Abstract
The modern world, swaddled in the benefits of civilization, has fostered the development of science and the introduction of products of technological progress. This has allowed serious individual health problems, including those associated with viral diseases, to become targets for prophylaxis, treatment, and even cure. Human immunodeficiency viruses, hepatitis viruses, coronaviruses, and influenza viruses are among the most disturbing infectious agents in the human experience. Influenza appears to be one of the oldest viruses known to man; these viruses were among the first to cause major epidemics and pandemics in human history, collectively causing up to 0.5 million deaths worldwide each year. The main problem in the fight against influenza viruses is that they mutate constantly, which leads to molecular changes in antigens, including outer membrane glycoproteins, which play a critical role in the creation of modern vaccines. Due to the constant microevolution of the virus, influenza vaccine formulas have to be reviewed and improved every year. Today, flu vaccines represent an eternal molecular race between a person and a virus, which neither entity seems likely to win.
1. Introduction
Of the more than 300 widely recognized human diseases, influenza is one of the most common [1,2]. Influenza viruses are considered to be quite successful human pathogens: their persistence in the human population and ability to cause sporadic pandemics make them a continuous public health threat. Seasonal influenza causes approximately 500,000 deaths worldwide [3,4,5,6]. Occasionally, and unpredictably, influenza sweeps the world, infecting 20 to 40% of the population in a single year [7].
The epidemiological picture of influenza is based on many factors such as changes in the nature of the antigenic properties of the virus, the transmissibility of the virus, and the susceptibility of the population [8]. In fact, everyone has had the flu at least once in their life. It is worth clarifying that there are pediatric populations that do not have influenza until they are naturally exposed, and some vaccinated individuals do not have a true flu infection until the vaccine fails to protect them against the current circulating strain. The first mention of the alleged influenza syndrome was noted as early as 412 BC in the writings of Hippocrates [9]. For many centuries, there have been global outbreaks of influenza epidemics, most notably in 1510 and 1580 [10], 1688 [11], and 1693 [12]. Three influenza virus pandemics of the 20th century have affected the entire globe: 1918-19 (‘Spanish’ flu H1N1)—50 million deaths [13,14,15,16], 1957 (‘Asian’ flu, H2N2)—1 million victims [1,17], 1968 (‘Hong Kong’ flu, H3N2)—0.5 million deaths in total [18,19,20], 2009 H1N1pdm09 > 280,000 deaths [21]. Few diseases can match the flu in terms of contagiousness and number of deaths per year.
2. The Virus Is a Cosmopolitan Being with Its Own Characteristics
Influenza viruses have no borders and are found on every continent and in every place where people live. For example, the GIBS database (the global influenza B study) examined 358,768 cases of influenza registered between 1999 and 2014; influenza was found in 29 countries around the world, 4 of which were in the Southern Hemisphere, 15 in the intertropical zone, and 10 in the Northern Hemisphere [22]. In temperate zones, influenza epidemics strongly favor winter. The most intense influenza activity occurs between November and December according to FluNet [22].
It is harder to predict how influenza activity will manifest in the tropics; outbreaks may occur sporadically throughout the year or at irregular intervals [23]. Tropical seasons differ from those of temperate regions, particularly with respect to humidity and temperature; as these factors fluctuate, influenza activity fluctuates as well [24,25]. This difference has led to speculation that influenza viruses are transmitted primarily as aerosols during the winter in temperate regions, whereas direct or indirect contact appears to be responsible for year-round transmission in the tropics [26].
Each year, temperate regions experience winter influenza epidemics [23]. It has been estimated that the mortality resulting from these annual outbreaks has led to deaths ranging from 0.004% to 0.02% per 100,000 people; most are caused by a variant of the A virus H1N1 [26,27,28]. Tropical countries have fared much worse, with some of the highest global rates of mortality observed due to respiratory diseases; statistics from influenza pandemics in Mexico, India, Bangladesh, Myanmar, Indonesia, and Guatemala bear this out [26], [29,30,31]. In addition, it is difficult to tease out the effects of influenza on its own, owing to a strong association between influenza and annual mortality from all causes. In Singapore, influenza associated with concomitant infection with pneumonia, as well with diseases of the circulatory and respiratory system, was found to be responsible for 14.8 deaths per 100,000 people [32].
3. Remember Me Forever
Influenza is not a shy creature; it does not pass without a trace and because it likes company, its complications can be dangerous. Bacteria infecting in tandem with influenza or later as a secondary infection has been documented in 11–35% of laboratory-confirmed influenza in patients of all ages [33,34]. The players most likely to cause influenza-associated secondary infection, which often leads to bacterial pneumonia, are Streptococcus pneumoniae, Haemophilus influenzae, and Staphylococcus aureus [35].
While influenza is transmitted most often among young people, the highest mortality rates are seen among older people. The high mortality and morbidity rates in the elderly have also been observed in those suffering from certain high-risk diseases, including cardiovascular disease [28], acute myocardial infarction (AMI) [36,37], heart failure [38], and metabolic diseases such as diabetes mellitus [8,39] and diabetic ketoacidosis [40]. In addition, influenza exacerbates chronic obstructive pulmonary disease, causing airway obstruction as a result of airway inflammation, mucus hypersecretion, mucosal edema and bronchospasm [41], obesity, and kidney disease [42]. In addition to increasing abnormal glucose levels by 75%, influenza disrupts the daily routine of diabetic patients by reducing sleep and interfering with physical activity [43].
Influenza may also elicit systemic effects via inflammatory cytokines and prothrombotic changes, and these effects are associated with population-level increases in cardiovascular hospitalizations and deaths from AMI, heart failure, and cerebrovascular events [33,44,45].
However, rare, neurological complications have also been observed in association with influenza virus infections. These include febrile seizures, influenza-like encephalitis or encephalopathy, Guillain–Barré syndrome, and exacerbations in patients with epilepsy [35,46,47]. In 1890, the viral pandemic later dubbed the ‘Russian’ or ‘Asian’ flu killed one million people. Neurological consequences, particularly chronic headaches that manifested months or years after the pandemic had ended, were well documented [48,49]. While chronic fatigue syndrome (myalgic encephalomyelitis) and/or fibromyalgia may be precursors of neurasthenia, some cases may actually be incidences of post-infectious new daily persistent headache (NDPH) [50].
4. Stratagems Designed to Elude Host Adaptation
Influenza belongs to the Orthomyxoviridae family of viruses [8]. Viruses in this family are characterized by negative-sense, single-stranded RNA genome contained within seven or eight segments; influenza A and B has eight, comprising a 13,500-letter genome [2,51]. Each RNA segment is assembled into a viral ribonucleoprotein complex (vRNP) consisting of viral RNA, several nucleoprotein (NP) molecules, and viral RNA-dependent RNA polymerase (RdRp). The surface of influenza A and B viruses, which have a spherical shape, is permeated with spikes of glycoproteins: the hemagglutinin trimmer (HA) and the neuraminidase tetramer (NA), as well as the membrane protein M2, which acts as an ion channel. The basis of the double lipid layer is the matrix protein M1 surrounding the core of the virion [6,52]. One reason that influenza has been so successful at infecting humans is the lack of proofreading activity in influenza RNA polymerase, leading to a mutation rate of approximately one error each time a genome replicates [53]. Each cell can infect the cells around it with 10,000 new viral mutants, which is key to influenza’s evolutionary strategy. By constantly changing its glycoprotein sequences, particularly those of the hemagglutinin (HA) protein responsible for attachment to host cells [54], the virus stays one step ahead of the human immune system. The virus also alters neuraminidase (NA), its second most important transmembrane glycoprotein (located in the viral envelope), which helps the virus gain entry to the cell and is responsible for viral egress [55].
Influenza viruses have two main mechanisms of antigenic evolution: antigenic drift and antigenic shift. Antigenic drift occurs when, due to the high error rate of RNA polymerase during replication, mutations accumulate at antigenic sites that in turn produce virus variants able to elude immune defenses. This frequent phenomenon is common to both influenza A and B viruses. Antigenic shift is a less frequent but more striking; the segmentation of influenza and other Orthomyxoviridae allows them to undergo reassortment, which can cause the virus to acquire antigenically novel HA [3]. On the surface of the virion, HA is presented as a trimeric glycoprotein with a globular head and a proximal region of the membrane stem. The globular head domain of the HA protein determines the host specificity and tissue tropism of the virus through its ability to interact with N-acetylneuraminic (sialic) acid receptors, although receptor distribution and oligosaccharide-associated specificity varies across tissues and hosts. Thus, the hemagglutinin of human strains of the virus binds to sialic acid, which forms α-2,6-linkages with galactose, while the protein of avian strains preferentially binds to an acid containing terminal α-2,3-linkages [56,57]. Seasonal influenza epidemics are driven by antigen drift variants in which minor antigenic changes in HA and NA accumulate over time, resulting from mutations in the HA and NA genes due to error-prone viral RNA polymerase and positive selection pressure caused by pre-existing immunities [58]. The general topology of the NA evolutionary tree is similar to that of the HA1 tree, showing typical ‘ladder’ gradual evolution, with old strains rapidly being replaced by newer ones. NA was found to have fewer nucleotide substitutions over 40 years of evolution compared to HA1; overall, the genetic distance from the tree root to the most recent cluster of CA04 strains was about 1.5 times greater for HA1 than for NA [59].
5. WHO and Influenza Viruses: Global Surveillance
The World Health Organization (WHO) estimates that influenza viruses cause 3 to 5 million cases of severe illness and 250,000 to 500,000 deaths annually worldwide [60,61,62]. At the moment, the influenza virus comprises four main groups [8]:
- A (H1N1pdm09) [63,64,65].
- A (H3N2) [51].
- B/Yamagata lineage [66].
- B/Victoria lineage [67,68].
According to WHO, vaccination represents the most effective way to prevent the disease. Global surveillance of circulating influenza viruses influences the choice of influenza antigens annually for inclusion in vaccines [69]. Each year during influenza season, 5–15% of the world’s population becomes infected [70]. The global surveillance program mentioned above, maintained by WHO, generates a large amount of sequence data from wild-type viral isolates [71]. These data are accessible via databases at the National Center for Biotechnology Information (NCBI) and the Global Initiative on Sharing All Influenza Data (GISAID) [55,72,73]. Safe and effective vaccines have been available and in use for over 60 years. WHO recommends annual vaccination for pregnant women at any stage of pregnancy, children aged 6 months to 5 years, the elderly (over 65 years), people with chronic diseases, and healthcare workers [74]. One of the most important things about the WHO recommendations (2022) [75] is that vaccination choice should be based on clear, reliable, general criteria. The need is clear, particularly as new indications for vaccination emerge. However, even as new vaccines become available, responding to the need is complicated by the limited economic resources of health care systems, even in developed countries [76]. Influenza vaccines target the production of strain-specific antibodies to the globular head (HA1) protein hemagglutinin. It is the HA1 domain that contains most of the antigenic sites that undergo changes during antigenic drift [77]. It is also possible to use other proteins as targets for the vaccine: neuraminidase and transmembrane protein M2 [78,79,80]. Currently, the two vaccine platforms used worldwide are inactivated vaccines and live-attenuated vaccines (Figure 1).
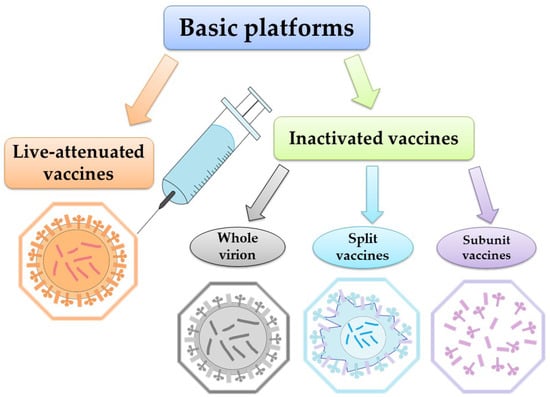
Figure 1.
Major influenza vaccine platforms.
Interesting approach is also realized in Flublok vaccine, obtained by culturing insect cells using the technology of baculovirus expression systems which produce exact analogues of the HA of circulating influenza viruses [81].
According to the Centers for Disease Control and Prevention (CDC), in 2019–2020, during the last flu season before the COVID-19 pandemic, influenza vaccination prevented approximately 7.5 million influenza cases, 3.7 million of medical visits, 105,000 influenza-related hospitalizations, and 6300 deaths. In addition, using statistical data, it was calculated that the flu vaccine reduced the risk of doctor visits for influenza by 40–60%. Moreover, studies carried out among children in 2022 showed a 75% reduction in the risk of a severe course of the disease [82]. The CDC suggests that influenza vaccines should be predominantly quadrivalent.
6. Where Did It Come from, Where Did It Go?
The molecular evolution of the influenza virus through reassortment and the accumulation of mutations allows us to conclude the origin of the virus. Thus, wild birds are recognized as the progenitors of influenza A virus, and avian viruses have contributed genetic material to most human-infecting viruses, including the H5N1 and H1N1 subtypes [1]. We suppose that the transmission of the influenza virus from wild and domestic animals and humans might be closely linked.
Scientists continue to debate to what extent the emergence and global spread of new pandemic human strains of influenza involve birds [83]; it is believed that several segments in the most recent pandemic strain of influenza (H1N1; swine influenza virus) are likely to have avian origins [1,84]. Since transmission of influenza, like all viruses, is affected by how the host community interacts with the environment as well as coevolution between host and pathogen [85], it is not surprising that the dynamics of influenza incidence among birds and mammals (including humans) is closely related.
The changes wrought in the environment by human beings affects everything, including viruses; the ecology and evolution of influenza viruses has had to adapt to the development of agriculture and other land use, globalization, and climate change. Climate change has altered the distribution, composition, and migratory behavior of wild birds, resulting in dramatic changes to the epidemiology of avian influenza [1,85], including the ability of the virus to survive in the environment.
Studies of viruses circulating in poultry markets have provided much useful information about the transmission of viruses and the processes by which they evolve [86,87,88]. For example, observation of domestic and wild birds and phylogenetic reconstruction led to the identification of H5N1 as a pandemic threat as early as 2002, when its distribution was limited to China and only a few people were infected [89]. Unfortunately, preventive measures did not completely eradicate the H5N1 virus, and it reappeared in 2003 and spread throughout Asia, Europe, and Africa over the next four years [90].
Influenza A viruses are more prone to antigenic evolution than influenza B viruses, whose genetic changes occur more slowly [91,92,93]. Influenza B virus was found in both pigs and humans throughout the 20th century; while pigs are naturally infected with influenza A and influenza C, it is believed that pigs may have initially contracted influenza B from humans [15].
7. Catch Me If You Can—Alternative Sources of Disease Outbreaks
The risk of an artificial pandemic as a result of a laboratory escape is not hypothetical. In the past, pathogens have been diverted from the laboratory with serious consequences, including transmission to others outside the laboratory staff [94,95,96,97,98,99]. Historical data from the pandemic influenza of 1977 provide evidence of laboratory outbreaks and deaths caused by pathogens. It was during this period that the first escape of the virus from the laboratory in the USSR was recorded. It is believed that the outbreak was not natural, and three possible sources have been suggested: an accident in the laboratory, a test escape with a live vaccine, or a deliberate release as a biological weapon [100]. The 2009 H1N1 flu brought the 1977 epidemic back to the forefront as debunked rumors soon surfaced that it was the result of a laboratory accident [101,102].
8. How to Emerge from the Molecular Race as a Winner
It seems clear that prevention of future influenza virus epidemics would benefit from the use of super conservative micro antigens, which change very little over time, to create vaccines (Figure 2).
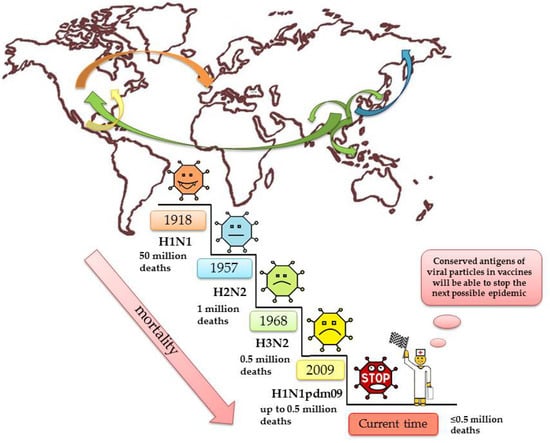
Figure 2.
The biggest flu epidemics in human history.
Systems biology approaches have been used to identify early molecular signatures that can be used to predict subsequent immune responses and better understand the mechanisms underlying immunogenicity [103,104,105]. A number of scientists have been engaged in factor analysis [106,107]; their efforts have provided important data on mRNA expression and the pattern of gene expression.
A truly universal vaccine that provides lifelong protection against any strain of influenza with one or more shots may not be achievable, but some variation of this concept should be considered [108]. As with any project with such a wide scope and complexity, problems exist that make it difficult to find a universal vaccine:
- The protruding loops of the HA head vary greatly among viral strains, and head-binding antibodies do not neutralize viruses that are not closely bound to the immunogen [109,110,111].
- Up until 2009, the amino acid sequences of the seasonal human H1 strain differed only by 50–60%; however, since 2009, they have been found to differ by as much as 80% [112].
- The emergence of new subtype sequences of archaic sequences from animal reservoirs poses a challenge for seasonal vaccine antigen selection.
- As the amino acid sequences of hemagglutinin continue to change [113], the seasonal inactivated influenza vaccine is reformulated every year in an attempt to match the strains of the virus predicted to circulate [79,114].
- The segmented influenza virus genome exchanges RNA segments between genotypically different influenza viruses, resulting in the formation of new strains and/or subtypes [83,115], which is called reassortment. Reassortment is an important mechanism for generating a ‘new’ virus [115,116].
To genuinely provide long-term protection, a universal vaccine must break the annual vaccination cycle. Long-term protection of at least 1 year, but preferably for several seasons, is one of the four criteria established for a universal influenza vaccine by the NIAID (National Institute of Allergy and Infectious Diseases) [117], but no one has yet discovered how to achieve this goal. Prosaic but important factors such as immunization schedules, formulations, doses, and adjuvants need to be taken into consideration [118]. Despite these inherent difficulties, many next-generation vaccines are currently under development, and WHO expects a universal influenza A vaccine to enter expanded clinical trials as early as 2027 [118,119].
Given that influenza viruses will continue to coexist with humans and other animals, consideration should be given to the study of long-lived immune responses to natural infection and to the translation of this knowledge into the development of vaccines that induce long-term, ideally life-long immunity. This can be achieved by using more advanced adjuvants to stimulate the desired innate immune sensors in precisely defined cell types, or by delivering vaccines in ways that enhance antigen and epitope integrity [120].
The increased availability of viral gene sequencing promises much wider distribution and the ability to rapidly detect the emergence and spread of new viral variants. Synthetic biology now allows digitally transmitted sequences to be quickly converted into genes that code for new flu variants. SAM® (self-amplifying mRNA) vaccine technology, a new technology under development, promises fully synthetic vaccines. The platform combines self-amplifying mRNA and a synthetic, liposomal, nonviral delivery system. SAM RNA is produced from a cell-free enzymatic transcription reaction, and it encodes the antigen of interest and an RNA-dependent RNA polymerase. The lipid delivery system provides efficient delivery of the RNA cargo to muscle cells at the site of injection and protection of the RNA from enzymatic degradation during delivery. After delivery, the mRNA directs the expression of the polymerase, which amplifies the RNA intra-cytoplasmically and launches the expression of a subgenomic message that directs high-level target antigen expression. The innate immunity perceives replicating RNA as if it were a virus, triggering a robust adaptive immune response [121,122]. If SAM technology lives up to its promises, pandemic vaccine batches could be produced using portable production devices deployed around the world based on electronic information transmitted from a synthetic virus reference laboratory. Combined, these technologies could greatly improve our ability to respond effectively to the continuous change in influenza viruses and their associated seasonal and pandemic disease threats [123].
As an alternative to creating vaccines based on influenza virus protein antigens, long-term vaccines are also being developed using conserved regions of the genome of RNA viruses. Indeed, as influenza viruses have shown us, it is impossible to predict the emergence of new mutations of RNA viruses. Modern vaccines are vulnerable to these unpredictable shifts in nucleotide sequences since the principle of their action and effectiveness depends on the presence of the certain specific extended amino acid sequences in the peptide chains of the pathogen that play the role of antigen. For this reason, the use of conserved amino acid sequences of RNA virus surface proteins, such HA and NA, has little chance of long-term success. In fact, these proteins are constantly changing. However, the nucleic acids of RNA viruses contain extended, highly conserved regions that hardly change over time, which represents an ideal target for therapeutic development. It has been impossible to hit this target so far because the concept has been poorly studied. However, these vaccines can be versatile and very effective. Oligonucleotides have been acting as excellent adjuvants for decades [123,124], and more and more data are accumulating that show that they can be antigens as well. We may be running out of options with modern vaccines, which depend on the amino acid sequences of RNA virus proteins, but we are well on the way to creating new, less limited options with universal oligonucleotide vaccines.
Two obstacles stand in the way of the development of oligonucleotide vaccines: the lack of knowledge concerning antibodies capable of penetrating cells and attacking the unique nucleic acid sequences of RNA viruses, and the unknown antigen presentation ability of dendritic cells with respect to nucleic acids. At the same time, there is no doubt that oligonucleotide vaccines based on highly conserved regions of the RNA virus genomes have great potential in the prevention of viral diseases, despite the fact that they do not yet fit within the framework of modern concepts of immunology [125].
9. Conclusions
Influenza viruses are among the most serious infectious agents circulating in the human population, periodically causing epidemics and even pandemics. Influenza viruses claim hundreds of thousands of lives every year, exacerbating old and new chronic diseases in survivors.
Due to their biology, influenza viruses constantly use antigenic drift and antigenic shift to change their genetic material, which changes the antigenic properties of viral particles. Each year, this leads to a revision of the formulas of inactivated and live-attenuated vaccines. In addition, animals, especially birds, act as natural reservoirs, which leads to the emergence and global the spread of the virus.
Today, a strategically important area of research is the development of vaccines with a long operational period of action. Such vaccines can be created on the basis of conserved influenza virus antigens. Traditional approaches using proteins as antigens have good potential for solving this problem but have not yet achieved significant results. No less promising is the development of oligonucleotide vaccines that use conservative regions of the influenza virus genome as antigens. It can be assumed that if influenza vaccines with an operational validity period of 10 years or more appear, then the race between the virus and the person will be won by the latter.
Author Contributions
Conceptualization: V.V.O.; writing—original draft preparation: V.V.O. and O.A.A.; writing—review and editing: V.V.O., O.A.A., E.E.A. and A.I.B.; drawing: V.V.O., O.A.A. and E.E.A. All authors are equally responsible for plagiarism, self-plagiarism or other ethical transgressions. All authors have read and agreed to the published version of the manuscript.
Funding
The research results are obtained within the framework of as state assignment V.I. Vernadsky Crimean Federal University for 2021 and the planning period of 2022–2023 No. FZEG-2021-0009 (‘Development of oligonucleotide constructs for making selective and highly effective preparations for medicine and agriculture’, registration number 121102900145-0).
Acknowledgments
We thank our many colleagues, too numerous to name, for the technical advances and lively discussions that have prompted us to write this review. We apologize to the many colleagues whose work has not been cited. We are very much indebted to all anonymous reviewers and our colleagues from the Lab on DNA technologies, PCR analysis and creation of DNA insecticides (V.I. Vernadsky Crimean Federal University, Institute of Biochemical Technologies, Ecology and Pharmacy, Department of Molecular Genetics and Biotechnologies), and OLINSCIDE BIOTECH LLC. for valuable comments on our manuscript. We are very thankful to Georgia Morgan for English language editing service.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Vandegrift, K.J.; Sokolow, S.H.; Daszak, P.; Kilpatrick, A.M. Ecology of avian influenza viruses in a changing world. Ann. N. Y. Acad. 2010, 1195, 113–128. [Google Scholar] [CrossRef]
- Cox, N.J.; Subbarao, K. Global epidemiology of influenza: Past and present. Annu. Rev. Med. 2000, 51, 407–421. [Google Scholar] [CrossRef] [PubMed]
- Wong, S.; Webby, R.J. Traditional and New Influenza Vaccines. Clin. Microbiol. Rev. 2013, 26, 476–492. [Google Scholar] [CrossRef] [PubMed]
- Centers for Disease Control and Prevention. Estimates of deaths associated with seasonal influenza—United States, 1976–2007. MMWR Morb. Mortal. Wkly. Rep. 2010, 59, 1057–1062. [Google Scholar]
- Thompson, W.W.; Shay, D.K.; Weintraub, E.; Brammer, L.; Bridges, C.B.; Cox, N.J.; Fukuda, K. Influenza-associated hospitalizations in the United States. JAMA 2004, 292, 1333–1340. [Google Scholar] [CrossRef] [PubMed]
- Eisfeld, A.J.; Neumann, G.; Kawaoka, Y. At the centre: Influenza A virus ribonucleoproteins. Nat. Rev. Microbiol. 2015, 13, 28–41. [Google Scholar] [CrossRef] [PubMed]
- Thompson, W.W.; Shay, D.K.; Weintraub, E.; Brammer, L.; Cox, N.; Anderson, L.J.; Fukuda, K. Mortality associated with influenza and respiratory syncytial virus in the United States. JAMA 2003, 289, 179–186. [Google Scholar] [CrossRef]
- Moghadami, M. A narrative review of influenza: A seasonal and pandemic disease. Iran. J. Med. Sci. 2017, 42, 2–13. [Google Scholar]
- Pappas, G.; Kiriaze, I.J.; Falagas, M.E. Insights into infectious disease in the era of Hippocrates. Int. J. Infect. Dis. 2008, 12, 347–350. [Google Scholar] [CrossRef]
- Beveridge, W. Influenza: The Last Great Plague, an Unfinished Story of Discovery; Prodist.: New York, NY, USA, 1977. [Google Scholar]
- Duffy, J. Epidemics in Colonial America; LSU Press: Baton Rouge, LA, USA, 1953; pp. 187–188. [Google Scholar]
- Pettit, D.A. A Cruel Wind: America Experiences the Pandemic Influenza. Ph.D. Dissertation, University of New Hampshire, Durham, NH, USA, 1976. Volume 32. pp. 1918–1920. [Google Scholar]
- Cheong, H.J.; Song, J.Y.; Heo, J.Y.; Noh, J.Y.; Choi, W.S.; Park, D.W.; Wie, S.H.; Kim, W.J. Immunogenicity and safety of the influenza A/H1N1 2009 inactivated split-virus vaccine in young and older adults: MF59-adjuvanted vaccine versus nonadjuvanted vaccine. Clin. Vaccine Immunol. 2011, 18, 1358–1364. [Google Scholar] [CrossRef]
- Shen, X.; Soderholm, J.; Lin, F.; Kobinger, G.; Bello, A.; Gregg, D.A.; Broderick, K.E.; Sardesai, N.Y. Influenza A vaccines using linear expression cassettes delivered via electroporation afford full protection against challenge in a mouse model. Vaccine 2012, 30, 6946–6954. [Google Scholar] [CrossRef] [PubMed]
- Taubenberger, J.K.; Morens, D.M. 1918 Influenza: The Mother of All Pandemics. Emerg. Infect. Dis. 2006, 12, 15–22. [Google Scholar] [CrossRef] [PubMed]
- Neumann, G.; Noda, T.; Kawaoka, Y. Emergence and pandemic potential of swine-origin H1N1 influenza virus. Nature 2009, 459, 931–939. [Google Scholar] [CrossRef]
- Simonsen, L.; Clarke, M.J.; Schonberger, L.B.; Arden, N.H.; Cox, N.J.; Fukuda, K. Pandemic versus epidemic influenza mortality: A pattern of changing age distribution. J. Infect. Dis. 1998, 178, 53–60. [Google Scholar] [CrossRef]
- Knobler, S.L.; Mack, A.; Mahmoud, A.; Lemon, S.M. The Threat of Pandemic Influenza: Are We Ready? Workshop Summary. The National Academies Collection: Reports Funded by National Institutes of Health; National Academies Press (US): Washington, DC, USA, 2005. [Google Scholar] [CrossRef]
- Webby, R.J.; Webster, R.G. Are we ready for pandemic influenza? Science 2003, 302, 1519–1522. [Google Scholar] [CrossRef] [PubMed]
- Johnson, N.P.; Mueller, J. Updating the accounts: Global mortality of the 1918–1920 “Spanish” influenza pandemic. Bull. Hist. Med. 2002, 76, 105–115. [Google Scholar] [CrossRef]
- Otte, A.; Marriott, A.; Dreier, C.; Dove, B.; Mooren, K.; Klingen, T.R.; Sauter, M.; Thompson, K.; Bennett, A.; Klingel, K.; et al. Evolution of 2009 H1N1 influenza viruses during the pandemic correlates with increased viral pathogenicity and transmissibility in the ferret model. Sci. Rep. 2016, 6, 28583. [Google Scholar] [CrossRef]
- Caini, S.; Spreeuwenberg, P.; Kusznierz, G.F.; Rudi, J.M.; Owen, R.; Pennington, K.; Wangchuk, S.; Gyeltshen, S.; Ferreira de Almeida, W.A.; Henriques, C.M.P.; et al. Distribution of influenza virus types by age using case-based global surveillance data from twenty-nine countries, 1999–2014. BMC Infect. Dis. 2018, 18, 269. [Google Scholar] [CrossRef]
- Viboud, C.; Alonso, W.J.; Simonsen, L. Influenza in tropical regions. PLoS Med. 2006, 3, 468–471. [Google Scholar] [CrossRef]
- Lowen, A.C.; Mubareka, S.; Steel, J.; Palese, P. Influenza virus transmission is dependent on relative humidity and temperature. PLoS Pathog. 2007, 3, 1470–1476. [Google Scholar] [CrossRef]
- Lowen, A.C.; Steel, J.; Mubareka, S.; Palese, P. High temperature (30 degrees C) blocks aerosol but not contact transmission of influenza virus. J. Virol. 2008, 82, 5650–5652. [Google Scholar] [CrossRef] [PubMed]
- Ng, S.; Gordon, A. Influenza burden and transmission in the tropics. Curr. Epidemiol. Rep. 2015, 2, 89–100. [Google Scholar] [CrossRef] [PubMed]
- Charu, V.; Simonsen, L.; Lustig, R.; Steiner, C.; Viboud, C. Mortality burden of the 2009-10 influenza pandemic in the United States: Improving the timeliness of influenza severity estimates using inpatient mortality records. Influenza Other Respir. Viruses 2013, 7, 863–871. [Google Scholar] [CrossRef] [PubMed]
- Nicoll, A.; Ciancio, B.C.; Lopez Chavarrias, V.; Mølbak, K.; Pebody, R.; Pedzinski, B.; Penttinen, P.; van der Sande, M.; Snacken, R.; Van Kerkhove, M.D. Influenza-related deaths—Available methods for estimating numbers and detecting patterns for seasonal and pandemic influenza in Europe. Euro Surveill. 2012, 17, 20162. [Google Scholar] [CrossRef]
- Dawood, F.S.; Iuliano, A.D.; Reed, C.; Meltzer, M.I.; Shay, D.K.; Cheng, P.-Y.; Bandaranayake, D.; Breiman, R.F.; Brooks, W.A.; Buchy, P.; et al. Estimated global mortality associated with the first 12 months of 2009 pandemic influenza a H1N1 virus circulation: A modelling study. Lancet Infect. Dis. 2012, 12, 687–695. [Google Scholar] [CrossRef]
- Simonsen, L.; Spreeuwenberg, P.; Lustig, R.; Taylor, R.J.; Fleming, D.M.; Kroneman, M.; Van Kerkhove, M.D.; Mounts, A.W.; Paget, W.J. Global mortality estimates for the 2009 influenza pandemic from the GLaMOR project: A modeling study. PLoS Med. 2013, 10, 1001558. [Google Scholar] [CrossRef]
- Nair, H.; Brooks, W.A.; Katz, M.; Roca, A.; Berkley, J.A.; Madhi, S.A.; Simmerman, J.M.; Gordon, A.; Sato, M.; Howie, S.; et al. Global burden of respiratory infections due to seasonal influenza in young children: A systematic review and meta-analysis. Lancet 2011, 378, 1917–1930. [Google Scholar] [CrossRef]
- Chow, A.; Ma, S.; Ling, A.E.; Chew, S.K. Influenza-associated deaths in tropical Singapore. United States: U.S. National Center for Infectious Diseases. Emerg. Infect. Dis. 2006, 12, 114–121. [Google Scholar] [CrossRef]
- Macias, A.E.; McElhaney, J.E.; Chaves, S.S.; Nealon, J.; Nunes, M.C.; Samson, S.I.; Seet, B.T.; Weinke, T.; Yu, H. The disease burden of influenza beyond respiratory illness. Vaccine 2021, 39, A6–A14. [Google Scholar] [CrossRef]
- Klein, E.Y.; Monteforte, B.; Gupta, A.; Jiang, W.; May, L.; Hsieh, Y.H.; Dugas, A. The frequency of influenza and bacterial coinfection: A systematic review and meta-analysis. Influenza Other Respir. Viruses 2016, 10, 394–403. [Google Scholar] [CrossRef]
- Morris, D.E.; Cleary, D.W.; Clarke, S.C. Secondary bacterial infections associated with influenza pandemics. Front. Microbiol. 2017, 8, 1041. [Google Scholar] [CrossRef] [PubMed]
- Kwong, J.C.; Schwartz, K.L.; Campitelli, M.A. Acute myocardial infarction after laboratory-confirmed influenza infection. N. Engl. J. Med. 2018, 378, 345–353. [Google Scholar] [CrossRef] [PubMed]
- Buchan, S.A.; Kwong, J.C. Trends in influenza vaccine coverage and vaccine hesitancy in Canada, 2006/07 to 2013/14: Results from cross-sectional survey data. CMAJ Open. 2016, 4, E455–E462. [Google Scholar] [CrossRef]
- Kadoglou, N.P.E.; Bracke, F.; Simmers, T.; Tsiodras, S.; Parissis, J. Influenza infection and heart failure-vaccination may change heart failure prognosis? Heart Fail. Rev. 2017, 22, 329–336. [Google Scholar] [CrossRef] [PubMed]
- Goeijenbier, C.M.; van Sloten, T.T.; Slobbe, L.; Mathieu, C.; van Genderen, P.; Beyer, W.E.P.; Osterhaus, A. Benefits of flu vaccination for persons with diabetes mellitus: A review. Vaccine 2017, 35, 5095–5101. [Google Scholar] [CrossRef] [PubMed]
- Moghadami, M.; Honarvar, B.; Sabaeian, B.; Zamiri, N.; Pourshahid, O.; Rismanchi, M.; Lankarani, K.B. H1N1 influenza infection complicated with diabetic ketoacidosis. Arch. Iran. Med. 2012, 15, 55–58. [Google Scholar] [PubMed]
- Wallick, C.; Toa, T.M.; Korom, S.; Masterset, H.; Hanania, N.A.; Moawad, D. Impact of influenza infection on the short- and long-term health of patients with chronic obstructive pulmonary disease. Infect. Dis. 2022, 25, 930–939. [Google Scholar] [CrossRef]
- Dixit, R.; Webster, F.; Booy, R.; Menzies, B. The role of chronic disease in the disparity of influenza incidence and severity between indigenous and non-indigenous Australian peoples during the 2009 influenza pandemic. BMC Public Health 2022, 22, 1295. [Google Scholar] [CrossRef]
- Samson, S.I.; Konty, K.; Lee, W.N.; Quisel, T.; Foschini, L.; Kerr, D.; Liska, J.; Mills, H.; Hollingsworth, R.; Greenberg, M.; et al. Quantifying the impact of influenza among persons with type 2 diabetes mellitus: A new approach to determine medical and physical activity impact. J. Diabetes Sci. Technol. 2019, 15, 44–52. [Google Scholar] [CrossRef]
- Fischer, W.A.; Gong, M.; Bhagwanjee, S.; Sevransky, J. Global burden of influenza as a cause of cardiopulmonary morbidity and mortality. Glob. Heart 2014, 9, 325–336. [Google Scholar] [CrossRef]
- Nguyen, J.L.; Yang, W.; Ito, K.; Matte, T.D.; Shaman, J.; Kinney, P.L. Seasonal influenza infections and cardiovascular disease mortality. JAMA Cardiol. 2016, 1, 274–281. [Google Scholar] [CrossRef] [PubMed]
- Sellers, S.A.; Hagan, R.S.; Hayden, F.G.; Fische, W.A. 2nd The hidden burden of influenza: A review of the extra-pulmonary complications of influenza infection. Influenza Other Respir. Viruses 2017, 11, 372–393. [Google Scholar] [CrossRef] [PubMed]
- Ekstrand, J.J. Neurologic complications of influenza. Semin. Pediatr. Neurol. 2012, 19, 96–100. [Google Scholar] [CrossRef] [PubMed]
- Rozen, T.D. Daily persistent headache after a viral illness during a worldwide pandemic may not be a new occurrence: Lessons from the 1890 Russian/Asiatic flu. Cephalalgia 2020, 40, 1406–1409. [Google Scholar] [CrossRef]
- Kempińska-Mirosławska, B.; Woźniak-Kosek, A. The influenza epidemic of 1889–90 in selected European cities—A picture based on the reports of two Poznań daily newspapers from the second half of the nineteenth century. Med. Sci. Monit. 2013, 19, 1131–1141. [Google Scholar] [CrossRef]
- Overholser, J.C.; Beale, E.E. Neurasthenia: Modern malady or historical relic? J. Nerv. Ment. Dis. 2019, 207, 731–739. [Google Scholar] [CrossRef]
- Yang, J.Y.; Yang, L.; Zhu, W.F.; Wang, D.; Shu, Y. Epidemiological and genetic characteristics of the H3 subtype avian influenza viruses in China. China CDC Wkly. 2021, 3, 929–936. [Google Scholar] [CrossRef]
- Palese, P.; Shaw, M.L. Orthomyxoviridae: The Viruses and their Replication. In Fields Virology, 5th ed.; Knipe, D.M., Howley, P.M., Eds.; Lippincott Williams & Wilkins: Philadelphia, PA, USA, 2007. [Google Scholar]
- Drake, J.W. Rates of spontaneous mutation among RNA viruses. Proc. Natl. Acad. Sci. USA 1993, 90, 4171–4175. [Google Scholar] [CrossRef]
- Boivin, S.; Cusack, S.; Ruigrok, R.W.; Hart, D.J. Influenza A virus polymerase: Structural insights into replication and host adaptation mechanisms. J. Biol. Chem. 2010, 285, 28411–28417. [Google Scholar] [CrossRef]
- Gamblin, S.J.; Skehel, J.J. Influenza hemagglutinin and neuraminidase membrane glycoproteins. J. Biol. Chem. 2010, 285, 28403–28409. [Google Scholar] [CrossRef]
- Couceiro, J.N.; Paulson, J.C.; Baum, L.G. Influenza virus strains selectively recognize sialyloligosaccharides on human respiratory epithelium; the role of the host cell in selection of hemagglutinin receptor specificity. Virus Res. 1993, 29, 155–165. [Google Scholar] [CrossRef] [PubMed]
- Matrosovich, M.N.; Matrosovich, T.Y.; Gray, T.; Roberts, H.D. Klenk Human and avian influenza viruses target different cell types in cultures of human airway epithelium. Proc. Natl. Acad. Sci. USA 2004, 101, 4620–4624. [Google Scholar] [CrossRef] [PubMed]
- Rudraraju, R.; Subbarao, K. Passive immunization with influenza haemagglutinin specific monoclonal antibodies. Hum. Vaccin. Immunother. 2018, 14, 2728–2736. [Google Scholar] [CrossRef] [PubMed]
- Westgeest, K.B.; de Graaf, M.; Fourment, M.; Bestebroer, T.M.; van Beek, R.; Spronken, M.I.J.; de Jong, J.C.; Rimmelzwaan, G.F.; Russell, C.A.; Osterhaus, A.D.M.E.; et al. Genetic evolution of the neuraminidase of influenza A (H3N2) viruses from 1968 to 2009 and its correspondence to haemagglutinin evolution. J. Gen. Virol. 2012, 93, 1996–2007. [Google Scholar] [CrossRef]
- Gasparini, R.; Amicizia, D.; Lai, P.L.; Panatto, D. Influenza vaccination: From epidemiological aspects and advances in research to dissent and vaccination policies. J. Prev. Med. Hyg. 2016, 57, 1–4. [Google Scholar] [CrossRef]
- Wright, P.F.; Neumann, G.; Kawaoka, Y. Orthomyxoviruses. In Fields Virology, 5th ed.; Knipe, D.M., Howley, P.M., Eds.; Lippincott Williams & Wilkins: Philadelphia, PA, USA, 2007; pp. 1691–1740. [Google Scholar]
- Taubenberger, J.K.; Morens, D.M. The Pathology of Influenza Virus Infections. Annu. Rev. Pathol. 2008, 3, 499–522. [Google Scholar] [CrossRef]
- Juvet, L.K.; Robertson, A.H.; Laake, I.; Mjaaland, S.; Trogstad, L. Safety of Influenza A H1N1pdm09 Vaccines: An Overview of Systematic Reviews. Front. Immunol. 2021, 12, 740048. [Google Scholar] [CrossRef]
- Minchole, E.; Figueredo, A.L.; Omeñaca, M.; Panadero, C.; Royo, L.; Vengoechea, J.J.; Fandos, S.; de Pablo, F.; Bello, S. Seasonal Influenza A H1N1pdm09 Virus and Severe Outcomes: A Reason for Broader Vaccination in Non-Elderly, At-Risk People. PLoS ONE 2016, 11, 16571. [Google Scholar] [CrossRef]
- Garten, R.J.; Davis, C.T.; Russell, C.A.; Shu, B.; Lindstrom, S.; Balish, A.; Sessions, W.M.; Xu, X.; Skepner, E.; Deyde, V.; et al. Antigenic and genetic characteristics of swine-origin 2009 A(H1N1) influenza viruses circulating in humans. Science 2009, 325, 197–201. [Google Scholar] [CrossRef]
- Yang, J.; Lau, Y.C.; Wu, P.; Feng, L.; Wang, X.; Chen, T.; Aki, S.T.; Peng, Z.; Fang, V.J.; Zhang, J.; et al. Variation in influenza B virus epidemiology by lineage, China. Emerg. Infect. Dis. J. 2018, 24, 1536–1540. [Google Scholar] [CrossRef]
- Langat, P.; Raghwani, J.; Dudas, G.; Bowden, T.A.; Edwards, S.; Gall, A.; Bedford, T.; Rambaut, A.; Daniels, R.S.; Russell, S.A.; et al. Genome-wide evolutionary dynamics of influenza B viruses on a global scale. PLoS Pathog. 2017, 13, e1006749. [Google Scholar] [CrossRef] [PubMed]
- Vijaykrishna, D.; Holmes, E.C.; Joseph, U.; Fourment, M.; Su, Y.C.F.; Halpin, R.; Lee, R.T.C.; Deng, Y.; Gunalan, V.; Lin, X.; et al. The contrasting phylodynamics of human influenza B viruses. eLife 2015, 16, e05055. [Google Scholar] [CrossRef] [PubMed]
- Ang, L.W.; Tien, W.S.; Lin, R.T.; Cui, L.; Cutter, J.; James, L.; Goh, K.T. Characterization of influenza activity based on virological surveillance of influenza-like illness in tropical Singapore 2010–2014. J. Med. Virol. 2016, 88, 2069–2077. [Google Scholar] [CrossRef]
- Petrova, V.N.; Russell, C.A. The evolution of seasonal influenza viruses. Nat. Rev. Microbiol. 2018, 16, 47–60. [Google Scholar] [CrossRef]
- McHardy, A.C.; Adams, B. The Role of Genomics in Tracking the Evolution of Influenza A Virus. PLoS Pathog. 2009, 5, e1000566. [Google Scholar] [CrossRef] [PubMed]
- Bao, Y.; Bolotov, P.; Dernovoy, D.; Kiryutin, B.; Zaslavsky, L.; Tatusova, T.; Ostell, J.; Lipman, D. The Influenza Virus Resource at the National Center for Biotechnology Information. J. Virol. 2008, 82, 596–601. [Google Scholar] [CrossRef] [PubMed]
- Enserink, M. DATA SHARING: New Swiss Influenza Database to Test Promises of Access. Science 2007, 315, 923. [Google Scholar] [CrossRef]
- World Health Organization (WHO). Influenza (Seasonal). Available online: http://www.who.int/mediacentre/factsheets/fs211/en/ (accessed on 15 February 2016).
- World Health Organization (WHO). Available online: https://www.who.int/news/item/25-02-2022-recommendations-announced-for-influenza-vaccine-composition-for-the-2022-2023-northern-hemisphere-influenza-season (accessed on 25 February 2022).
- Gasparini, R.; Mennini, F.S.; Panatto, D.; Bonanni, P.; Bechini, A.; Ricciardi, W.; De Waure, C.; Marcellusi, A.; Cicchetti, A.; Ruggeri, M.; et al. How can the results of Health Technology Assessment (HTA) evaluations applied to vaccinations be communicated to decision-makers and stakeholders? The ISPOR Rome Chapter Project. J. Prev. Med. Hyg. 2015, 56, E150–E154. [Google Scholar]
- Tapia, R.; Torremorell, M.; Culhane, M.; Medina, R.A.; Neira, V. Antigenic characterization of novel H1 influenza A viruses in swine. Sci. Rep. 2020, 10, 4510. [Google Scholar] [CrossRef]
- Tosh, P.K.; Jacobson, R.M.; Poland, G.A. Influenza vaccines: From surveillance through production to protection. Mayo Clin. Proc. 2010, 85, 257–273. [Google Scholar] [CrossRef]
- Betakova, T. M2 protein-a proton channel of influenza A virus. Curr. Pharm. Des. 2007, 13, 3231–3235. [Google Scholar] [CrossRef] [PubMed]
- Tompkins, S.M.; Zhao, Z.S.; Lo, C.Y.; Misplon, J.A.; Liu, T.; Ye, Z.; Hogan, R.J.; Wu, Z.; Benton, K.A.; Tumpey, T.M.; et al. Matrix protein 2 vaccination and protection against influenza viruses, including subtype H5N1. Emerg. Infect. Dis. 2007, 13, 426–435. [Google Scholar] [CrossRef] [PubMed]
- Cox, M.M.; Izikson, R.; Post, P.; Dunkle, L. Safety, efficacy, and immunogenicity of Flublok in the prevention of seasonal influenza in adults. Ther. Adv. Vaccines 2015, 3, 97–108. [Google Scholar] [CrossRef] [PubMed]
- Centers for Disease Control and Prevention. Available online: https://www.cdc.gov/flu/vaccines-work/vaccineeffect.htm (accessed on 1 May 2023.).
- Reid, A.H.; Taubenberger, J.K. The origin of the 1918 pandemic influenza virus: A continuing enigma. J. Gen. Virol. 2003, 84, 2285–2292. [Google Scholar] [CrossRef] [PubMed]
- Fraser, C.; Donnelly, C.A.; Cauchemez, S.; Hanage, W.P.; Kerkhove, M.D.V.; Hollingsworth, T.D.; Griffin, J.; Baggaley, R.F.; Jenkins, H.E.; Lyons, E.J.; et al. Pandemic potential of a strain of influenza A (H1N1): Early findings. Science 2009, 324, 1557–1561. [Google Scholar] [CrossRef]
- Gilbert, M.; Slingenbergh, J.; Xiao, X. Climate change and avian influenza. Rev. Sci. Et Tech. Off. Int. Des Epizoot. 2008, 27, 459–466. [Google Scholar] [CrossRef]
- Chen, H.; Smith, G.; Li, K.; Wang, J.; Fan, X.H.; Rayner, J.M.; Vijaykrishna, D.; Zhang, J.X.; Zhang, L.J.; Guo, C.T.; et al. Establishment of multiple sublineages of H5N1 influenza virus in Asia: Implications for pandemic control. Proc. Natl. Acad. Sci. USA 2006, 103, 2845–2850. [Google Scholar] [CrossRef]
- Takano, R.; Nidom, C.A.; Kiso, M.; Muramoto, M.; Yamada, S.; Sakai-Tagawa, Y.; Macken, C.; Kawaoka, Y. Phylogenetic characterization of H5N1 avian influenza viruses isolated in Indonesia from 2003–2007. Virology 2009, 390, 13–21. [Google Scholar] [CrossRef]
- Guan, Y.; Smith, G.J.D.; Webby, R.; Webster, R.G. Molecular epidemiology of H5N1 avian influenza. Rev. Sci. Et Tech. Off. Int. Des Epizoot. 2009, 28, 39–47. [Google Scholar] [CrossRef]
- Guan, Y.; Peiris, J.S.M.; Lipatov, A.S.; Ellis, T.M.; Dyrting, K.C.; Krauss, S.; Zhang, L.J.; Webster, R.G.; Shortridge, K.F. Emergence of multiple genotypes of H5N1 avian influenza viruses in Hong Kong SAR. Proc. Natl. Acad. Sci. USA 2002, 99, 8950–8955. [Google Scholar] [CrossRef]
- Kilpatrick, A.M.; Chmura, A.A.; Gibbons, D.W.; Fleischer, R.C.; Marra, P.P.; Daszak, P. Predicting the global spread of H5N1 avian influenza. Proc. Natl. Acad. Sci. USA 2006, 103, 19368–19373. [Google Scholar] [CrossRef] [PubMed]
- Chen, R.; Rubing, C.; Holmes, E.C. The Evolutionary Dynamics of Human Influenza B Virus. J. Mol. Evol. 2008, 66, 655–663. [Google Scholar] [CrossRef] [PubMed]
- McCullers, J.A.; Wang, G.C.; He, S.; Webster, R.G. Reassortment and insertion-deletion are strategies for the evolution of influenza B viruses in nature. J. Virol. 1999, 73, 7343–7348. [Google Scholar] [CrossRef] [PubMed]
- Dudas, G.; Bedford, T.; Lycett, S.; Rambaut, A. Reassortment between Influenza B Lineages and the Emergence of a Coadapted PB1–PB2–HA Gene Complex. Mol. Biol. Evol. 2015, 32, 162–172. [Google Scholar] [CrossRef] [PubMed]
- Klotz, L.C.; Sylvester, E.J. The consequences of a lab escape of a potential pandemic pathogen. Front. Public Health 2014, 2, 116. [Google Scholar] [CrossRef]
- Furmanski, M. Lab Escapes and “Self-Fulfilling Prophecy” Epidemics. Center for Arms Control and Nonproliferation. 2014. Available online: http://armscontrolcenter.org/Escaped_Viruses-final_2-17-14.pdf (accessed on 1 May 2023).
- Furmanski, M. Threatened Pandemics and Lab Escapes: Self-Fulfilling Prophecies. Bull. Atom. Sci. 2014. Available online: http://thebulletin.org/threatened-pandemics-and-laboratory-escapes-self-fulfilling-prophecies7016 (accessed on 1 May 2023).
- Furmanski, M. The 1977 H1N1 influenza virus reemergence demonstrated gain-of-function hazards. mBio 2015, 6, 1. [Google Scholar] [CrossRef]
- U.S. Department of Defense 2015. Review Committee Report: Inadvertent Shipment of Live Bacillus Anthracis Spores by DoD. Available online: http://www.defense.gov/Portals/1/features/2015/0615_lab-stats/Review-Committee-Report-Final.pdf (accessed on 1 May 2023).
- Kaiser, J. Escape of dangerous bacterium leads to halt of risky studies at Tulane. Sci. News. 2015. [Google Scholar] [CrossRef]
- Rozo, M.; Gronvall, G.K. The reemergent 1977 H1N1 strain and the gain-of-function debate. MBio 2015, 6, e01013-15. [Google Scholar] [CrossRef]
- Trifonov, V.; Khiabanian, H.; Rabadan, R. Geographic dependence, surveillance, and origins of the 2009 influenza A (H1N1) virus. N. Engl. J. Med. 2009, 361, 115–119. [Google Scholar] [CrossRef]
- McNeil, D.G. Swine Flu Not an Accident from a Lab, W.H.O. New York Times. 2009, A12. New York, NY, USA. 2009. Available online: http://www.nytimes.com/2009/05/15/health/policy/15flu.html?_r=0 (accessed on 1 May 2023).
- Bucasas, K.L.; Franco, L.M.; Shaw, C.A.; Bray, M.S.; Wells, J.M.; Niño, D.; Arden, N.; Quarles, J.M.; Couch, R.B.; Belmont, J.W. Early patterns of gene expression correlate with the humoral immune response to influenza vaccination in humans. J. Infect. Dis. 2011, 203, 921–929. [Google Scholar] [CrossRef]
- Nakaya, H.I.; Wrammert, J.; Lee, E.K.; Racioppi, L.; Marie-Kunze, S.; Haining, W.N.; Means, A.R.; Kasturi, S.P.; Khan, N.; Li, G.M.; et al. Systems biology of vaccination for seasonal influenza in humans. Nat. Immunol. 2011, 12, 786–795. [Google Scholar] [CrossRef] [PubMed]
- Gomez Lorenzo, M.M.; Fenton, M.J. Immunobiology of influenza vaccines. Chest 2013, 143, 502–510. [Google Scholar] [CrossRef] [PubMed]
- Zaas, A.K.; Chen, M.; Varkey, J.; Veldman, T.; Hero, A.O., 3rd; Lucas, J.; Huang, Y.; Turner, R.; Gilbert, A.; Lambkin-Williams, R.; et al. Gene expression signatures diagnose influenza and other symptomatic respiratory viral infections in humans. Cell Host Microbe 2009, 6, 207–217. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, C.M.; Chang, J.; Lucas, J.E.; Nevins, J.R.; Wang, Q.; West, M. High-Dimensional Sparse Factor Modeling: Applications in Gene Expression Genomics. J. Am. Stat. Assoc. 2008, 103, 1438–1456. [Google Scholar] [CrossRef] [PubMed]
- Lambert, L.C.; Fauci, A.S. Influenza Vaccines for the Future. N. Engl. J. Med. 2010, 363, 2036–2044. [Google Scholar] [CrossRef]
- Yu, X.; Tsibane, T.; McGraw, P.A.; House, F.S.; Keefer, C.J.; Hicar, M.D.; Tumpey, T.M.; Pappas, C.; Perrone, L.A.; Martinez, O.; et al. Neutralizing antibodies derived from the B cells of 1918 influenza pandemic survivors. Nature 2008, 455, 532–536. [Google Scholar] [CrossRef]
- Hensley, S.E.; Das, S.R.; Bailey, A.L.; Schmidt, L.M.; Hickman, H.D.; Jayaraman, A.; Viswanathan, K.; Raman, R.; Sasisekharan, R.; Bennink, J.R.; et al. Hemagglutinin receptor binding avidity drives influenza A virus antigenic drift. Science 2009, 326, 734–736. [Google Scholar] [CrossRef]
- Skountzou, I.; Koutsonanos, D.G.; Kim, J.H.; Powers, R.; Satyabhama, L.; Masseoud, F.; Weldon, W.C.; Martin Mdel, P.; Mittler, R.S.; Compans, R.; et al. Immunity to pre-1950 H1N1 influenza viruses confers cross-protection against the pandemic swine-origin 2009 A (H1N1) influenza virus. J. Immunol. 2010, 185, 1642–1649. [Google Scholar] [CrossRef]
- Xu, R.; Ekiert, D.C.; Krause, J.C.; Hai, R.; Crowe, J.E., Jr.; Wilson, I.A. Structural basis of preexisting immunity to the 2009 H1N1 pandemic influenza virus. Science 2010, 328, 357–360. [Google Scholar] [CrossRef]
- Han, T.; Marasco, W.A. Structural basis of influenza virus neutralization. Ann. N. Y. Acad. Sci. 2011, 1217, 178–190. [Google Scholar] [CrossRef]
- Mossad, S.B. 2008–2009 Influenza update: A better vaccine match. Clevel. Clin. J. Med. 2008, 75, 865–870. [Google Scholar] [CrossRef]
- Shao, W.; Li, X.; Goraya, M.U.; Wang, S.; Chen, J.L. Evolution of Influenza A Virus by Mutation and Re-Assortment. Int. J. Mol. Sci. 2017, 18, 1650. [Google Scholar] [CrossRef]
- Vergaraalert, J.; Busquets, N.; Ballester, M.; Chaves, A.J.; Rivas, R.; Dolz, R.; Wang, Z.; Pleschka, S.; Majó, N.; Rodríguez, F. The NS segment of H5N1 avian influenza viruses (AIV) enhances the virulence of an H7N1 AIV in chickens. Vet. Res. 2014, 45, 7. [Google Scholar] [CrossRef]
- Erbelding, E.J.; Post, D.J.; Stemmy, E.J.; Roberts, P.C.; Augustine, A.D.; Ferguson, S.; Paules, C.I.; Graham, B.S.; Fauci, A.S. A universal influenza vaccine: The strategic plan for the National Institute of Allergy and Infectious Diseases. J. Infect. Dis. 2018, 218, 347–354. [Google Scholar] [CrossRef]
- Chen, J.R.; Liu, Y.M.; Tseng, Y.C.; Ma, C. Better influenza vaccines: An industry perspective. J. Biomed. Sci. 2020, 27, 33. [Google Scholar] [CrossRef]
- World Health Organization (WHO). WHO Preferred Product Characteristics for Next Generation Influenza Vaccines. Available online: https://www.who.int/publications/i/item/9789241512466 (accessed on 1 May 2017).
- Krammer, F. The human antibody response to influenza A virus infection and vaccination. Nat. Rev. Immunol. 2019, 19, 383–397. [Google Scholar] [CrossRef]
- Dormitzer, P.R. Rapid production of synthetic influenza vaccines. Curr. Top. Microbiol. Immunol. 2015, 386, 237–273. [Google Scholar] [CrossRef]
- Deering, R.P.; Kommareddy, S.; Ulmer, J.B.; Brito, L.A.; Geall, A.J. Nucleic acid vaccines: Prospects for non-viral delivery of mRNA vaccines. Expert. Opin. Drug. Deliv. 2014, 11, 885–899. [Google Scholar] [CrossRef]
- Oberemok, V.V.; Laikova, K.V.; Yurchenko, K.A.; Marochkin, N.A.; Fomochkina, I.I.; Kubyshkin, A.V. SARS-CoV-2 will constantly sweep its tracks: A vaccine containing CpG motifs in ‘lasso’ for the multi-faced virus. Inflamm. Res. 2020, 69, 801–812. [Google Scholar] [CrossRef]
- Kenney, R.T.; Cross, A.S. Adjuvants for the future. In New Generation Vaccines; Levine, M.M., Dougan, G., Good, M.F., Liu, M.A., Nabel, G.J., Nataro, J.P., Rappuoli, R., Eds.; Informa Healthcare USA, Inc.: New York, NY, USA, 2010; pp. 250–262. [Google Scholar]
- Oberemok, V.V.; Andreeva, O.A.; Laikova, K.V.; Novikov, I.A.; Kubyshkin, A.V. Post-genomic platform for development of oligonucleotide vaccines against RNA viruses: Diamond cuts diamond. Inflamm. Res. 2022, 71, 729–739. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).