and the root mean square deviation (RMSD) was calculated using Chimera 1.17.3 [28]. The lowest energy conformation with the lowest RMSD value with respect to the crystallographic structures for each protein was selected for illustrative purposes.
2.5. Molecular Dynamics Simulations (MDS)
The protein–ligand complexes that showed the best binding energy in the molecular docking calculations underwent 100 ns molecular dynamics simulations (MDS) using the Gromacs program version 2020.2 [32]; this was carried out to assess their stability during the simulation. The force fields used for the protein and the ligand were the CHARMM force field [33] and the CHARMM General Force Field (CGenFF) [34], respectively. The complexes were immersed in a cubic periodic box, and each complex was solvated with TIP3P water under periodic boundary conditions [35]. The systems were neutralized, and the ionic strength (0.1 mol/L) of the medium was adjusted by adding Na+ and Cl− ions while keeping the number of particles constant. The systems’ energy was minimized until convergence was achieved. Subsequently, an equilibrium phase was carried out, maintaining constant pressure and temperature (NVT and NPT ensemble) at 300 K and 1.0 bar, respectively. The equilibration periods lasted 1.0 ns. The production runs extended for 100 ns, and trajectories were saved every 0.01 ns. Molecular dynamics results were used to calculate the root mean square deviation (RMSD) and the root mean square fluctuation (RMSF), and hydrogen bond (Hbond) to analyze the flexibility and stability of the complex over time. The conformations obtained from the simulations were used to compute the free binding energy.
The MMPBSA.py script [36] in the AMBER 21 suite was used to predict the free binding energies of these protein–ligand complexes. To apply the MMPBSA.py script, the topologic file, coordinate file, and production file generated in Gromacs were converted to their counterparts in AMBER. The interaction energy and solvation free energy for the complex, receptor, and ligand were used to estimate the binding free energy according to MM/GBSA approaches [37].
3. Results
3.1. Virtual Screening by Automatic Docking Generation
From the automatic docking performed on a set of proteins from our Automatic Docking Generation database (ADGEN AV/SM) and the ligands of interest (benzo(a)pyrene, acetaminophen, AM404, NAPQI, and acetaminophen glucuronide conjugate), 256 complexes with the highest absolute affinities were obtained (Table S1). The PDB codes of 71 proteins exhibiting the highest absolute affinity values for docking with both benzo(a)pyrene and acetaminophen (or its metabolites) were compiled and input into the STRING tool. As a result, a protein–protein interaction (PPI) network comprising 37 nodes connected to each other was generated, with a PPI value of <1.0 × 10−16 (Figure 2).

Figure 2.
Protein–protein interaction networks of proteins with highest absolute value of affinity for docking with both benzo(a)pyrene and acetaminophen (or its metabolites). Network nodes and edges represent proteins and protein–protein associations. Line colors indicate different interactions as follows: light blue, interactions from curated databases; magenta, relationships experimentally determined; green, gene neighborhood; red, gene fusions; dark blue, gene co-occurrence; yellow, text mining; black, co-expression; purple, protein homology.
The protein–protein interaction (PPI) network of proteins with the highest absolute value of affinity revealed that 5 out of the 37 proteins connected in the network are involved in at least three biological processes (1P8D, 3NMQ, 3O96, 4ACG, and 5J20). A functional analysis using the biological process of gene ontology (GO) is shown in Table 1.

Table 1.
Functional analysis of selected proteins for molecular docking.
The PPI network of selected proteins from the initial screening process reveals that proteins with a high affinity for both B[a]P and acetaminophen (or its metabolites) are involved in crucial biological processes. These include the assembly of the telomerase holoenzyme complex, which is essential for genomic stability [38], the positive regulation of tau protein kinase activity, which is implicated in neurodegenerative diseases [39], and the retinoic acid receptor signaling pathway, which plays a critical role in cellular differentiation and pancreatic cell development [40]. Additionally, some of the proteins represented in the interaction network are involved in the positive regulation of glycogen biosynthetic processes, fatty acid biosynthesis, and cholesterol efflux, which are all vital for cellular homeostasis [41,42,43]. These proteins are also associated with cellular signaling pathways related to cell proliferation, including DNA biosynthesis and the negative regulation of the extrinsic apoptotic signaling pathway [44,45]. Furthermore, the interaction network highlights the role of some selected proteins in mechanisms controlling cellular redox balance, such as oxidative demethylation and the negative regulation of cytochrome c release from mitochondria [46,47].
3.2. Docking Analysis
The results of molecular docking for B[a]P and APAP metabolites with human proteins selected through virtual screening are shown in Table 2.

Table 2.
Docking scores for selected proteins.
All selected proteins demonstrated the best affinity score for B[a]P, followed by Am404 and the APAP-glucuronide conjugate; therefore, the best complex for B[a]P and Am404 was subjected to molecular dynamics simulations to study the dynamic behavior of the protein–ligand complex, including conformational changes, ligand binding, and protein folding [48].
The proteins that exhibited the best affinity scores for each ligand were as follows: the Oxysterol receptor LXR-beta (1P8D) with a binding affinity of −12.17 kcal/mol for B[a]P and −9.0 kcal/mol for APAP-glucuronide; RAC-alpha serine/threonine protein kinase (3O96) with an affinity of −8.9 kcal/mol for B[a]P and heat shock protein HSP 90-beta (3NMQ), which showed affinities of −11.7 kcal/mol for B[a]P and −8.3 kcal/mol for AM404 (Table 2).
According to the protein–protein interaction analysis, all selected proteins play a significant role in the regulation of gene expression [49,50,51,52]. Specifically, proteins with PDB IDs 3NMQ and 5J20 share biological functions associated with maintaining genetic stability. They are involved in the assembly of the telomerase holoenzyme complex, the positive regulation of kinase activity, and the positive regulation of the DNA biosynthetic process [53].
Proteins with PDB IDs 3O96 and 4ACG are involved in the regulation of cellular proliferation processes [54,55], such as the regulation of type B pancreatic cell development [56,57], the positive regulation of the DNA biosynthetic process [58,59], the positive regulation of proteasomal ubiquitin-dependent protein catabolic processes [52,60], and the negative regulation of the extrinsic apoptotic signaling pathway [61,62]. Protein 1P8D, in addition to its role in gene expression regulation, plays a significant role in the positive regulation of cholesterol efflux [63] and the retinoic acid receptor signaling pathway [64].
3.3. The Nature of the Interactions of Protein–Ligand Complexes
The interactions between B[a]P and AM404 at the protein binding sites are illustrated in Figure 3 and Figure 4, respectively.
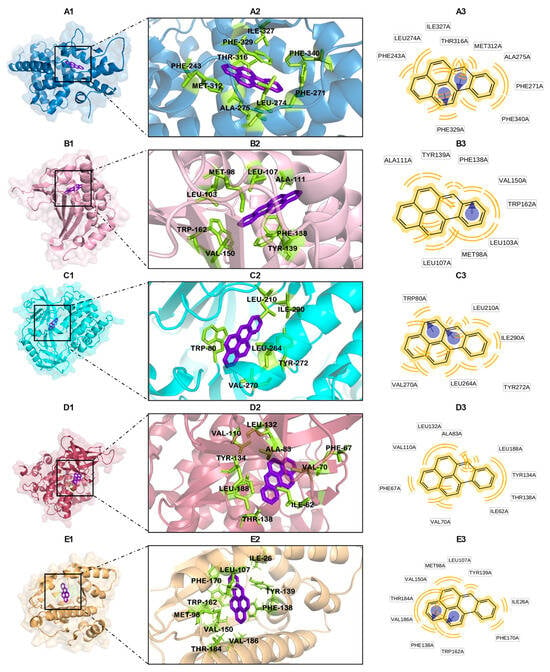
Figure 3.
Interactions between B[a]P and active site of proteins with lowest binding affinities (kcal/mol): 1P8D (A), 3NMQ (B), 3O96 (C), 4ACG (D), and 5J20 (E). Protein–ligand complex (A1–E1); binding site (A2–E2); and ligand–residue interactions (two-dimensional view) (A3–E3).

Figure 4.
A three-dimensional view of the five best complexes with AM404: 1P8D (A), 3NMQ (B), 3O96 (C), 4ACG (D), and 5J20 (E). Protein–ligand complex (A1–E1); binding site (A2–E2); and ligand–residue interactions (two-dimensional view) (A3–E3).
Figure 3 demonstrates how B[a]P interacts with the protein binding sites primarily through hydrophobic interactions. In the case of the LXR-beta protein, these interactions involve residues Phe-243, Phe-271, Leu-274, Thr-316, Ile-327, Phe-329, Met-312, and Phe-340 (Figure 3A and Figure S1; Table S2). Additionally, π-stacking interactions are observed with the amino acid Phe-329.
For heat shock protein HSP 90-beta, B[a]P forms hydrophobic interactions with amino acids Ala-111, Tyr-139, Phe-138, Val-150, Trp-162, Leu-103, Met-98, and Leu-107, also establishing π-stacking interactions with Phe-138 (Figure 3B and Figure S2; Table S3).
In the case of the RAC-alpha serine/threonine protein kinase, B[a]P interacts with amino acids Trp-80 (which also forms π-stacking interactions), Leu-210, Ile-290, Tyr-272, Leu-264, and Val-270 (Figure 3C and Figure S3; Table S4).
For glycogen synthase kinase-3 beta, B[a]P exhibits hydrophobic interactions with amino acids Ile-62, Phe-67, Val-70, Leu-132, Ala-83, Val-110, Leu-188, Tyr-134, and Thr-138 (Figure 3D and Figure S4; Table S5). Additionally, B[a]P shows hydrophobic interactions with Leu-107, Tyr-139, Ile-26, Phe-170, Trp-162, and Phe-138 (which also forms π-stacking interactions), as well as with Val-186, Thr-184, Val-150, and Met-98 in heat shock protein HSP 90-alpha (Figure 3E and Figure S5; Table S6).
Figure 4 illustrates the predicted interactions in AM404/protein complexes, which include hydrogen bonds and hydrophobic interactions. For LXR-beta, key interactions include a hydrogen bond donor interaction with Glu-281 and a hydrogen acceptor with Thr-316. Additionally, there are hydrophobic interactions with Phe-243, Leu-442, Phe-268, Ala-343, Phe-271, Leu-345, and Thr-272 (Figure 4A and Figure S6; Table S7).
For heat shock protein HSP 90-beta, AM404 establishes hydrogen bond donor interactions with Ile-104 and Tyr-139, while hydrophobic interactions involve Phe-22, Val-150, Thr-184, Met-98, Phe-138, Val-186, Ile-26, and Phe-170 (Figure 4B and Figure S7; Table S8).
In the case of the RAC-alpha serine/threonine protein kinase, AM404 shows hydrogen donor interactions with Ser-56, Ala-58, and Asp-119, while hydrophobic interactions involve Thr-82 and Met-118 (Figure 4C and Figure S8; Table S9).
For glycogen synthase kinase-3 beta, AM404 exhibits hydrogen bond acceptor interactions with Arg-144 and hydrophobic interactions with Phe-67, Val-70, Tyr-140, and Thr-138 (Figure 4D and Figure S9; Table S10). Finally, AM404 establishes hydrogen bond donor interactions with Ile-104 and Gln-23 as well as hydrophobic interactions with Phe-138, Tyr-139, Leu-107, Phe-170, and Ile-26 at the binding site of heat shock protein HSP 90-alpha (Figure 4E and Figure S10; Table S11).
3.4. Docking Validation
The docking validation demonstrated that the predicted poses for these ligands (co-crystallized ligands) were optimally reproduced compared to the experimental binding modes, yielding satisfactory results.
The calculated RMSD values for each native ligand ranged from 1.02 to 2.10 Å (see Table S12), and the nature of the interactions is detailed in Tables S13–S17. Figure 5 shows the RMSD values and binding affinities for the re-docked native ligands. The results from the AutoDock Vina docking protocol were satisfactory, with the RMSD values consistently being below 2.2 Å, as depicted in Figure 5. These results confirm the ability of AutoDock Vina to accurately reproduce the conformation of ligands in its experimental complexes [65].
3.5. Molecular Dynamics Simulations
Considering that interactions in protein–ligand complexes are dynamic, it is crucial to analyze molecular dynamics simulations (MDS) to gain a deeper understanding of the chemical microenvironment that ligands encounter at a receptor’s binding site. The complexes formed between each of the five proteins with ligands B[a]P and Am404 underwent 100 ns molecular dynamics simulations (MDS) to assess their flexibility and stability over time. Additionally, MDS was performed on proteins with their respective co-crystallized ligands to establish a benchmark for conformational stability under identical conditions. RMSD, RMSF, and Hb were used for comparison, and free energy calculations were performed via MMGBSA for each complex.
3.5.1. Root Mean Square Deviations (RSMD)
One important factor to evaluate in a protein–ligand complex is the RMSD of the backbone atoms of the protein, which indicates its overall conformational stability in a dynamic state throughout the simulation. The system reaches equilibrium and stability when it shows low RMSD values with steady fluctuations throughout the simulation; on the contrary, greater fluctuations suggest a lack of stability.
The backbone RMSD calculated for the 1P8D protein bound to the ligands AM404 and B[a]P and its cocrystallized ligand (CO1) is shown in Figure 6(A1).The RMSD results indicate that the backbone atoms of the 1P8D-CO1, 1P8D-AM404, and 1P8D-B[a]P complexes exhibited minimal structural fluctuations, with average RMSD values of 0.15 ± 0.015 nm, 0.19 ± 0.016 nm, and 0.18 ± 0.021 nm for the 1P8D-CO1, 1P8D-AM404, and 1P8D-B[a]P complexes, respectively. This suggests that the protein maintains high conformational stability with three ligands during the 100 ns of MDS. Figure 6(A2) shows the RMSD graph for CO1, AM404, and B[a]P. The average RMSD values are 0.061 ± 0.014 nm, 0.26 ± 0.019 nm, and 0.015 ± 0.004 nm, respectively. These low values of the RMSD suggest that the ligands that interact with 1PD8 do not undergo abrupt changes in their positions throughout the simulation.
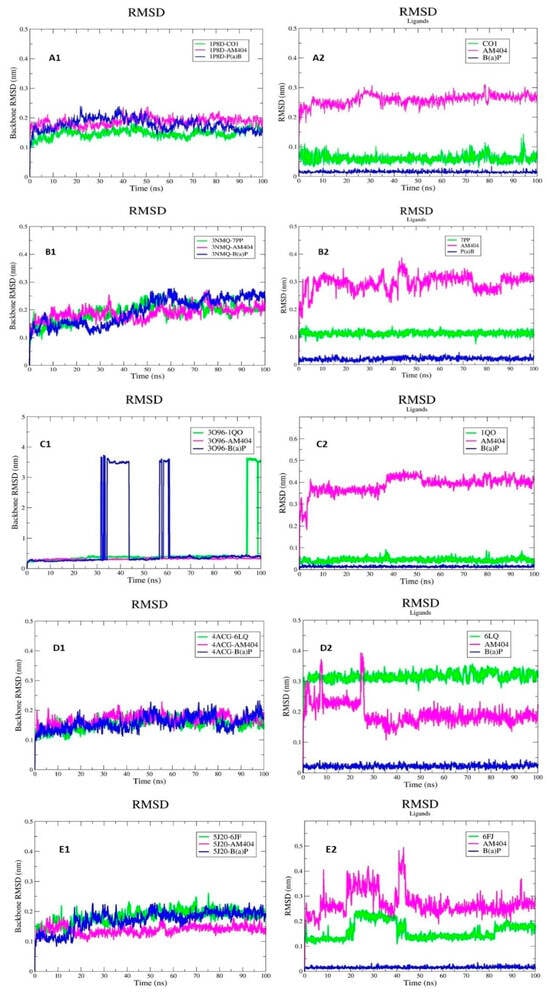
Figure 6.
RMSD. 1P8D (A), 3NMQ (B), 3O96 (C), 4ACG (D), and 5J20 (E). The backbone RMSD calculated for the proteins bound to the ligands AM404 and B[a]P and its cocrystallized ligands (A1–E1). The RMSD values of the ligands (A2–E2).
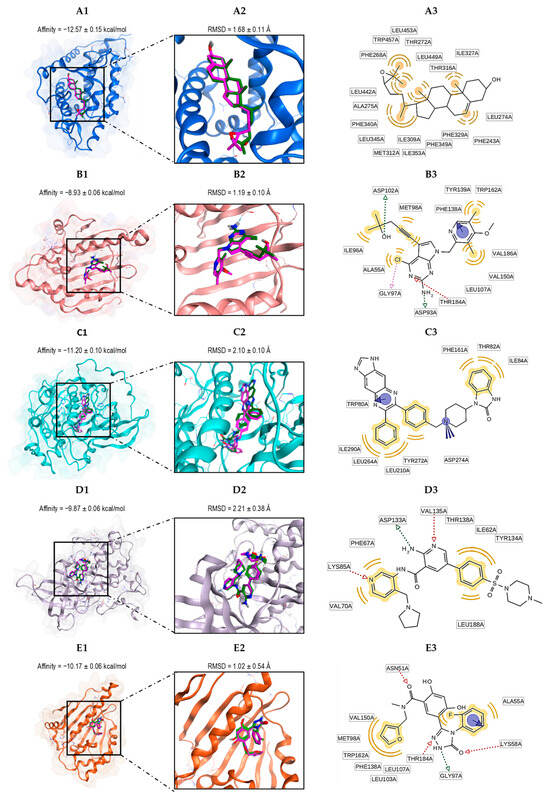
Figure 5.
The alignment between the docked ligand (magenta) and the ligand present in the crystal structure (green) of the proteins with the lowest binding affinities (kcal/mol): 1P8D (A), 3NMQ (B), 3O96 (C), 4ACG (D), and 5J20 (E). Protein–ligand complex (A1–E1); binding site (A2–E2); and ligand–residue interactions (two-dimensional view) (A3–E3).
The backbone RMSD calculated for the 3NMQ protein bound to the ligands AM404 and B[a]P and its cocrystallized ligand (7PP) is shown in Figure 6(B1). The RMSD results indicate that the backbone atoms of the 3NMQ-7PP, 3NMQ-AM404, and 3NMQ-B[a]P complexes exhibited minimal structural fluctuations with average RMSD values of 0.19 ± 0.030 nm, 0.19 ± 0.025 nm, and 0.20 ± 0.050 nm for the 3NMQ-7PP, 3NMQ-AM404, and 3NMQ-B[a]P complexes, respectively. This suggests that the protein maintains high conformational stability with three ligands during the 100 ns of MDS, and a very similar pattern was observed in the three proteins.
Figure 6(B2) shows the RMSD graph for 7PP, AM404, and B[a]P. The average RMSD values are 0.11 ± 0.009 nm, 0.29 ± 0.03 nm, and 0.21 ± 0.006 nm, respectively. In the RMSD graph of the ligands, it can be seen that 7PP and B[a]P present an almost constant behavior throughout the simulation, while Am404 presents fluctuations throughout the simulation.
The backbone RMSD calculated for the 3O96 protein bound to the ligands AM404 and B[a]P and its cocrystallized ligand (6FJ) is shown in Figure 6(C1). The average RMSD values for the 3O96-6FJ, 3O96-AM404, and 3O96-B[a]P complexes are 0.55 ± 0.075 nm, 0.32 ± 0.024 nm, and 0.77 ± 1.10 nm, respectively. The 3O96-B[a]P complex presents several fluctuations during the simulation, specifically between 33 ns and 45 ns, and afterward, 60 ns equilibrium is reached. Figure 6(C2) shows the RMSD graph for 6FJ, AM404, and B[a]P. The average RMSD values are 0.044 ± 0.009 nm, 0.40 ± 0.04 nm, and 0.014 ± 0.003 nm, respectively.
The backbone RMSD calculated for the 4ACG protein bound to the ligands AM404 and B[a]P and its cocrystallized ligand (6LQ) is shown in Figure 6(D1). The RMSD results indicate that the backbone atoms of the 4ACG-6LQ, 3NMQ-AM404, and 3NMQ-B[a]P complexes exhibited minimal structural fluctuations with average RMSD values of 0.15 ± 0.019 nm, 0.17 ± 0.019 nm, and 0.16 ± 0.025 nm for the 3NMQ-6LQ, 3NMQ-AM404, and 3NMQ-B[a]P complexes, respectively. This suggests that the protein maintains high conformational stability with three ligands during the 100 ns of MDS. Figure 6(D2) shows the RMSD graph for 6LQ, AM404, and B[a]P. The average RMSD values are 0.32 ± 0.018 nm, 0.19 ± 0.040 nm, and 0.022 ± 0.006 nm, respectively. In the RMSD graph of the ligands, it can be seen that 6LQ and B[a]P present an almost constant behavior throughout the simulation, while Am404 presents fluctuations during the first 30 ns of MDS.
The backbone RMSD calculated for the 5J20 protein bound to the ligands AM404 and B[a]P and its cocrystallized ligand (1QO) is shown in Figure 6(E1). The RMSD results indicate that the backbone atoms of the 5J20-1QO, 5J20-AM404, and 5J20-B[a]P complexes exhibited minimal structural fluctuations with average RMSD values of 0.18 ± 0.024 nm, 0.14 ± 0.015 nm, and 0.17 ± 0.03 nm for the 5J20-1QO, 5J20-AM404, and 5J20-B[a]P complexes, respectively. This suggests that the protein maintains high conformational stability with three ligands during the 100 ns of MDS. Figure 6(E2) shows the RMSD graph for 6LQ, AM404, and B[a]P. The average RMSD values are 0.16 ± 0.035 nm, 0.27 ± 0.048 nm, and 0.016 ± 0.004 nm, respectively. In the RMSD graph of the ligands, it can be seen that 1QO and AM404 present fluctuations during the first 50 ns of MDS.
3.5.2. Root Mean Square Fluctuation (RMSF)
The mobility of residues during the molecular dynamics simulations (MDS) was evaluated using root mean square fluctuation (RMSF). The RMSF profiles for all complexes are displayed in Figure 7. In general, the complexes display minimal fluctuations over the 100 ns simulations, with the RMSF graphs nearly overlapping and average RMSF values around 0.1 nm ± 0.05 nm.

Figure 7.
The RMSF plots calculated for the proteins bound to the ligands AM404 and B[a]P and its cocrystallized ligands: 1P8D (A), 3NMQ (B), 3O96 (C), 4ACG (D), and 5J20 (E).
However, the RMSF plots for complexes involving 3O96 show greater fluctuations, with values of approximately 0.21 and 1.27 nm and an average RMSF value of 0.58 ± 0.23 nm for the 3O96-6FJ complex and variations between 0.31 and 1.46 nm and an average RMSF value of 0.70 ± 0.25 nm in the case of the 3O96-AM404 complex; the greatest fluctuation in RMSF values was found for the 3O96-B[a]P complex, where the RMSF values varied from 0.7 to 3.3 nm with an average RMSF value of 1.8 ± 0.65 nm (see Figure 7C), which is an indication of the great mobility that it presents when this unit is attached to the B[a]P ligand.
3.5.3. Hydrogen Bond Analysis
The intermolecular hydrogen bond interactions between the protein and ligand are crucial for stabilizing the protein–ligand complexes. An increase in the number of hydrogen bonds indicates greater stability in the protein–ligand complex.
The B[a]P ligand does not form hydrogen bonds with any of the receptors because its chemical structure allows only for hydrophobic interactions or stacking interactions. In contrast, the AM404 ligand forms between two and three hydrogen bonds with each of the proteins studied (see Figure S11). These interactions are maintained for a high percentage of the simulation time, indicating that the complexes between this ligand and the five proteins are stable. These results agree with those obtained in the molecular docking calculations shown in Figure 4. In some cases, AM404 even forms more hydrogen bonds than the co-crystallized ligand of each protein.
3.5.4. Molecular Mechanics Energies Combined with Surface Area Continuum Solvation (MMGBSA)
The total binding free energy of the proteins bound to AM404 and B[a]P and its cocrystallized ligand could be estimated using MGBSA calculations. Total binding free energy values that are positive indicate unfavorable binding, whereas values that are negative indicate favorable binding. The overall binding energy of each protein bound to AM404 and B[a]P and its co-crystallized ligand is shown in Table 3, along with a breakdown of the various energy components for each complex under investigation.

Table 3.
MMGBSA-based total binding free energies along with standard error of mean (SEM).
The ligand that presented the best binding energy with all of the proteins was AM404. Among the studied complexes, 3NMQ-AM404, 3O96-AM404, and 5J20-AM404 exhibited more favorable binding free energies: −40.52 ± 0.77 kcal/mol, −38.28 ± 0.58 kcal/mol, and −40.66 ± 0.61 kcal/mol, respectively. These values are even better than those observed by those proteins with their co-crystallized ligands. The binding free energy of the 3O96-B[a]P complex was −20.46 ± 0.20 kcal/mol, which is significantly lower than the −37.97 ± 0.39 kcal/mol value for the 3O96-1QO complex and −38.28 ± 0.58 kcal/mol for the 3O96-AM404 complex. This result is consistent with the data reported in the RMSD and RMSF graphs.
In all of the complexes, the energy component related to van der Waals interactions is greater than the energy component related to electrostatic interactions. This disparity is especially noticeable in the complexes involving the B[a]P ligand and the studied proteins (see Figure S12), where the energy related to electrostatic interactions is notably low compared to other energy components. This is largely attributed to the structural characteristics of the B[a]P ligand.
4. Discussion
Co-exposure to acetaminophen (APAP) and benzo(a)pyrene (B[a]P), both prevalent in the environment and human use, poses significant health risks [21]. APAP, commonly used as a pain reliever, can cause liver damage through its metabolite NAPQI [66], while B[a]P, a known carcinogen, disrupts antioxidant defenses and enhances the expression of cytochrome P450 enzymes via aryl hydrocarbon receptor (AHR) activation [17,67,68]. This interaction may potentiate APAP toxicity, thereby increasing the risk of liver damage and other adverse effects. Previous studies have demonstrated that prior exposure to B[a]P makes mice more susceptible to APAP-induced liver injury [21]. Therefore, this study aims to investigate the interactions between benzo(a)pyrene (B[a]P), acetaminophen (APAP) metabolites, and various human proteins using molecular docking.
Our results reveal that B[a]P and AM404 exhibit strong binding affinities with several human proteins, suggesting that these compounds may modulate protein functions critical to cellular processes. Specifically, the oxysterol receptor LXR-beta (1P8D) demonstrated the highest binding affinity for B[a]P with a value of −12.7 kcal/mol, and the heat shock protein HSP90-beta (3NMQ) also showed high affinity with a value of −11.7 kcal/mol (Table 2). These findings underscore B[a]P’s significant capacity to interact with these proteins, potentially influencing their activity.
The analysis of interactions between B[a]P and AM404 predominantly reveals hydrophobic interactions. B[a]P, known for its hydrophobic nature, binds to protein binding sites such as LXR-beta, HSP90, RAC-alpha serine/threonine protein kinase, and glycogen synthase kinase-3 beta mainly through hydrophobic interactions and π-stacking (Figure 3). A comparison of the interaction residues reported with co-crystallized ligands for these proteins suggests that exposure to B[a]P induces a conformational rearrangement in the protein’s active site, which maximizes the exposure of the nonpolar surface formed by hydrophobic amino acids. This rearrangement could potentially enhance the binding affinity compared to that observed with native ligands. However, these interactions are crucial for stabilizing protein–ligand complexes and may influence the biological activity of the proteins [69]. In contrast, AM404 exhibited a combination of hydrogen bonding and hydrophobic interactions (Figure 4). For instance, with LXR-beta, AM404 formed hydrogen bonds with Glu-281 and hydrophobic interactions with Phe-243, Phe-271, and Leu-442 (Figure 4A). These interactions were also observed with potent endogenous activators of LXR in the liver (CO1) [70]. Additionally, AM404 can form hydrogen bond acceptor interactions with Thr-316, similar to the interactions established by 28-homobrassinolide, a transactivator of LXR genes [65].
LXR-beta plays a critical role in the expression of cholesterol transporters ABCA1 and ABCG1 [71]. Our findings suggest that, despite its hydrophobic nature and inability to form hydrogen bonds, B[a]P exhibits a high affinity for LXR-beta. This high binding affinity contributes to significant stability in the protein, which could potentially lead to competition with endogenous ligands for the protein’s binding site, thereby affecting its function. This could lead to the inhibition of ABCA1 and ABCG1 cholesterol transporters, resulting in the accumulation of cholesterol ester lipid droplets, as observed in EA.hy926 cells [72]. Additionally, LXR has been implicated in the resistance to acetaminophen hepatotoxicity, which is mediated through the suppression of pro-toxic P450s and the induction of anti-toxic Phase II enzymes such as glutathione S-transferase (Gst) and sulfotransferase (Sult), which enhance APAP clearance [73]. Therefore, exposure to B[a]P could disrupt LXR-beta’s regulatory role in protecting against acetaminophen-induced hepatotoxicity. This may be one of the molecular mechanisms through which B[a]P exacerbates acetaminophen-induced liver injury in mice at non-hepatotoxic doses, as previously demonstrated [21].
In addition to affecting LXR-beta, B[a]P and AM404 interact with another crucial protein, heat shock protein 90 (HSP90). HSP90 consists of three domains: the N-terminal domain (which contains an ATP binding site and a client protein binding site), the C-terminal domain (containing a site for protein dimerization and another for calmodulin binding), and the middle domain (which is essential for ATP hydrolysis to ADP) [74]. Studies on HSP90 inhibition and function indicate that interaction with Asp-93 is critical for stable nucleotide binding [75]. Mutations at this position can disrupt HSP90 function and prevent nucleotide binding [76]. Our molecular docking results show that AM404 can interact with the N-terminal ATP-binding domains of HSP90α and HSP90β [77] through hydrogen bonds with residues Ile-104 and Gln-23 of HSP90α and Ile-104 and Tyr-139 of HSP90β (not seen with their respective inhibitors 7PP and 6FJ), as well as hydrophobic interactions (also established with its respective inhibitors) (Figure 4B,E). In contrast, B[a]P interacts only through hydrophobic interactions (also seen with its inhibitor 7PP) and π-stacking (Figure 3B,E). The interactions between HSP90 and AM404/B[a]P do not involve hydrogen bonds with Asp-93 unlike several HSP90α and HSP90β inhibitors [78].
HSP90 is a chaperone protein involved in the proper folding, activation, and assembly of many proteins, including key mediators of signal transduction, cell cycle control, and transcriptional regulation [79,80,81]. Its overexpression has been linked to tumor development [82,83], prompting recent studies to focus on inhibiting HSP90 as a strategy for anticancer therapy [74,84,85,86].
It is well established that in the absence of a ligand, heat shock protein 90 (HSP90) resides in the cytoplasm, forming a complex with co-chaperones and AHR, a transcription factor activated by xenobiotic binding, including B[a]P [87]. Recent studies suggested that B[a]P and APAP upregulate the expression of HSP90 [88,89]. Our study indicates that the interaction of B[a]P and AM404 with HSP90α and HSP90β isoforms may not significantly affect their function as they do not contact Asp-93, a critical residue for maintaining their structural conformation. Additionally, by promoting the synthesis of these chaperone proteins, B[a]P and AM404 could contribute to tumor growth, as previously reported [90].
The docking analysis of AM404 with RAC-alpha serine/threonine protein kinase (AKT1) shows that AM404, despite forming three hydrogen bonds with residues Ser-56, Ala-58, and Asp-119, exhibits lower binding affinity compared to the allosteric inhibitor IQO, which forms only one hydrogen bond but binds more effectively. One of the strongest hydrogen bonds established by AM404 with AKT1 is with residue Asp-119, featuring an angle of 155.12° (Table S9) and a distance shorter than that with IQO. IQO forms a hydrogen bond with residue Ser-205, which has an angle of 104° (Table S15). Although the hydrogen bond between AM404 and Asp-119 has a geometry closer to the ideal (180°) [91] and a shorter distance, its overall binding affinity remains lower than that of IQO. These results suggest that factors such as the hydrogen bond number, angle, and distance are not the sole determinants of binding affinity; rather, hydrophobic interactions and π-stacking might also play significant roles [92,93].
Conversely, B[a]P shows higher binding affinity with AKT1 compared to the allosteric inhibitor IQO (Table 2 and Figure 5C). B[a]P interacts exclusively through hydrophobic forces and π-stacking with residue Trp-80 (Figure 3C). In contrast, although IQO forms a hydrogen bond with residue Ser-205, it exhibits lower binding affinity. Nevertheless, the 3O96-B[a]P complex showed significant fluctuations during the simulation, which may indicate low conformational stability in the protein–ligand complex. This instability could potentially lead to alterations in the pathways regulated by AKT1 depending on various factors, including the type of cells being treated. These findings are consistent with studies showing that the exposure of HL-7702 human normal liver cells and trophoblast cell lines to B[a]P blocks the PI3K/Akt signaling pathway, inducing pyroptotic and autophagic cell death [94], and downregulates AKT phosphorylation in trophoblast cell lines [95]. This underscores the role of AKT1 in regulating cell proliferation, survival, and metabolism [96]. Conversely, in tumor cells, B[a]P exposure appears to promote AKT1 phosphorylation, thereby enhancing cell proliferation through the EGFR signaling pathway [97]. Additionally, AKT1 plays a significant role in survival mechanisms through the AMPK/Akt/GSK3β pathway in response to APAP toxicity [98]. Thus, its interaction with compounds that can alter its regulation may impact the survival of cells experiencing APAP toxicity.
Glycogen synthase kinase-3 beta (GSK3β) is a kinase involved in the aforementioned cellular survival mechanisms (via AMPK/Akt/GSK3β) and plays a crucial role in neuronal functions. It has been associated with the abnormal phosphorylation of the tau protein, a process believed to cause neurofibrillary tangles in Alzheimer’s disease [99]. Our docking results reveal that B[a]P binds to the same site as several GSK3β inhibitors [100], sharing hydrophobic interactions with most of the residues reported for its inhibitor, 6LQ (Val-110, Leu-132, Ala-83, Leu-188, Tyr-134, Ile-62, and Val-70) (Figure 3D) [101]. The high affinity of B[a]P for GSK3β suggests that its interaction could potentially disrupt GSK3β’s normal function without requiring hydrogen bond formation. This disruption could alter the cellular signaling pathways involving GSK3β, including the induction of apoptotic cascades through mitochondrial dysfunction [102,103], likely contributing to an environment conducive to APAP-induced cellular damage by impairing hepatic regeneration pathways [104].
In general, the RMSD values of the protein backbone exhibit minimal fluctuations throughout the molecular dynamics simulations, indicating that the system maintains equilibrium during the simulation time. However, this is not the case for the 3O96 protein when it is bound to the B[a]P ligand; fluctuations of up to 4 nm can be seen in the RMSD graph, as shown in Figure 6(C1). Regarding the RMSD calculated for the B[a]P and AM404 ligands, it can be observed that the B[a]P ligand shows low mobility, while the AM404 ligand shows high mobility. This difference in behavior can be attributed to the different structural characteristics of each ligand. The AM404 ligand presents many rotatable bonds, which allows for a greater generation of conformations during the simulation; on the contrary, the B[a]P ligand presents low conformational freedom due to its planarity.
The RMSF results allow us to infer that the backbone of the proteins bound with the ligands show few fluctuations throughout the 100 ns of simulation, suggesting low flexibility and intensity of movement of the amino acids throughout the simulation process, except for protein 3O96 bound with B[a]P, which presents RMSF values that are high compared to those presented by that same protein bound with the ligands AM404 and 1QO (see Figure 7C), and values that are much higher compared to the complexes formed by the other proteins.
The hydrogen bond interactions between the protein and ligand are essential for maintaining the stability of the protein–ligand complexes. The chemical structure of the AM404 ligand enables it to establish multiple hydrogen bonds with each of the proteins studied. These interactions persist for a significant portion of the simulation time, suggesting that the complexes formed with this ligand and the five proteins are stable. These findings corroborate the results from the molecular docking calculations. Although the B[a]P ligand cannot form hydrogen bonds with proteins due to its chemical structure, the results of the docking calculations are very favorable, indicating that intermolecular interactions of the hydrophobic type govern the stability of the complexes formed by this ligand.
The MMGBSA calculations support the findings from the docking calculations, suggesting that the complexes are thermodynamically stable. Among the complexes under observation, those formed by the AM404 ligand exhibit the most favorable binding free energy. The average binding free energy for the complexes formed by AM404 and the proteins under study is around −40 kcal/mol. In contrast, the average binding free energy for the complexes formed by B[a]P and the proteins under study is approximately −20 kcal/mol. In general, van der Waals energy was observed to be the primary contributor to the interactions of the two ligands, although in AM404, the electrostatic component is significantly higher than that in B[a]P.
5. Conclusions
Molecular docking revealed that benzo(a)pyrene (B[a]P) and AM404 exhibit high affinity for the proteins LXR-β, HSP90α, HSP90β, and AKT1. Specifically, B[a]P predominantly interacts with these proteins through hydrophobic and π-stacking interactions, whereas AM404, a metabolite of acetaminophen, primarily forms hydrogen bonds in addition to hydrophobic interactions. The RMSD and RMSF analyses indicated that most protein–ligand complexes demonstrate minimal mobility, suggesting that the protein residues are relatively stable and exhibit low flexibility during the simulations. However, the 3O96-B[a]P complex exhibited increased mobility. The AM404 ligand’s chemical structure facilitates the formation of multiple hydrogen bonds with the studied proteins, enhancing the stability of these complexes and corroborating the docking results. MMGBSA calculations confirm the thermodynamic stability of the complexes, as evidenced by negative total binding free energy values that favor protein–ligand binding. Our findings provide detailed insights into the molecular interactions between benzo(a)pyrene (B[a]P), AM404, and key proteins, highlighting potential health implications, particularly regarding the combined exposure to these compounds. Both B[a]P and AM404 exhibit significant affinities for crucial human proteins, including LXR-β, HSP90α, HSP90β, and AKT1, suggesting possible alterations in their essential biological functions.
In particular, the high affinity of B[a]P for LXR-β could lead to competition with endogenous ligands, potentially disrupting its role in regulating cholesterol transporters such as ABCA1 and ABCG1. This disruption may result in cholesterol accumulation and an increased risk of acetaminophen-induced liver damage.
Furthermore, although B[a]P and AM404 interact with HSP90 through different mechanisms, our simulations suggest that these interactions might affect the protein’s stability. This could have implications for tumor growth and cellular stress responses.
Finally, the high affinity of B[a]P for AKT1 and GSK3β suggests a potential disruption of cellular signaling pathways related to cell proliferation and the response to acetaminophen-induced liver damage. These effects could be significant in the context of combined exposure to these compounds.
Overall, our findings provide valuable insights into how B[a]P and AM404 may influence essential protein functions, offering significant implications for understanding their toxic effects and the mechanisms underlying their combined exposure. This study underscores the importance of considering the combined effects of B[a]P and acetaminophen (APAP) in environmental and health risk assessments. Since both compounds interact with critical proteins and impact key cellular functions, their co-exposure could lead to compounded toxic effects. Understanding these interactions helps elucidate the potential mechanisms of increased toxicity and provides a basis for developing strategies to mitigate risks associated with simultaneous exposure to these substances.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/scipharm92040055/s1, Figures S1–S5: Two-dimensional image of interaction pattern of B(a)P against active site of LXR-beta, HSP 90-beta, AKT, GSK3, and LXR-alfa, respectively; Figures S6–S10: Two-dimensional image of interaction pattern of AM404 against active site of LXR-beta, HSP 90-beta, AKT, GSK3, and LXR-alfa, respectively; Figure S11: Hydrogen bond profiles of proteins bound to ligands; Figure S12: The binding free energy terms obtained from MM-GBSA; Table S1: Two hundred fifty-six complexes with the highest absolute affinities from docking; Tables S2–S6: The nature of the interactions of B(a)P with LXR-beta, HSP 90-beta, AKT, GSK3, and LXR-alfa; Tables S7–S11: The nature of the interactions of AM404 with LXR-beta, HSP 90-beta, AKT, GSK3, and LXR-alfa; Table S12: Affinity and RMSD values for co-crystallized ligands; Tables S13–S17: The nature of the interactions of LXR-beta, HSP 90-beta, AKT, GSK3, and LXR-alfa with their co-crystallized ligands.
Author Contributions
Conceptualization, Y.M.-P., J.O.-V., N.P.-C. and N.C.-P.; methodology, Y.M.-P., M.A.-M., N.P.-C. and N.C.-P.; software, Y.M.-P. and M.A.-M.; validation, M.A.-M.; formal analysis, Y.M.-P. and M.A.-M.; resources, Y.M.-P., M.A.-M., N.P.-C. and N.C.-P.; data curation, Y.M.-P. and M.A.-M.; writing—original draft preparation, Y.M.-P., M.A.-M. and J.O.-V.; writing—review and editing, Y.M.-P., M.A.-M. and J.O.-V.; visualization, Y.M.-P. and M.A.-M.; supervision, J.O.-V.; funding acquisition, J.O.-V. All authors have read and agreed to the published version of the manuscript.
Funding
The authors are thankful for the support from the University of Cartagena (Plan to Support Research Groups and Doctoral Programs (2020–2023) and MinCiencias (Doctoral formation, 647/2014 and 727/2015).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors on request.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Mallet, C.; Desmeules, J. An Updated Review on the Metabolite (AM404)-Mediated Central Mechanism of Action of Paracetamol (Acetaminophen): Experimental Evidence and Potential Clinical Impact. J. Pain. Res. 2023, 16, 1081–1094. [Google Scholar] [CrossRef]
- Hurwitz, J.; Sands, S.; Davis, E.; Nielsen, J.; Warholak, T. Patient knowledge and use of acetaminophen in over-the-counter medications. J. Am. Pharm. Assoc. 2014, 54, 19–26. [Google Scholar] [CrossRef]
- Chidiac, A.S.; Buckley, N.A.; Noghrehchi, F.; Cairns, R. Paracetamol (acetaminophen) overdose and hepatotoxicity: Mechanism, treatment, prevention measures, and estimates of burden of disease. Expert. Opin. Drug Metab. Toxicol. 2023, 19, 297–317. [Google Scholar] [CrossRef]
- Mazaleuskaya, L.L.; Sangkuhl, K.; Thorn, C.F.; FitzGerald, G.A.; Altman, R.B.; Klein, T.E. PharmGKB summary: Pathways of acetaminophen metabolism at the therapeutic versus toxic doses. Pharmacogenet Genom. 2015, 25, 416–426. [Google Scholar] [CrossRef]
- Ben-Shachar, R.; Chen, Y.; Luo, S.; Hartman, C.; Reed, M.; Nijhout, H.F. The biochemistry of acetaminophen hepatotoxicity and rescue: A mathematical model. Theor. Biol. Med. Model. 2012, 9, 55. [Google Scholar] [CrossRef]
- Gloor, Y.; Schvartz, D.; Samer, C.F. Old problem, new solutions: Biomarker discovery for acetaminophen liver toxicity. Expert. Opin. Drug Metab. Toxicol. 2019, 15, 659–669. [Google Scholar] [CrossRef] [PubMed]
- Luo, G.; Huang, L.; Zhang, Z. The molecular mechanisms of acetaminophen-induced hepatotoxicity and its potential therapeutic targets. Exp. Biol. Med. 2023, 248, 412–424. [Google Scholar] [CrossRef]
- Sampaio, G.R.; Guizellini, G.M.; da Silva, S.A.; de Almeida, A.P.; Pinaffi-Langley, A.C.C.; Rogero, M.M.; de Camargo, A.C.; Torres, E. Polycyclic Aromatic Hydrocarbons in Foods: Biological Effects, Legislation, Occurrence, Analytical Methods, and Strategies to Reduce Their Formation. Int. J. Mol. Sci. 2021, 22, 6010. [Google Scholar] [CrossRef]
- Bukowska, B.; Mokra, K.; Michałowicz, J. Benzo[a]pyrene-Environmental Occurrence, Human Exposure, and Mechanisms of Toxicity. Int. J. Mol. Sci. 2022, 23, 6348. [Google Scholar] [CrossRef] [PubMed]
- IARC Working Group on the Evaluation of Carcinogenic Risks to Humans. Chemical Agents and Related Occupations. Lyon (FR): International Agency for Research on Cancer; (IARC Monographs on the Evaluation of Carcinogenic Risks to Humans, No. 100F.) Benzo[a]pyrene. Available online: https://www.ncbi.nlm.nih.gov/books/NBK304415/ (accessed on 4 October 2018).
- Carlson, E.A.; Li, Y.; Zelikoff, J.T. Benzo[a]pyrene-induced immunotoxicity in Japanese medaka (Oryzias latipes): Relationship between lymphoid CYP1A activity and humoral immune suppression. Toxicol. Appl. Pharmacol. 2004, 201, 40–52. [Google Scholar] [CrossRef] [PubMed]
- Usal, M.; Veyrenc, S.; Darracq-Ghitalla-Ciock, M.; Regnault, C.; Sroda, S.; Fini, J.B.; Canlet, C.; Tremblay-Franco, M.; Raveton, M.; Reynaud, S. Transgenerational metabolic disorders and reproduction defects induced by benzo[a]pyrene in Xenopus tropicalis. Env. Pollut. 2021, 269, 116109. [Google Scholar] [CrossRef]
- Kim, J.T.; Park, J.E.; Lee, S.J.; Yu, W.J.; Lee, H.J.; Kim, J.M. Benzo[a]pyrene Cytotoxicity Tolerance in Testicular Sertoli Cells Involves Aryl-hydrocarbon Receptor and Cytochrome P450 1A1 Expression Deficiencies. Dev. Reprod. 2021, 25, 15–24. [Google Scholar] [CrossRef]
- Yang, Y.; Zhou, Y.; Pan, L.; Xu, R.; Li, D. Benzo[a]pyrene exposure induced reproductive endocrine-disrupting effects via the steroidogenic pathway and estrogen signaling pathway in female scallop Chlamys farreri. Sci. Total Env. 2020, 726, 138585. [Google Scholar] [CrossRef]
- Ayoub, S.S. Paracetamol (acetaminophen): A familiar drug with an unexplained mechanism of action. Temperature 2021, 8, 351–371. [Google Scholar] [CrossRef]
- Bukowska, B.; Duchnowicz, P. Molecular Mechanisms of Action of Selected Substances Involved in the Reduction of Benzo[a]pyrene-Induced Oxidative Stress. Molecules 2022, 27, 1379. [Google Scholar] [CrossRef]
- Deng, C.; Dang, F.; Gao, J.; Zhao, H.; Qi, S.; Gao, M. Acute benzo[a]pyrene treatment causes different antioxidant response and DNA damage in liver, lung, brain, stomach and kidney. Heliyon 2018, 4, e00898. [Google Scholar] [CrossRef]
- Fu, C.; Li, Y.; Xi, H.; Niu, Z.; Chen, N.; Wang, R.; Yan, Y.; Gan, X.; Wang, M.; Zhang, W.; et al. Benzo(a)pyrene and cardiovascular diseases: An overview of pre-clinical studies focused on the underlying molecular mechanism. Front. Nutr. 2022, 9, 978475. [Google Scholar] [CrossRef]
- Stevens, E.A.; Mezrich, J.D.; Bradfield, C.A. The aryl hydrocarbon receptor: A perspective on potential roles in the immune system. Immunology 2009, 127, 299–311. [Google Scholar] [CrossRef]
- Schuran, F.A.; Lommetz, C.; Steudter, A.; Ghallab, A.; Wieschendorf, B.; Schwinge, D.; Zuehlke, S.; Reinders, J.; Heeren, J.; Lohse, A.W.; et al. Aryl Hydrocarbon Receptor Activity in Hepatocytes Sensitizes to Hyperacute Acetaminophen-Induced Hepatotoxicity in Mice. Cell Mol. Gastroenterol. Hepatol. 2021, 11, 371–388. [Google Scholar] [CrossRef] [PubMed]
- Montero-Pérez, Y.; Olivero-Verbel, J. Exposure to Benzo(a)pyrene Enhances Acetaminophen-Induced Liver Injury in Mice at Non-Hepatotoxic Doses. Sci. Pharm. 2024, 92, 30. [Google Scholar] [CrossRef]
- Eberhardt, J.; Santos-Martins, D.; Tillack, A.F.; Forli, S. AutoDock Vina 1.2.0: New Docking Methods, Expanded Force Field, and Python Bindings. J. Chem. Inf. Model. 2021, 61, 3891–3898. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Kirsch, R.; Koutrouli, M.; Nastou, K.; Mehryary, F.; Hachilif, R.; Gable, A.L.; Fang, T.; Doncheva, N.T.; Pyysalo, S.; et al. The STRING database in 2023: Protein-protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 2023, 51, D638–D646. [Google Scholar] [CrossRef] [PubMed]
- Dennington, R.D.; Keith, T.A.; Millam, J.M. GaussView 5.0; Gaussian Inc.: Wallingford, CT, USA, 2008; p. 340. [Google Scholar]
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Mennucci, B.; Petersson, G.A. Gaussian 09; Gaussian Inc.: Wallingford, CT, USA, 2009. [Google Scholar]
- O'Boyle, N.M.; Banck, M.; James, C.A.; Morley, C.; Vandermeersch, T.; Hutchison, G.R. Open Babel: An open chemical toolbox. J. Cheminformatics 2011, 3, 33. [Google Scholar] [CrossRef] [PubMed]
- Jo, S.; Kim, T.; Iyer, V.G.; Im, W. CHARMM-GUI: A web-based graphical user interface for CHARMM. J. Comput. Chem. 2008, 29, 1859–1865. [Google Scholar] [CrossRef] [PubMed]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef]
- Wolber, G.; Langer, T. LigandScout: 3-D Pharmacophores Derived from Protein-Bound Ligands and Their Use as Virtual Screening Filters. J. Chem. Inf. Model. 2005, 45, 160–169. [Google Scholar] [CrossRef]
- Maldonado-Rojas, W.; Olivero-Verbel, J.; Marrero-Ponce, Y. Computational fishing of new DNA methyltransferase inhibitors from natural products. J. Mol. Graph. Model. 2015, 60, 43–54. [Google Scholar] [CrossRef]
- Salentin, S.; Schreiber, S.; Haupt, V.J.; Adasme, M.F.; Schroeder, M. PLIP: Fully automated protein-ligand interaction profiler. Nucleic Acids Res. 2015, 43, W443–W447. [Google Scholar] [CrossRef]
- Abraham, M.J.; Murtola, T.; Schulz, R.; Páll, S.; Smith, J.C.; Hess, B.; Lindahl, E. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 2015, 1–2, 19–25. [Google Scholar] [CrossRef]
- MacKerell, A.D., Jr.; Bashford, D.; Bellott, M.; Dunbrack, R.L., Jr.; Evanseck, J.D.; Field, M.J.; Fischer, S.; Gao, J.; Guo, H.; Ha, S.; et al. All-Atom Empirical Potential for Molecular Modeling and Dynamics Studies of Proteins. J. Phys. Chem. B 1998, 102, 3586–3616. [Google Scholar] [CrossRef]
- Vanommeslaeghe, K.; Hatcher, E.; Acharya, C.; Kundu, S.; Zhong, S.; Shim, J.; Darian, E.; Guvench, O.; Lopes, P.; Vorobyov, I.; et al. CHARMM general force field: A force field for drug-like molecules compatible with the CHARMM all-atom additive biological force fields. J. Comput. Chem. 2010, 31, 671–690. [Google Scholar] [CrossRef]
- Mark, P.; Nilsson, L. Structure and Dynamics of the TIP3P, SPC, and SPC/E Water Models at 298 K. J. Phys. Chem. A 2001, 105, 9954–9960. [Google Scholar] [CrossRef]
- Miller, B.R., III.; McGee, T.D., Jr.; Swails, J.M.; Homeyer, N.; Gohlke, H.; Roitberg, A.E. MMPBSA.py: An Efficient Program for End-State Free Energy Calculations. J. Chem. Theory Comput. 2012, 8, 3314–3321. [Google Scholar] [CrossRef]
- Kollman, P.A.; Massova, I.; Reyes, C.; Kuhn, B.; Huo, S.; Chong, L.; Lee, M.; Lee, T.; Duan, Y.; Wang, W.; et al. Calculating Structures and Free Energies of Complex Molecules: Combining Molecular Mechanics and Continuum Models. Acc. Chem. Res. 2000, 33, 889–897. [Google Scholar] [CrossRef]
- Collins, K. The biogenesis and regulation of telomerase holoenzymes. Nat. Rev. Mol. Cell Biol. 2006, 7, 484–494. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Zhi, W.; Wang, L. Role of Tau Protein in Neurodegenerative Diseases and Development of Its Targeted Drugs: A Literature Review. Molecules 2024, 29, 2812. [Google Scholar] [CrossRef] [PubMed]
- Lorberbaum, D.S.; Kishore, S.; Rosselot, C.; Sarbaugh, D.; Brooks, E.P.; Aragon, E.; Xuan, S.; Simon, O.; Ghosh, D.; Mendelsohn, C.; et al. Retinoic acid signaling within pancreatic endocrine progenitors regulates mouse and human β cell specification. Development 2020, 147, dev189977. [Google Scholar] [CrossRef]
- Roach, P.J.; Depaoli-Roach, A.A.; Hurley, T.D.; Tagliabracci, V.S. Glycogen and its metabolism: Some new developments and old themes. Biochem. J. 2012, 441, 763–787. [Google Scholar] [CrossRef]
- Ameur, A.; Enroth, S.; Johansson, A.; Zaboli, G.; Igl, W.; Johansson, A.C.; Rivas, M.A.; Daly, M.J.; Schmitz, G.; Hicks, A.A.; et al. Genetic adaptation of fatty-acid metabolism: A human-specific haplotype increasing the biosynthesis of long-chain omega-3 and omega-6 fatty acids. Am. J. Hum. Genet. 2012, 90, 809–820. [Google Scholar] [CrossRef]
- Duan, Y.; Gong, K.; Xu, S.; Zhang, F.; Meng, X.; Han, J. Regulation of cholesterol homeostasis in health and diseases: From mechanisms to targeted therapeutics. Signal Transduct. Target. Ther. 2022, 7, 265. [Google Scholar] [CrossRef]
- Madhavan, H. Simple Laboratory methods to measure cell proliferation using DNA synthesis property. J. Stem Cells Regen. Med. 2007, 3, 12–14. [Google Scholar] [CrossRef]
- Wiman, K.G.; Zhivotovsky, B. Understanding cell cycle and cell death regulation provides novel weapons against human diseases. J. Intern. Med. 2017, 281, 483–495. [Google Scholar] [CrossRef] [PubMed]
- Garrido, C.; Galluzzi, L.; Brunet, M.; Puig, P.E.; Didelot, C.; Kroemer, G. Mechanisms of cytochrome c release from mitochondria. Cell Death Differ. 2006, 13, 1423–1433. [Google Scholar] [CrossRef] [PubMed]
- Cyr, A.R.; Domann, F.E. The redox basis of epigenetic modifications: From mechanisms to functional consequences. Antioxid. Redox Signal 2011, 15, 551–589. [Google Scholar] [CrossRef]
- Hollingsworth, S.A.; Dror, R.O. Molecular Dynamics Simulation for All. Neuron 2018, 99, 1129–1143. [Google Scholar] [CrossRef]
- Mouzat, K.; Baron, S.; Marceau, G.; Caira, F.; Sapin, V.; Volle, D.H.; Lumbroso, S.; Lobaccaro, J.-M. Emerging roles for LXRs and LRH-1 in female reproduction. Mol. Cell. Endocrinol. 2013, 368, 47–58. [Google Scholar] [CrossRef]
- Prodromou, C. Mechanisms of Hsp90 regulation. Biochem. J. 2016, 473, 2439–2452. [Google Scholar] [CrossRef]
- Huo, X.; Sun, H.; Liu, Q.; Ma, X.; Peng, P.; Yu, M.; Zhang, Y.; Cao, D.; Shen, K. Clinical and Expression Significance of AKT1 by Co-expression Network Analysis in Endometrial Cancer. Front. Oncol. 2019, 9, 1147. [Google Scholar] [CrossRef]
- Beurel, E.; Grieco, S.F.; Jope, R.S. Glycogen synthase kinase-3 (GSK3): Regulation, actions, and diseases. Pharmacol. Ther. 2015, 148, 114–131. [Google Scholar] [CrossRef] [PubMed]
- Wei, H.; Zhang, Y.; Jia, Y.; Chen, X.; Niu, T.; Chatterjee, A.; He, P.; Hou, G. Heat shock protein 90: Biological functions, diseases, and therapeutic targets. MedComm 2024, 5, e470. [Google Scholar] [CrossRef]
- Duggal, S.; Jailkhani, N.; Midha, M.K.; Agrawal, N.; Rao, K.V.S.; Kumar, A. Defining the Akt1 interactome and its role in regulating the cell cycle. Sci. Rep. 2018, 8, 1303. [Google Scholar] [CrossRef]
- Cao, Q.; Lu, X.; Feng, Y.-J. Glycogen synthase kinase-3β positively regulates the proliferation of human ovarian cancer cells. Cell Res. 2006, 16, 671–677. [Google Scholar] [CrossRef] [PubMed]
- Tuttle, R.L.; Gill, N.S.; Pugh, W.; Lee, J.P.; Koeberlein, B.; Furth, E.E.; Polonsky, K.S.; Naji, A.; Birnbaum, M.J. Regulation of pancreatic beta-cell growth and survival by the serine/threonine protein kinase Akt1/PKBalpha. Nat. Med. 2001, 7, 1133–1137. [Google Scholar] [CrossRef] [PubMed]
- Elmadbouh, O.H.M.; Pandol, S.J.; Edderkaoui, M. Glycogen Synthase Kinase 3β: A True Foe in Pancreatic Cancer. Int. J. Mol. Sci. 2022, 23, 14133. [Google Scholar] [CrossRef] [PubMed]
- Codenotti, S.; Zizioli, D.; Mignani, L.; Rezzola, S.; Tabellini, G.; Parolini, S.; Giacomini, A.; Asperti, M.; Poli, M.; Mandracchia, D.; et al. Hyperactive Akt1 Signaling Increases Tumor Progression and DNA Repair in Embryonal Rhabdomyosarcoma RD Line and Confers Susceptibility to Glycolysis and Mevalonate Pathway Inhibitors. Cells 2022, 11, 2859. [Google Scholar] [CrossRef]
- Liu, H.; Remedi, M.S.; Pappan, K.L.; Kwon, G.; Rohatgi, N.; Marshall, C.A.; McDaniel, M.L. Glycogen Synthase Kinase-3 and Mammalian Target of Rapamycin Pathways Contribute to DNA Synthesis, Cell Cycle Progression, and Proliferation in Human Islets. Diabetes 2009, 58, 663–672. [Google Scholar] [CrossRef]
- Noguchi, M.; Hirata, N.; Suizu, F. The links between AKT and two intracellular proteolytic cascades: Ubiquitination and autophagy. Biochim. Biophys. Acta (BBA) Rev. Cancer 2014, 1846, 342–352. [Google Scholar] [CrossRef]
- Kim, R.; Kin, T.; Beck, W.T. Impact of Complex Apoptotic Signaling Pathways on Cancer Cell Sensitivity to Therapy. Cancers 2024, 16, 984. [Google Scholar] [CrossRef]
- Beurel, E.; Jope, R.S. The paradoxical pro- and anti-apoptotic actions of GSK3 in the intrinsic and extrinsic apoptosis signaling pathways. Prog. Neurobiol. 2006, 79, 173–189. [Google Scholar] [CrossRef]
- Edwards, P.A.; Kennedy, M.A.; Mak, P.A. LXRs; oxysterol-activated nuclear receptors that regulate genes controlling lipid homeostasis. Vasc. Pharmacol. 2002, 38, 249–256. [Google Scholar] [CrossRef] [PubMed]
- Komati, R.; Spadoni, D.; Zheng, S.; Sridhar, J.; Riley, K.E.; Wang, G. Ligands of Therapeutic Utility for the Liver X Receptors. Molecules 2017, 22, 88. [Google Scholar] [CrossRef]
- Premalatha, R.; Srikumar, K.; Vijayalaksmi, D.; Kumar, G.N.; Mathur, P.P. 28-Homobrassinolide: A novel oxysterol transactivating LXR gene expression. Mol. Biol. Rep. 2014, 41, 7447–7461. [Google Scholar] [CrossRef] [PubMed]
- Cai, X.; Cai, H.; Wang, J.; Yang, Q.; Guan, J.; Deng, J.; Chen, Z. Molecular pathogenesis of acetaminophen-induced liver injury and its treatment options. J. Zhejiang Univ. Sci. B 2022, 23, 265–285. [Google Scholar] [CrossRef] [PubMed]
- Salamat, N.; Derakhshesh, N. Oxidative stress in liver cell culture from mullet, Liza klunzingeri, induced by short-term exposure to benzo[a]pyrene and nonylphenol. Fish. Physiol. Biochem. 2020, 46, 1183–1197. [Google Scholar] [CrossRef]
- Guo, B.; Feng, D.; Xu, Z.; Qi, P.; Yan, X. Acute benzo[a]pyrene exposure induced oxidative stress, neurotoxicity and epigenetic change in blood clam Tegillarca granosa. Sci. Rep. 2021, 11, 18744. [Google Scholar] [CrossRef] [PubMed]
- Kumar, R.; Lathwal, A.; Nagpal, G.; Kumar, V.; Raghav, P.K. Chapter 1—Impact of chemoinformatics approaches and tools on current chemical research. In Chemoinformatics and Bioinformatics in the Pharmaceutical Sciences; Sharma, N., Ojha, H., Raghav, P.K., Goyal, R.K., Eds.; Academic Press: Cambridge, MA, USA, 2021; pp. 1–26. [Google Scholar]
- Wong, J.; Quinn, C.M.; Brown, A.J. Synthesis of the oxysterol, 24(S), 25-epoxycholesterol, parallels cholesterol production and may protect against cellular accumulation of newly-synthesized cholesterol. Lipids Health Dis. 2007, 6, 10. [Google Scholar] [CrossRef]
- Gerin, I.; Dolinsky, V.W.; Shackman, J.G.; Kennedy, R.T.; Chiang, S.H.; Burant, C.F.; Steffensen, K.R.; Gustafsson, J.A.; MacDougald, O.A. LXRbeta is required for adipocyte growth, glucose homeostasis, and beta cell function. J. Biol. Chem. 2005, 280, 23024–23031. [Google Scholar] [CrossRef]
- Duan, J.; Chen, C.; Li, H.; Ju, G.; Gao, A.; Sun, Y.; Zhang, W. Multifaceted Protective Effects of Hesperidin by Aromatic Hydrocarbon Receptor in Endothelial Cell Injury Induced by Benzo[a]Pyrene. Nutrients 2022, 14, 574. [Google Scholar] [CrossRef]
- Saini, S.P.; Zhang, B.; Niu, Y.; Jiang, M.; Gao, J.; Zhai, Y.; Hoon Lee, J.; Uppal, H.; Tian, H.; Tortorici, M.A.; et al. Activation of liver X receptor increases acetaminophen clearance and prevents its toxicity in mice. Hepatology 2011, 54, 2208–2217. [Google Scholar] [CrossRef]
- Li, Z.N.; Luo, Y. HSP90 inhibitors and cancer: Prospects for use in targeted therapies. Oncol. Rep. 2023, 49, 6. [Google Scholar] [CrossRef]
- Obermann, W.M.; Sondermann, H.; Russo, A.A.; Pavletich, N.P.; Hartl, F.U. In vivo function of Hsp90 is dependent on ATP binding and ATP hydrolysis. J. Cell Biol. 1998, 143, 901–910. [Google Scholar] [CrossRef]
- Zuehlke, A.D.; Moses, M.A.; Neckers, L. Heat shock protein 90: Its inhibition and function. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2018, 373, 20160527. [Google Scholar] [CrossRef] [PubMed]
- Khandelwal, A.; Kent, C.N.; Balch, M.; Peng, S.; Mishra, S.J.; Deng, J.; Day, V.W.; Liu, W.; Subramanian, C.; Cohen, M.; et al. Structure-guided design of an Hsp90β N-terminal isoform-selective inhibitor. Nat. Commun. 2018, 9, 425. [Google Scholar] [CrossRef] [PubMed]
- Hoxie, R.S.; Street, T.O. Hsp90 chaperones have an energetic hot-spot for binding inhibitors. Protein Sci. 2020, 29, 2101–2111. [Google Scholar] [CrossRef] [PubMed]
- Isaacs, J.S.; Xu, W.; Neckers, L. Heat shock protein 90 as a molecular target for cancer therapeutics. Cancer Cell 2003, 3, 213–217. [Google Scholar] [CrossRef]
- Kamal, A.; Boehm, M.F.; Burrows, F.J. Therapeutic and diagnostic implications of Hsp90 activation. Trends Mol. Med. 2004, 10, 283–290. [Google Scholar] [CrossRef] [PubMed]
- Abdul, N.S.; Ahmad Alrashed, N.; Alsubaie, S.; Albluwi, H.; Badr Alsaleh, H.; Alageel, N.; Ghaleb Salma, R. Role of Extracellular Heat Shock Protein 90 Alpha in the Metastasis of Oral Squamous Cell Carcinoma: A Systematic Review. Cureus 2023, 15, e38514. [Google Scholar] [CrossRef]
- Galam, L.; Hadden, M.K.; Ma, Z.; Ye, Q.Z.; Yun, B.G.; Blagg, B.S.; Matts, R.L. High-throughput assay for the identification of Hsp90 inhibitors based on Hsp90-dependent refolding of firefly luciferase. Bioorg Med. Chem. 2007, 15, 1939–1946. [Google Scholar] [CrossRef] [PubMed]
- Birbo, B.; Madu, E.E.; Madu, C.O.; Jain, A.; Lu, Y. Role of HSP90 in Cancer. Int. J. Mol. Sci. 2021, 22, 10317. [Google Scholar] [CrossRef]
- Abbasi, M.; Sadeghi-Aliabadi, H.; Amanlou, M. Prediction of new Hsp90 inhibitors based on 3,4-isoxazolediamide scaffold using QSAR study, molecular docking and molecular dynamic simulation. Daru 2017, 25, 17. [Google Scholar] [CrossRef]
- Rezvani, S.; Ebadi, A.; Razzaghi-Asl, N. In silico identification of potential Hsp90 inhibitors via ensemble docking, DFT and molecular dynamics simulations. J. Biomol. Struct. Dyn. 2022, 40, 10665–10676. [Google Scholar] [CrossRef]
- Magwenyane, A.M.; Ugbaja, S.C.; Amoako, D.G.; Somboro, A.M.; Khan, R.B.; Kumalo, H.M. Heat Shock Protein 90 (HSP90) Inhibitors as Anticancer Medicines: A Review on the Computer-Aided Drug Discovery Approaches over the Past Five Years. Comput. Math. Methods Med. 2022, 2022, 2147763. [Google Scholar] [CrossRef] [PubMed]
- Gruszczyk, J.; Grandvuillemin, L.; Lai-Kee-Him, J.; Paloni, M.; Savva, C.G.; Germain, P.; Grimaldi, M.; Boulahtouf, A.; Kwong, H.-S.; Bous, J.; et al. Cryo-EM structure of the agonist-bound Hsp90-XAP2-AHR cytosolic complex. Nat. Commun. 2022, 13, 7010. [Google Scholar] [CrossRef]
- Su, M.; Zhou, S.; Li, J.; Lin, N.; Chi, T.; Zhang, M.; Lv, X.; Hu, Y.; Bai, T.; Chang, F. Benzo(a)pyrene regulates chaperone-mediated autophagy via heat shock protein 90. Toxicol. Lett. 2023, 383, 121–127. [Google Scholar] [CrossRef] [PubMed]
- Wu, K.; Guo, C.; Su, M.; Wu, X.; Li, R. Biocharacterization of Heat Shock Protein 90 in Acetaminophen-Treated Livers Without Conspicuous Drug Induced Liver Injury. Cell Physiol. Biochem. 2017, 43, 1562–1570. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Liu, T.; Zhang, M.; Chen, Q.; Su, M.; Bai, T.; Hu, Y.; Li, J.; Chang, F.; Huangfu, W. Study on molecular mechanism of benzo (ɑ) pyrene on CMA by HSP90ɑ and HIF-1ɑ. Toxicol. Vitr. 2022, 83, 105372. [Google Scholar] [CrossRef]
- Milovanović, M.R.; Stanković, I.M.; Živković, J.M.; Ninković, D.B.; Hall, M.B.; Zarić, S.D. Water: New aspect of hydrogen bonding in the solid state. IUCrJ 2022, 9, 639–647. [Google Scholar] [CrossRef]
- Vaidyanathan, R.; Murugan Sreedevi, S.; Ravichandran, K.; Vinod, S.M.; Hari Krishnan, Y.; Babu, L.K.; Parthiban, P.S.; Basker, L.; Perumal, T.; Rajaraman, V.; et al. Molecular docking approach on the binding stability of derivatives of phenolic acids (DPAs) with Human Serum Albumin (HSA): Hydrogen-bonding versus hydrophobic interactions or combined influences? JCIS Open 2023, 12, 100096. [Google Scholar] [CrossRef]
- Van Lommel, R.; Bettens, T.; Barlow, T.M.A.; Bertouille, J.; Ballet, S.; De Proft, F. A Quantum Chemical Deep-Dive into the π-π Interactions of 3-Methylindole and Its Halogenated Derivatives-Towards an Improved Ligand Design and Tryptophan Stacking. Pharmaceuticals 2022, 15, 935. [Google Scholar] [CrossRef]
- Li, Q.; Gao, C.; Deng, H.; Song, Q.; Yuan, L. Benzo[a]pyrene induces pyroptotic and autophagic death through inhibiting PI3K/Akt signaling pathway in HL-7702 human normal liver cells. J. Toxicol. Sci. 2019, 44, 121–131. [Google Scholar] [CrossRef]
- Ye, Y.; Jiang, S.; Du, T.; Ding, M.; Hou, M.; Mi, C.; Liang, T.; Zhong, H.; Xie, J.; Xu, W.; et al. Correction: Environmental Pollutant Benzo[a]pyrene Upregulated Long Non-coding RNA HZ07 Inhibits Trophoblast Cell Migration by Inactivating PI3K/AKT/MMP2 Signaling Pathway in Recurrent Pregnancy Loss. Reprod. Sci. 2023, 30, 728. [Google Scholar] [CrossRef]
- Martorana, F.; Motta, G.; Pavone, G.; Motta, L.; Stella, S.; Vitale, S.R.; Manzella, L.; Vigneri, P. AKT Inhibitors: New Weapons in the Fight Against Breast Cancer? Front. Pharmacol. 2021, 12, 662232. [Google Scholar] [CrossRef] [PubMed]
- Kometani, T.; Yoshino, I.; Miura, N.; Okazaki, H.; Ohba, T.; Takenaka, T.; Shoji, F.; Yano, T.; Maehara, Y. Benzo[a]pyrene promotes proliferation of human lung cancer cells by accelerating the epidermal growth factor receptor signaling pathway. Cancer Lett. 2009, 278, 27–33. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Zhang, S.; Cheng, H.; Lv, H.; Cheng, G.; Ci, X. Nrf2-mediated liver protection by esculentoside A against acetaminophen toxicity through the AMPK/Akt/GSK3β pathway. Free Radic. Biol. Med. 2016, 101, 401–412. [Google Scholar] [CrossRef] [PubMed]
- Hanger, D.P.; Hughes, K.; Woodgett, J.R.; Brion, J.P.; Anderton, B.H. Glycogen synthase kinase-3 induces Alzheimer's disease-like phosphorylation of tau: Generation of paired helical filament epitopes and neuronal localisation of the kinase. Neurosci. Lett. 1992, 147, 58–62. [Google Scholar] [CrossRef]
- Zhu, J.; Wu, Y.; Xu, L.; Jin, J. Theoretical Studies on the Selectivity Mechanisms of Glycogen Synthase Kinase 3β (GSK3β) with Pyrazine ATP-competitive Inhibitors by 3DQSAR, Molecular Docking, Molecular Dynamics Simulation and Free Energy Calculations. Curr. Comput. Aided Drug Des. 2020, 16, 17–30. [Google Scholar] [CrossRef]
- Berg, S.; Bergh, M.; Hellberg, S.; Högdin, K.; Lo-Alfredsson, Y.; Söderman, P.; von Berg, S.; Weigelt, T.; Ormö, M.; Xue, Y.; et al. Discovery of novel potent and highly selective glycogen synthase kinase-3β (GSK3β) inhibitors for Alzheimer's disease: Design, synthesis, and characterization of pyrazines. J. Med. Chem. 2012, 55, 9107–9119. [Google Scholar] [CrossRef]
- Dendelé, B.; Tekpli, X.; Sergent, O.; Dimanche-Boitrel, M.T.; Holme, J.A.; Huc, L.; Lagadic-Gossmann, D. Identification of the couple GSK3α/c-Myc as a new regulator of hexokinase II in benzo[a]pyrene-induced apoptosis. Toxicol. Vitr. 2012, 26, 94–101. [Google Scholar] [CrossRef]
- Pastorino, J.G.; Hoek, J.B.; Shulga, N. Activation of glycogen synthase kinase 3beta disrupts the binding of hexokinase II to mitochondria by phosphorylating voltage-dependent anion channel and potentiates chemotherapy-induced cytotoxicity. Cancer Res. 2005, 65, 10545–10554. [Google Scholar] [CrossRef]
- Bhushan, B.; Poudel, S.; Manley, M.W., Jr.; Roy, N.; Apte, U. Inhibition of Glycogen Synthase Kinase 3 Accelerated Liver Regeneration after Acetaminophen-Induced Hepatotoxicity in Mice. Am. J. Pathol. 2017, 187, 543–552. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Published by MDPI on behalf of the Österreichische Pharmazeutische Gesellschaft. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).