A New DNA Repair-Related Platform for Pharmaceutical Outlook in Cancer Therapies: Ultrashort Single-Stranded Polynucleotides
Abstract
1. Introduction
2. Materials and Methods
2.1. Reagents and Disposal Materials
2.2. Ligands
2.3. Enzymes
2.4. Enzyme Activity Estimation
2.5. DNA and Protein Measurements
2.6. Radioactivity Measurements
2.7. Ligand–Enzyme Binding Measurements
2.8. Statistics
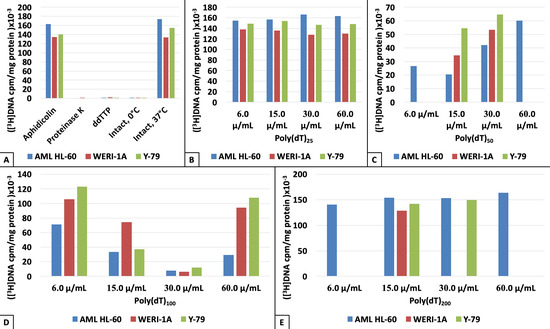
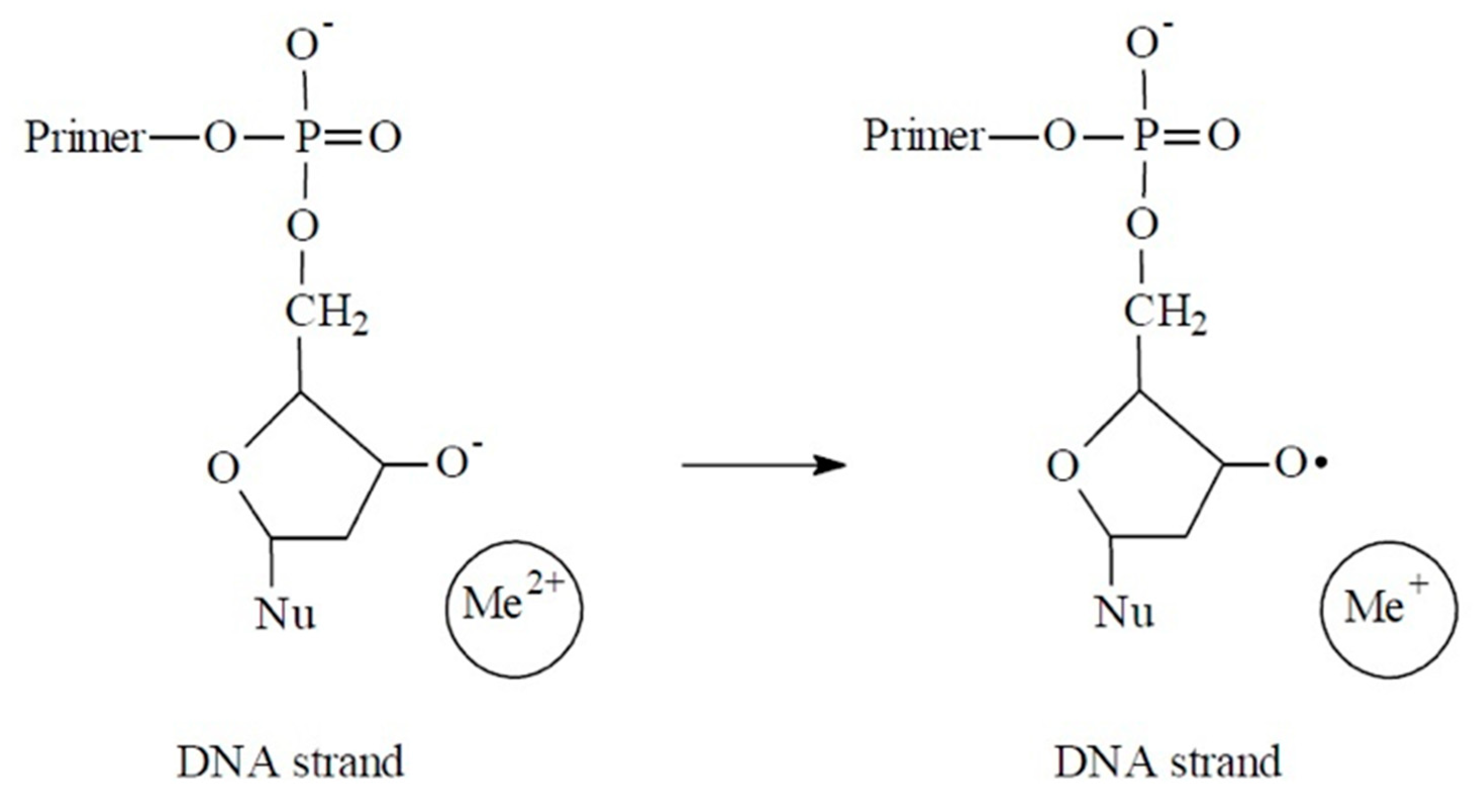
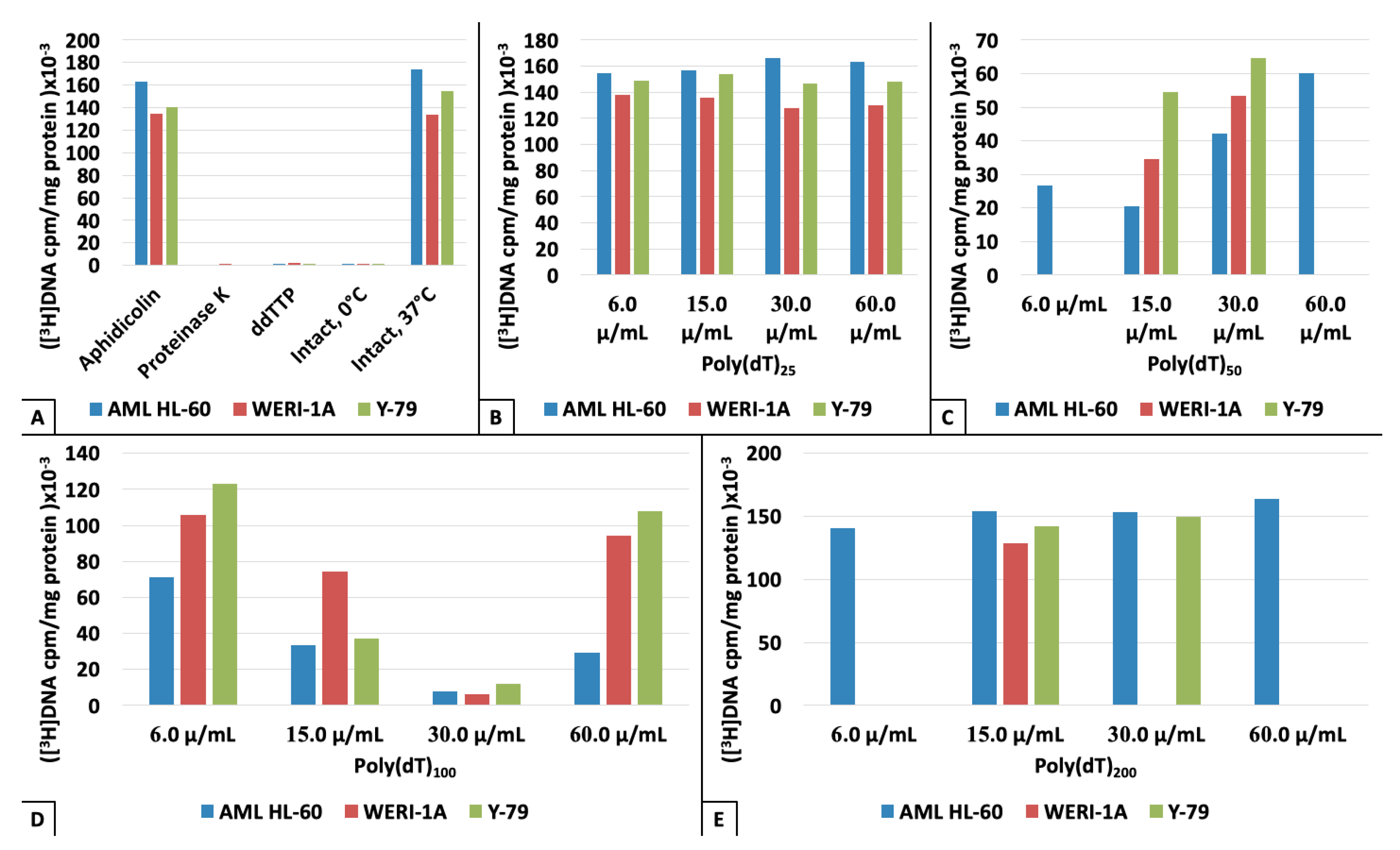
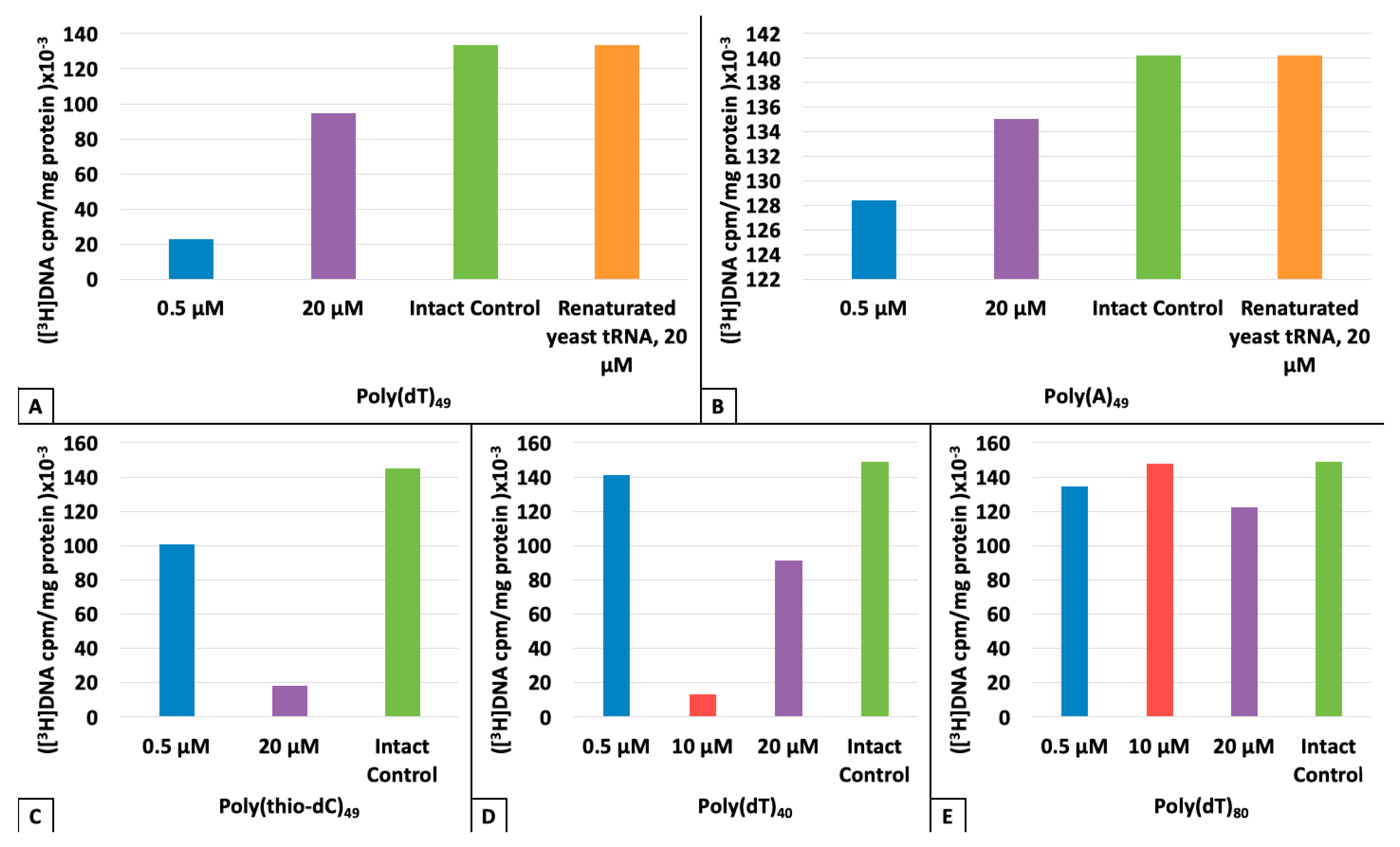
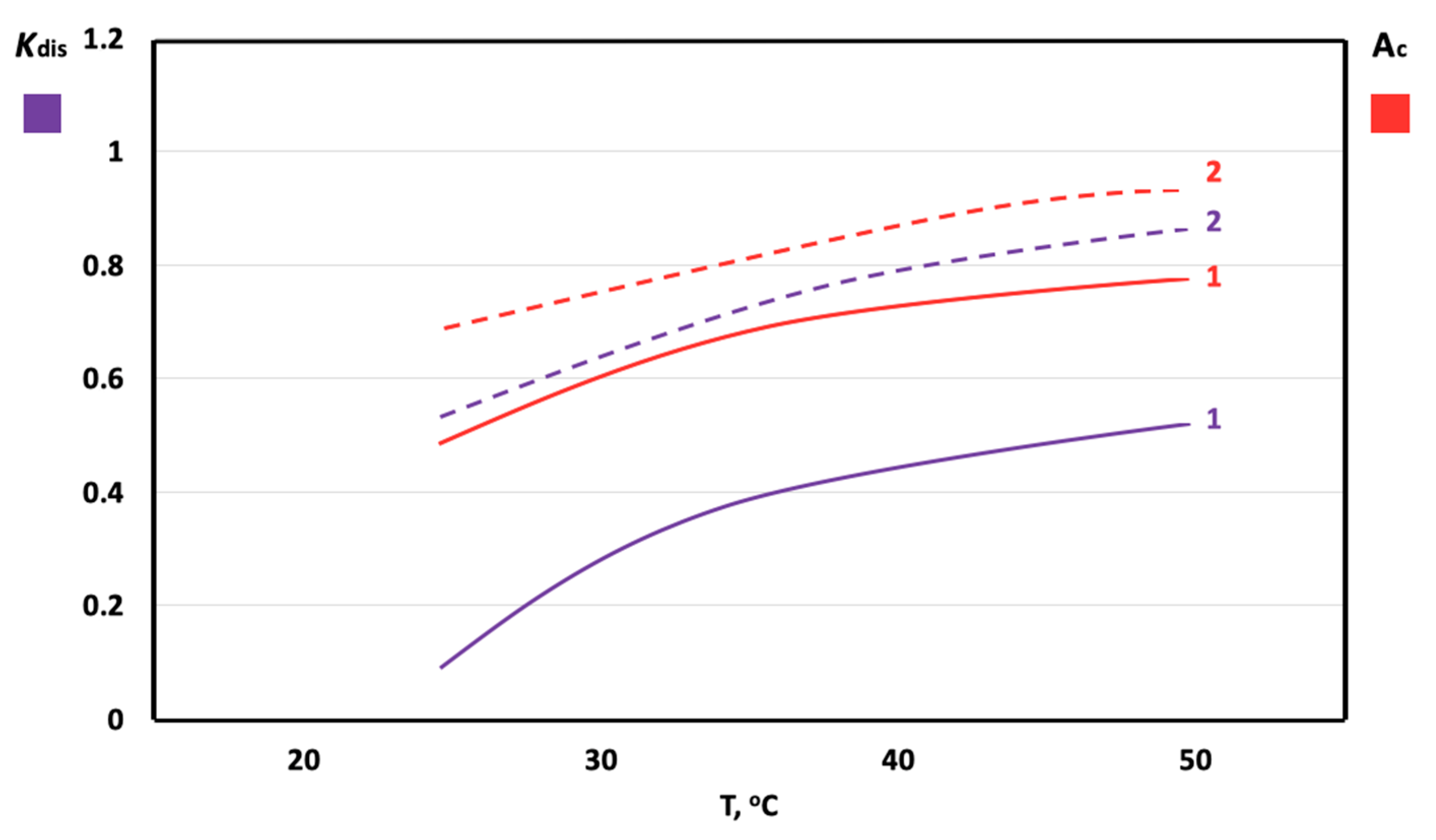
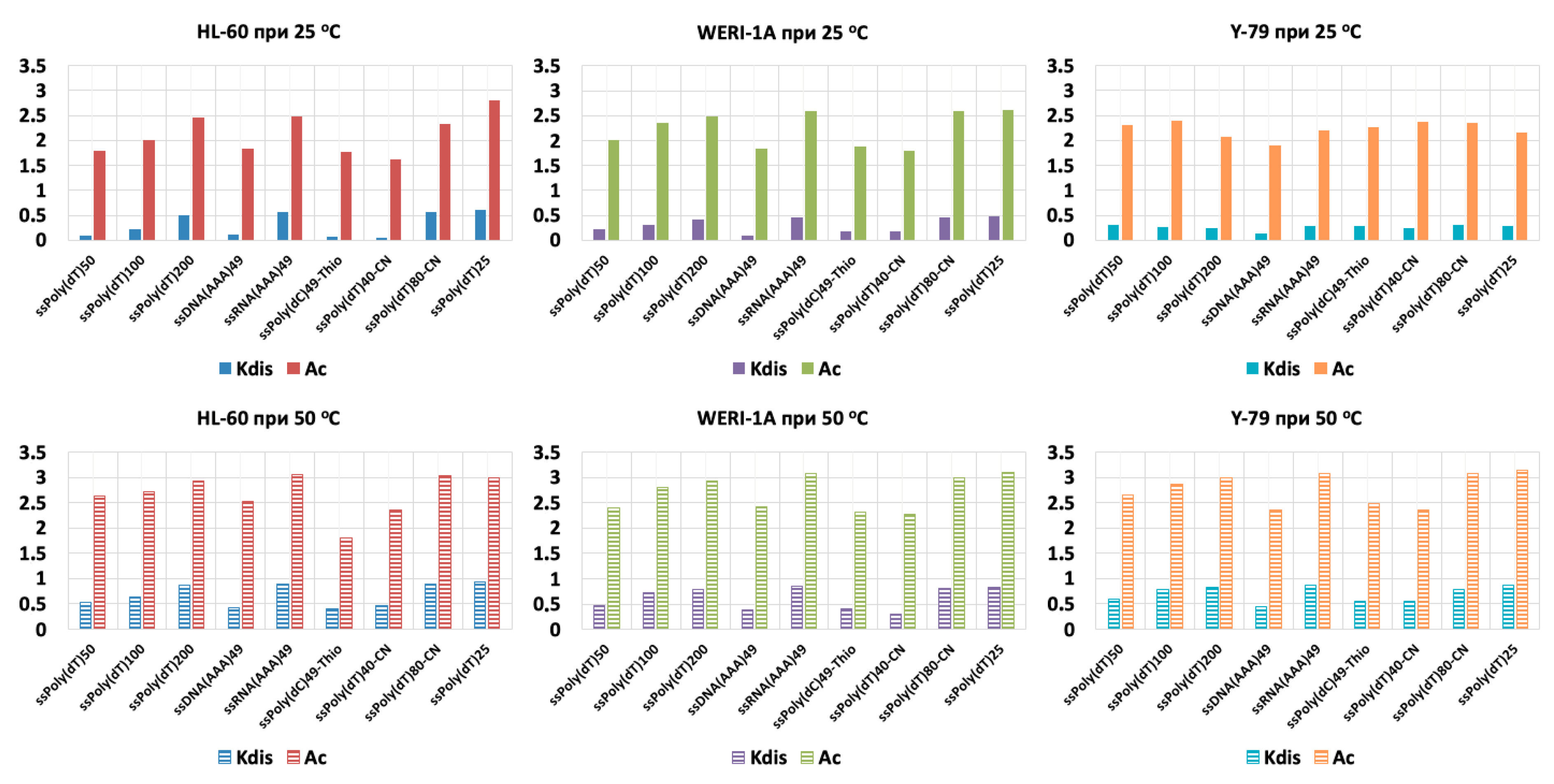
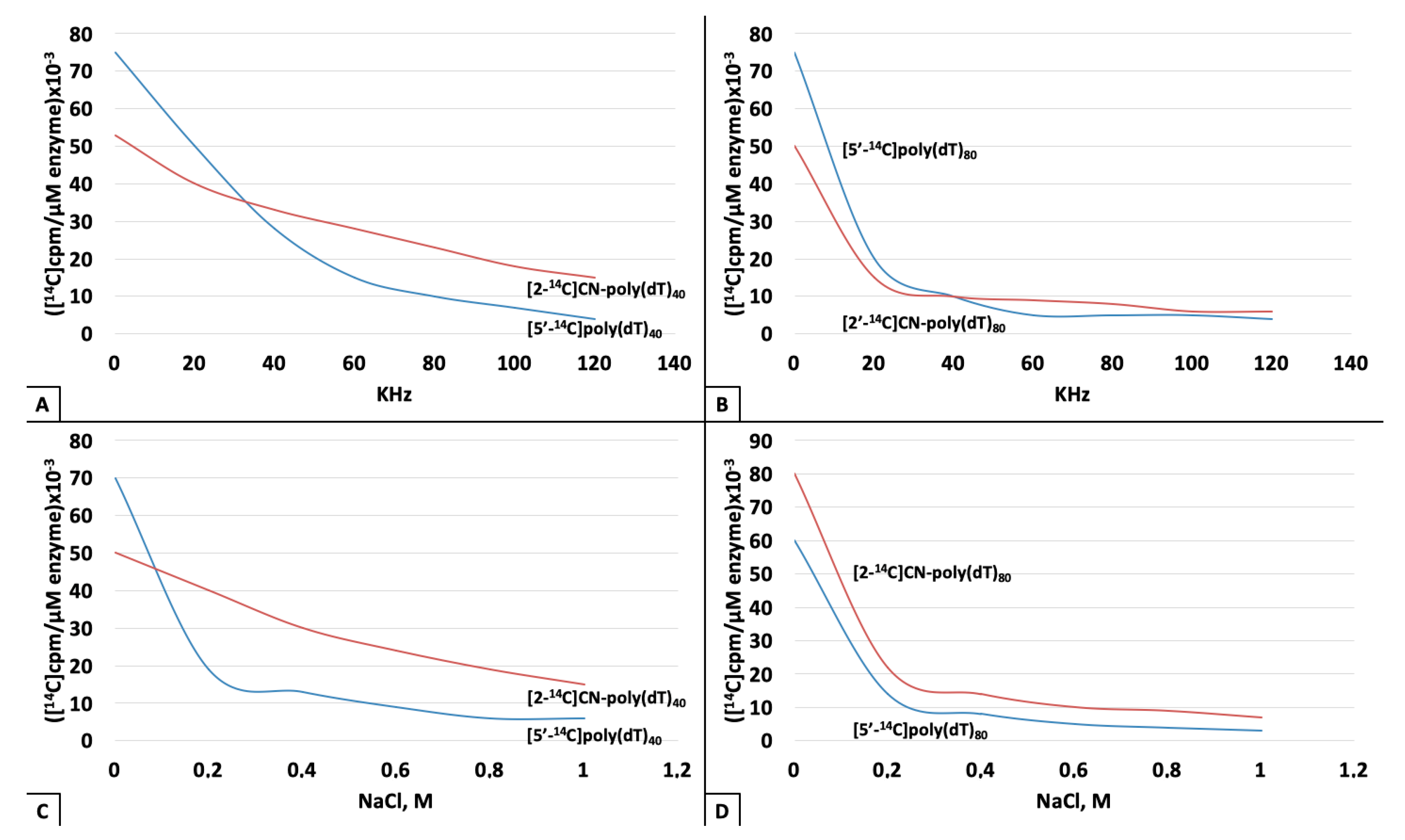
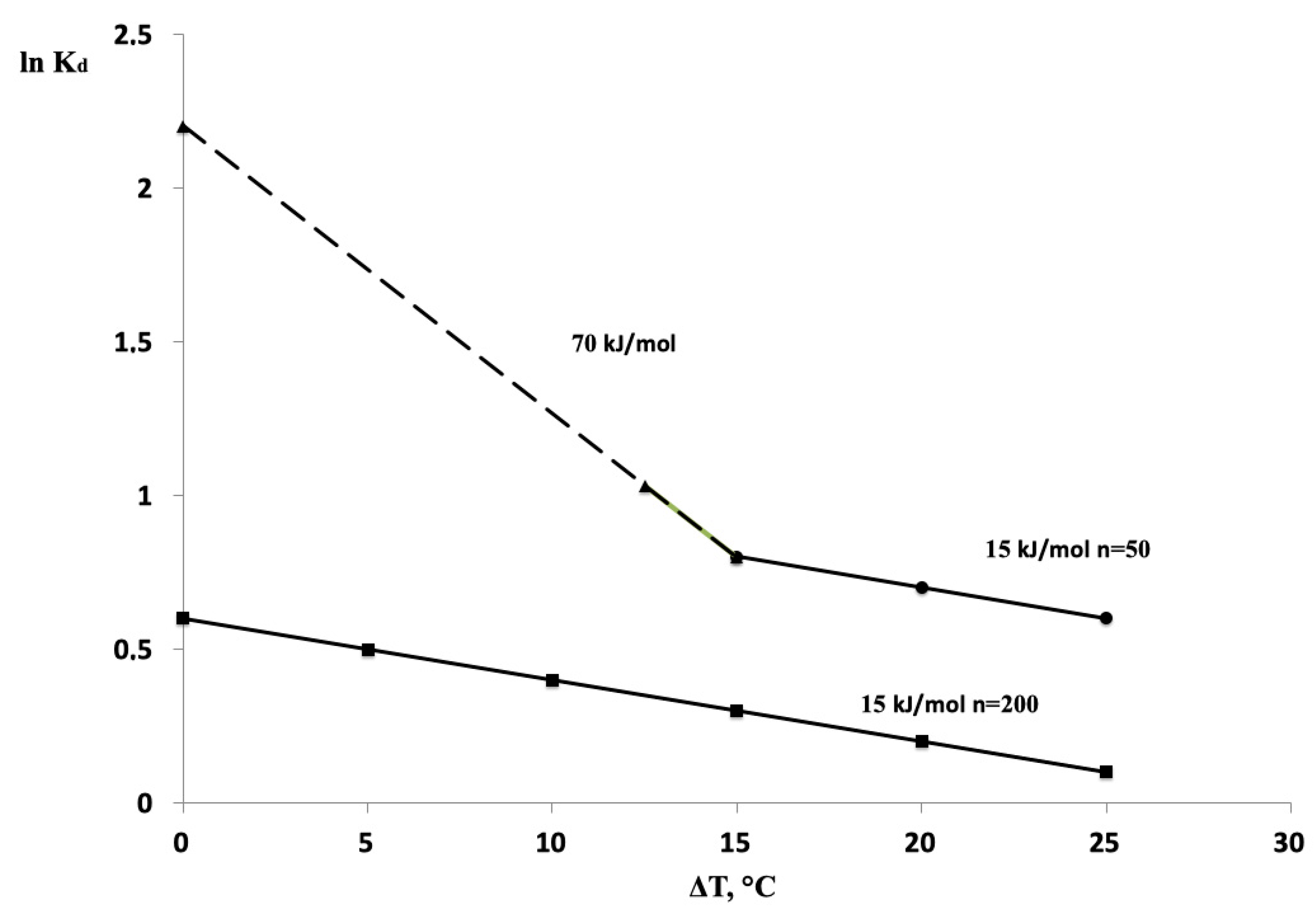
3. Results
4. Discussion
5. Conclusions
Author Contributions
Acknowledgments
Conflicts of Interest
Abbreviations
| ssDNA | single-stranded DNA |
| DNApolβ | DNA polymerase beta |
| PDRN | polydeoxyribonucleotides |
| MIE | magnetic isotope effects |
| AML | acute mueloid leukemia |
| RB | retinoblastoma |
References
- Buchachenko, A.L.; Bukhvostov, A.A.; Ermakov, K.V.; Kuznetsov, D.A. Nuclear spin selectivity in enzymatic catalysis. A caution for applied biophysics. Arch. Biochem. Biophys. 2019, 667, 30–35. [Google Scholar] [CrossRef] [PubMed]
- Bukhvostov, A.A.; Dvornikov, A.S.; Ermakov, K.V.; Kurapov, P.B.; Kyznetsov, D.A. Retinoblastoma: Magnetic isotope effects might make a difference in the current anti-cancer research strategy. Acta Medica (Hradec Kralove) 2017, 60, 93–96. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Bukhvostov, A.A.; Dvornikov, A.S.; Ermakov, K.V.; Kuznetsov, D.A. Retinoblastoma case: Shall we get a paramagnetic trend in chemotherapy? Arch. Cancer Res. 2017, 5, 158–162. [Google Scholar] [CrossRef]
- Chen, S.J. Site-specific binding of non-site-specific ions. Biophys. J. 2019, 116, 38–46. [Google Scholar] [CrossRef]
- Muller, W.E.; Obermeyer, J.; Totsuka, A. Influence of template inactivators on the binding of DNA polymerases to DNA. Nucl. Acids Res. 1974, 1, 63–69. [Google Scholar] [CrossRef]
- Corces, M.R.; Granja, J.M.; Shams, S.; Louie, B.H.; Seoane, J.A.; Zhou, W.; Silva, T.C.; Chang, H.V. The chromatin accessibility landscape of primary human cancers. Science 2018, 362, 16–28. [Google Scholar] [CrossRef]
- Ashley, J.S.; Kronenberg, G.H.; Lipman, K.R.; Bielka, H. Coupling the DNA turnover enzymes. Poly(dNMP) ligands. In Methods in Macromolecular Docking Research; Pitot, H., Van Nijemen, J.T., Eds.; University of Ghent Press: Ghent-Antwerp, Belgium, 2019; pp. 216–229. [Google Scholar]
- Turgeon, M.O.; Perry, N.J.S.; Poulogiannis, G. DNA damage, repair and cancer metabolism. Front. Oncol. 2018, 8, 15–22. [Google Scholar] [CrossRef]
- Breitenbach, M.; Hoffmann, J. Cancer Models. Front. Oncol. 2018, 8, 401–419. [Google Scholar] [CrossRef]
- Jain, R.; Aggarwal, A.K.; Rechkoblit, O. Eukaryotic DNA polymerases. Curr. Opinion Struct. Biol. 2018, 53, 77–87. [Google Scholar] [CrossRef]
- Bukhvostov, A.A.; Shatalov, O.A.; Orlov, A.P.; Kuznetsov, D.A. An atypical DNA polymerase beta overexpressed in human AML/HL-60 malignant cells. J. Cancer Sci. Therap. 2013, 5, 94–99. [Google Scholar] [CrossRef]
- Katoh, R. Analytical Techniques in Biochemistry and Molecular Biology; Springer Publ GmbH: Berlin/Heidelberg, Germany, 2011. [Google Scholar]
- Dewar, J.M.; Lyndall, D. Simple non-radioactive measurement of single-stranded DNA. In DNA Repair Protocols (Methods in Molecular Biology); Bjergbaek, L., Ed.; Humana Press, Inc.: Totowa, NJ, USA, 2012; Volume 920, pp. 341–349. [Google Scholar]
- Fukami, T.; Uchiyama, K.; Yoshimura, Y. Ultramicroanalysis by use of light-scanning photoacoustic densitometry of electrophoresed protein. Analyt. Biochem. 1996, 238, 60–67. [Google Scholar] [CrossRef] [PubMed]
- Rastogi, C.; Rube, H.T.; Kribelbauer, J.F.; Crocker, J.; Locker, R.E.; Martini, G.D.; Laptenko, O. Accurate and sensitive quantification of protein-DNA binding affinity. Proc. Nat. Acad. Sci. USA 2018, 115, E3692–E3701. [Google Scholar] [CrossRef] [PubMed]
- Laurent, C.; Neurath, K.J.; Baglioni, G.; Sieliwanowicz, B. Advances in multiwave UV-spectrophotometry: RNP/DNP integrity studies. In Physics of Biopolymers; Lindsey, A.L., Cohen, K., Eds.; Adler & Adler Publisher Inc.: Perth, Australia, 2019; pp. 41–56. [Google Scholar]
- Chowdhury, N.; Bagchi, A. An overview of DNA-protein Interactions. Curr. Chem. Biol. 2015, 9, 71–83. [Google Scholar] [CrossRef]
- Telashima, Y. Revival of Oligonucleotide Therapy Development; KOKEN Publ.: Tokyo-Nagoya, Japan, 2019. [Google Scholar]
- Byron, W.M.; Hollander, B.; Hollander, M. Statistics. A Biomedical Introduction; Wiley-Blackwell Publishers Inc.: Oxford, UK; Weinheim, Germany; Hoboken, NJ, USA, 2007. [Google Scholar]
- Ueda, S.; Watanabe, I.; Taichi, K. Nucleic Acid-Based Drugs; Sumitomo Chemical Co. Ltd.: Utajima-Osaka, Japan, 2019. [Google Scholar]
- Lavrik, I.N. Systems biology of death receptor networks: Live and let die. Cell Death Dis. 2014, 5, e1259–e1266. [Google Scholar] [CrossRef] [PubMed]
- Peterson, L.E. Small molecule docking of DNA repair proteins associated with cancer survival following PCNA metagene adjustment. A potential novel class of repair inhibitors. Molecules 2019, 24, 645. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Lu, J.; Kang, T. Human single-stranded DNA binding proteins: Guardians of genome stability. Acta Biochimica et Biophysica Sinica. 2016, 48, 671–677. [Google Scholar] [CrossRef]
- Finkelsten, A.V.; Ptitsyn, O.V. Physics of Proteins; Nova Science Publishers Inc.: Princeton, NJ, USA, 2012. [Google Scholar]
- Buchachenko, A.L. Magneto-Biology and Medicine; Nova Science Publishers Inc.: Princeton, NY, USA, 2015. [Google Scholar]
- Morten, M.J.; Gamsjaejer, R.; Cubeddu, L.; Kariawasam, R.; Peregrina, J.; Penedo, J.C.; White, M.F. High affinity RNA binding by a hyperthermophilic single-stranded DNA-binding protein. Extremophiles 2017, 21, 369–379. [Google Scholar] [CrossRef]
- Lyu, J.; Wang, S.; Balius, T.E.; Singh, I.; Levit, A.; Moroz, Y.S.; O’Meara, M.J. Ultra-large library docking for discovering new chemotypes. Nature 2019, 566, 224–229. [Google Scholar] [CrossRef]
- Udvardi, L.; Lakatos, S. Aptamers. Towards the Pharmacological Breakthrough; Alba Regia: Szegen-Budapest, Hungary, 2019. [Google Scholar]
- Zavyalova, E.; Samoylenkova, N.; Revishchin, A.; Turashev, A.; Gordeychuk, I.; Golovin, A.; Kopylov, A.; Pavlova, G. The evaluation of pharmacodynamics and pharmacokinetics of anti-thrombin DNA aptamer RA-36. Front Pharmacol. 2017, 8, 922–930. [Google Scholar] [CrossRef]
- Squadrito, F.; Bitto, A.; Irrera, N.; Pizzino, G.; Pallio, G.; Minutoli, L.; Altavilla, D. Pharmacological activity and clinical use of PDRN (polydeoxyribonucleotides). Front. Pharmacol. 2017, 8, 224–233. [Google Scholar] [CrossRef]
- Ashton, N.W.; Bolderson, E.; Cubeddu, L.; O’Byrne, K.J.; Richard, D.J. Human single-stranded DNA binding proteins are essential for maintaining genomic stability. BMC Mol. Biol. 2013, 14, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Bozic, I.; Nowak, M.A. Resisting resistance. Ann. Rev. Cancer Biol. 2017, 1, 203–221. [Google Scholar] [CrossRef][Green Version]
- Ansari, A.S.; Santerre, P.J.; Uludag, H. Biomaterials for polynucleotide delivery to anchorage-independent cells. J. Mater. Chem. B. 2017, 5, 7238–7261. [Google Scholar] [CrossRef]
- Svistunov, A.A.; Napolov, Y.K.; Bukhvostov, A.A.; Shatalov, O.A.; Alayutdin, R.N.; Kuznetsov, D.A. The mitochondria free iron content to limit an isotope effect of 25Mg2+ in ATP synthesis: A caution. Cell Biochem. Biophys. 2013, 66, 417–419. [Google Scholar] [CrossRef]
- Mouliere, F.; Chandrananda, D.; Piskorz, A.M.; Moore, E.K.; Morris, J.; Ahlborn, L.B.; Rosenfeld, N. Enhanced detection of circulating tumor DNA by fragment size analysis. Science Transl. Med. 2018, 10, 117–129. [Google Scholar] [CrossRef]
- Catachura, S.C.; Leys, N.; Mastroleo, J. Quorum sensing in life support systems. In Quorum Sensing; Kalia, V.C., Ed.; Springer Nature, Ltd.: Singapore, 2018; pp. 249–260. [Google Scholar]

© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Stovbun, S.; Ermakov, K.; Bukhvostov, A.; Vedenkin, A.; Kuznetsov, D. A New DNA Repair-Related Platform for Pharmaceutical Outlook in Cancer Therapies: Ultrashort Single-Stranded Polynucleotides. Sci. Pharm. 2019, 87, 25. https://doi.org/10.3390/scipharm87040025
Stovbun S, Ermakov K, Bukhvostov A, Vedenkin A, Kuznetsov D. A New DNA Repair-Related Platform for Pharmaceutical Outlook in Cancer Therapies: Ultrashort Single-Stranded Polynucleotides. Scientia Pharmaceutica. 2019; 87(4):25. https://doi.org/10.3390/scipharm87040025
Chicago/Turabian StyleStovbun, Sergey, Kirill Ermakov, Alexander Bukhvostov, Alexander Vedenkin, and Dmitry Kuznetsov. 2019. "A New DNA Repair-Related Platform for Pharmaceutical Outlook in Cancer Therapies: Ultrashort Single-Stranded Polynucleotides" Scientia Pharmaceutica 87, no. 4: 25. https://doi.org/10.3390/scipharm87040025
APA StyleStovbun, S., Ermakov, K., Bukhvostov, A., Vedenkin, A., & Kuznetsov, D. (2019). A New DNA Repair-Related Platform for Pharmaceutical Outlook in Cancer Therapies: Ultrashort Single-Stranded Polynucleotides. Scientia Pharmaceutica, 87(4), 25. https://doi.org/10.3390/scipharm87040025