Anomalous Separation of Small Y-Chromosomal DNA Fragments on Microchip Electrophoresis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Microchip System
2.2. Samples
- (1)
- Modified sY638: CAGCAG has been modified to be GTCCAG, and it is called: 1-Modified sY638
- (2)
- Modified sY638: TGTG has been modified to be ATTG, and it is called: 2-Modified sY638
- (3)
- Modified sY638: The third modification contains both previous modifications, and it is called: 3-Modified sY638 (Table 1).
3. Results and Discussion
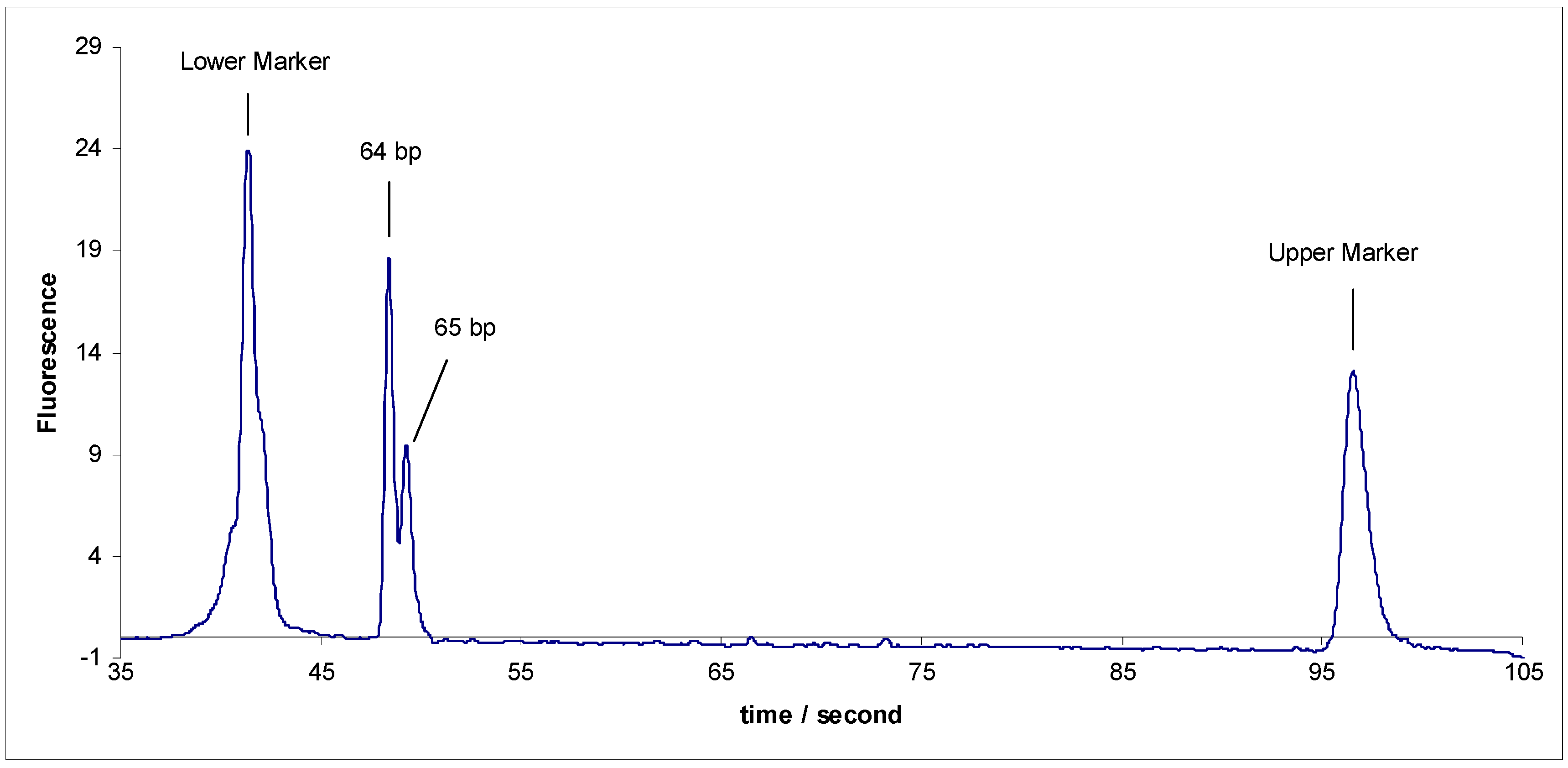
- (1)
- Separation of sY592 (65 bp) from 1-Modified sY638 (64 bp), the separation did not occur.
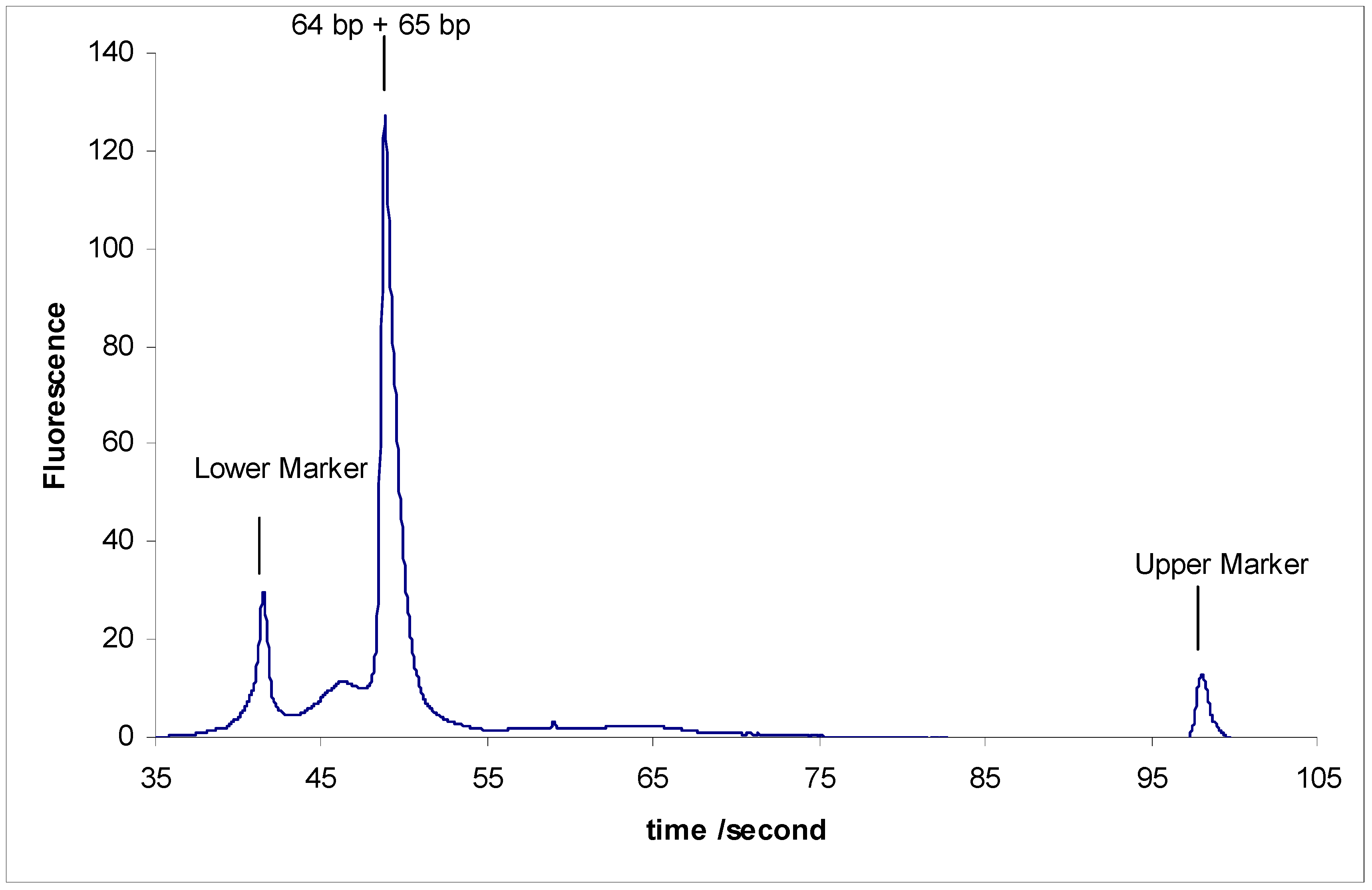
- (2)
- Separation of sY592 (65 bp) from 2-Modified sY638 (64 bp), the separation did not occur.
- (3)
- Separation of sY592 (65 bp) from 3-Modified sY638 (64 bp), the separation did not occur either (Figure 3).
- -
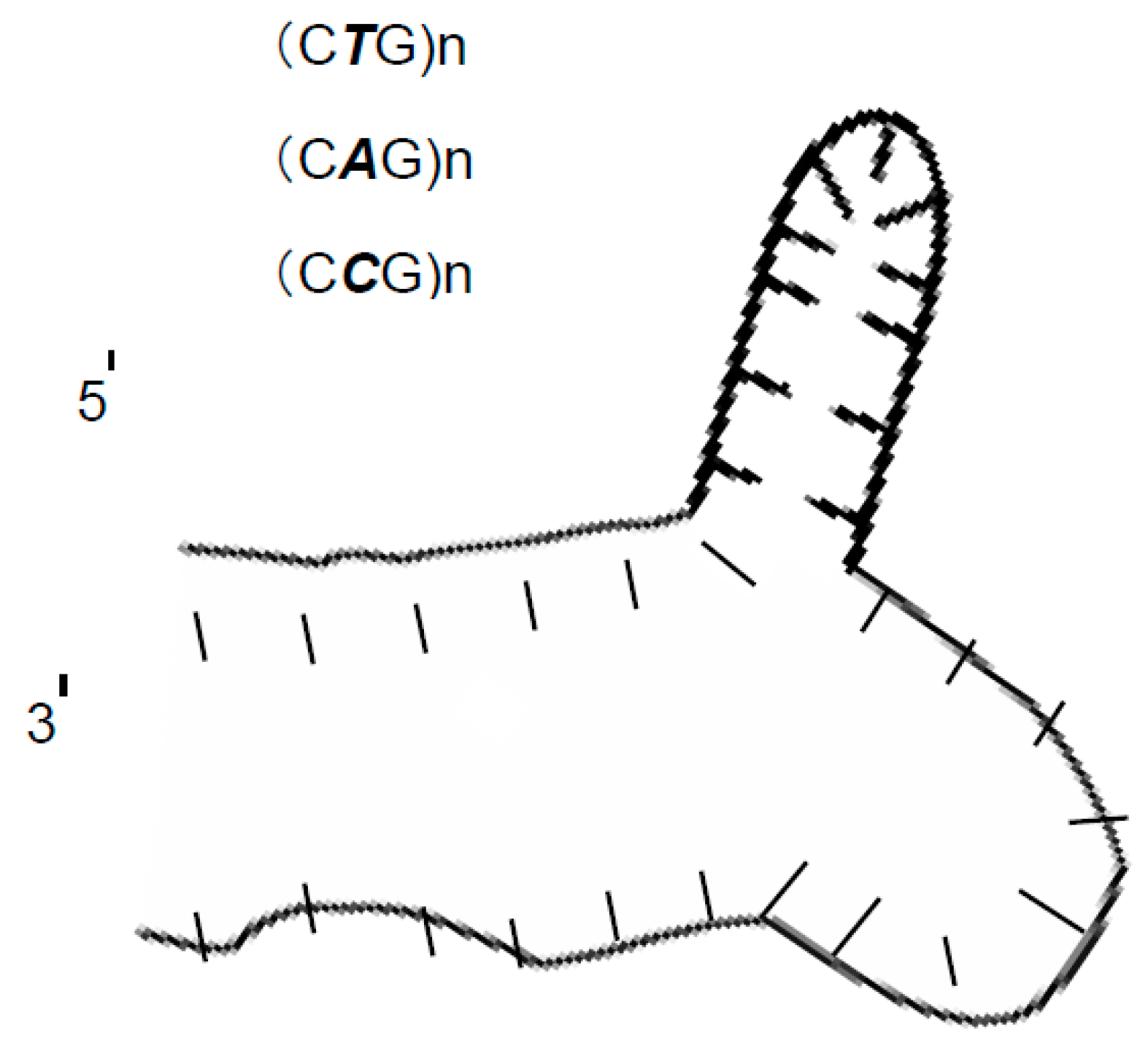
- The inability to separate sY610 (61 bp) from any of the modified fragments of sY638 (64 bp) supports a previous study result that 5 bp is the minimum length difference for separating two DNA marker fragments on the microchip electrophoresis system [21].
- -
- This anomalous separation can be a guiding example for the researcher in the separation field that if any research study resulted in separating two DNA markers with a very limited difference between the two DNA markers This result must not be accepted directly and should be deeply studied by taking into consideration that an anomalous DNA structure might be behind that.
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Balkwill, G.D.; Garner, T.P.; Searle, M.S. Folding of single-stranded DNA quadruplexes containing an autonomously stable mini-hairpin loop. Mol. Biosyst. 2009, 5, 542–547. [Google Scholar] [CrossRef] [PubMed]
- Kawakami, J.; Okabe, S.; Tanabe, Y.; Sugimoto, N. Recognition of a flipped base in a hairpinloop DNA by a small peptide. Nucleosides Nucleotides Nucleic Acids 2008, 27, 292–308. [Google Scholar] [CrossRef] [PubMed]
- Tautz, D.; Renz, M. Simple sequences are ubiquitous repetitive components of eukaryotic genomes. Nucleic Acids Res. 1984, 12, 4127–4138. [Google Scholar] [CrossRef] [PubMed]
- Tautz, D.; Trick, M.; Dover, G. Cryptic simplicity in DNA is a major source of genetic variation. Nature 1986, 322, 652–656. [Google Scholar] [CrossRef] [PubMed]
- Kuznetsov, S.V.; Ren, C.C.; Woodson, S.A.; Ansari, A. Loop dependence of the stability and dynamics of nucleic acid hairpins. Nucleic Acids Res. 2008, 36, 1098–1112. [Google Scholar] [CrossRef] [PubMed]
- Levinson, G.; Gutman, G.A. Slipped-strand mispairing: A major mechanism for DNA sequence evolution. Mol. Biol. Evol. 1987, 4, 203–221. [Google Scholar] [PubMed]
- Varani, G. Exceptionally stable nucleic acid hairpins. Annu. Rev. Biophys. Biomol. Struc. 1995, 24, 379–404. [Google Scholar] [CrossRef] [PubMed]
- Wells, R.D.; Goodman, T.C.; Hillen, W.; Horn, G.T.; Klein, R.D.; Larson, J.E.; Muller, U.R.; Neuendorf, S.K.; Panayotatos, N.; Stirdivant, S.M. DNA structure and gene regulation. Prog. Nucleic Acid Res. Mol. Biol. 1980, 25, 167–267. [Google Scholar]
- Schultz, G.E., Jr.; Drake, J.W. Templated mutagenesis in bacteriophage T4 involving imperfect direct or indirect sequence repeats. Genetics 2008, 178, 661–673. [Google Scholar] [CrossRef] [PubMed]
- Stalker, D.M.; Thomas, C.M.; Helinski, D.R. Nucleotide sequence of the region of the origin of replication of the broad host range plasmid RK2. Mol. Gen. Genet. 1981, 181, 8–12. [Google Scholar] [CrossRef] [PubMed]
- Blommers, M.J.; Walter, J.A.; Haasnoot, C.A.; Aelen, J.M.; van der Marel, G.A.; van Boom, J.H.; Hilber, C.W. Effects of base sequence on the loop folding in DNA hairpins. Biochemistry 1989, 28, 7491–7498. [Google Scholar] [CrossRef] [PubMed]
- Kamashev, D.; Balandina, A.; Mazur, A.K.; Arimondo, P.B.; Rouviere-Yaniv, J. HU binds and folds single-stranded DNA. Nucleic Acids Res. 2008, 36, 1026–1036. [Google Scholar] [CrossRef] [PubMed]
- Blommers, M.J.; van de Ven, F.J.; van der Marel, G.A.; van Boom, J.H.; Hilber, C.W. The three-dimensional structure of a DNA hairpin in solution two-dimensional NMR Studies and structural analysis of d(ATCCTATTTATAGGAT). Eur. J. Biochem. 1991, 201, 33–51. [Google Scholar] [CrossRef] [PubMed]
- Lim, C.S.; Jabrane-Ferrat, N.; Fontes, J.D.; Okamoto, H.; Garovoy, M.R.; Peterlin, B.M.; Hunt, C.A. Sequence-independent inhibition of RNA transcription by DNA dumbbells and other decoys. Nucleic Acids Res. 1997, 25, 575–581. [Google Scholar] [CrossRef] [PubMed]
- Van Dongen, M.J.; Wijmenga, S.S.; van der Marel, G.A.; van Boom, J.H.; Hilbers, C.W. The transition from a neutral-pH double helix to a low-pH triple helix induces a Conformational switch in the CCCG tetraloop closing a Watson-Crick stem. J. Mol. Biol. 1996, 263, 715–729. [Google Scholar] [CrossRef] [PubMed]
- Davison, A.; Leach, D.R. Two-base DNA hairpin-loop structures in vivo. Nucleic Acids Res. 1994, 22, 4361–4363. [Google Scholar] [CrossRef] [PubMed]
- Escaja, N.; Gomez-Pinto, I.; Rico, M.; Pedroso, E.; Gonzalez, C. Structures and stabilities of small DNA dumbbells with Watson-Crick and Hoogsteen base pairs. Chem. Biol. Chem. 2003, 4, 623–632. [Google Scholar] [CrossRef] [PubMed]
- Performance Characteristics of the High Sensitivity DNA Kit for the Agilent 2100 Bioanalyzer System. Available online: http://www.agilent.com/cs/library/technicaloverviews/public/5990-4417EN.pdf (accessed on 2 March 2016).
- Blanco, P.; Shlumukova, M.; Sargent, C.A.; Jobling, M.A.; Affara, N.; Hurles, M.E. Divergent outcomes of intrachromosomal recombination on the human Y chromosome: Male infertility and recurrent polymorphism. J. Med. Genet. 2000, 37, 752–758. [Google Scholar] [CrossRef] [PubMed]
- Jabasini, M.; Zhang, L.; Dang, F.; Xu, F.; Almofti, M.R.; Ewis, A.A.; Lee, J.; Nakahori, Y.; Baba, Y. Analysis of DNA polymorphisms on the human Y-chromosome by microchip electrophoresis. Electrophoresis 2002, 23, 1537–1542. [Google Scholar] [CrossRef]
- Jabasini, M.; Xu, F.; Dang, F.; Shinka, T.; Nakahori, Y.; Baba, Y. Range of separation of potential tool for bioseparation, microchip electrophoresis system, for DNA polymorphisms on the human Y-chromosome. Anal. Sci. 2003, 19, 175–176. [Google Scholar] [CrossRef] [PubMed]
- Nachamkin, I.; Panaro, N.J.; Li, M.; Ung, H.; Yuen, P.K.; Kricka, L.J.; Wilding, P. Agilent 2100 bioanalyzer for restriction fragment length polymorphism analysis of the Campylobacter jejuni flagellin gene. J. Clin. Microbiol. 2001, 39, 754–757. [Google Scholar] [CrossRef] [PubMed]
- Jabasini, M.; Ewis, A.A.; Xu, F.; Ping, G.; Fouad, M.; Shinka, T.; Nakahori, Y.; Ishikawa, M.; Baba, Y. Ultrafast diagnosis of the genetic-related disorders using the combined technologies of multiplex PCR and multichannel microchip electrophoresis. Anal. Sci. 2005, 21, 1537–1539. [Google Scholar] [CrossRef] [PubMed]
- Sinden, R.R. Biological implications of the DNA structures associated with disease-causing triplet repeats. Am. J. Hum. Genet. 1999, 64, 346–353. [Google Scholar] [CrossRef] [PubMed]
- Lah, J.; Drobnak, I.; Dolinar, M.; Vesnaver, G. What drives the binding of minor groove-directed ligands to DNA hairpins. Nucleic Acids Res. 2008, 36, 897–904. [Google Scholar] [CrossRef] [PubMed]
- Johnson, A.T.; Wiest, O. Structure and dynamics of poly(T) single-strand DNA: Implications toward CPD formation. J. Phys. Chem. B 2007, 111, 14398–14404. [Google Scholar] [CrossRef] [PubMed]
- Grzechnik, P.; Tan-Wong, S.M.; Proudfoot, N.J. Terminate and make a loop: Regulation of transcriptional directionality. Treds. Biochem. Sci. 2014, 39, 319–327. [Google Scholar] [CrossRef] [PubMed]
- Baltierra-Jasso, L.E.; Morten, M.J.; Laflör, L.; Quinn, S.D.; Magennis, S.W. Crowding-Induced Hybridization of Single DNA Hairpins. J. Am. Chem. Soc. 2015, 137, 16020–16023. [Google Scholar] [CrossRef] [PubMed]

| The Fragment | The Sequecing |
|---|---|
| SY638 (64 bp) without modification | 5′-GACCACAAGA AAACTGTGAG TGGCTTTCAG AAACTTGAGA AACTGGACCC TATTGCAGCA GATC-3′ |
| SY638 (64 bp) without modification, complementary | 5′-GATCTGCTGCAATA GGGTCCAGTT TCTCAAGTTT CTGAAAGCCA CTCACAGTTTTCTTGTGGTC-3′ |
| 1-Modified SY638 (64 bp) | 5′-GACCACAAGA AAACTGTGAG TGGCTTTCAG AAACTTGAGA AACTGGACCC TATTGGTCCA GATC-3′ |
| 1-Modified SY638 (64 bp) complementary | 5′-GATCTGGACC AATAGGGTCC AGTTTCTCAA GTTTCTGAAA GCCACTCACA GTTTTCTTGT GGTC-3′ |
| 2-Modified SY638 (64 bp) | 5′-GACCACAAGA AAACATTGAG TGGCTTTCAG AAACTTGAGA AACTGGACCC TATTGCAGCA GATC-3′ |
| 2-Modified SY638 (64 bp) complementary | 5′-GATCTGCTGC AATAGGGTCC AGTTTCTCAA GTTTCTGAAA GCCACTCAAT GTTTTCTTGT GGTC-3′ |
| 3-Modified SY638 (64 bp) | 5′-GACCACAAGA AAACATTGAG TGGCTTTCAG AAACTTGAGA AACTGGACCC TATTGGTCCA GATC-3′ |
| 3-Modified SY638 (64 bp) complementary | 5′-GATCTGGACC AATAGGGTCC AGTTTCTCAA GTTTCTGAAA GCCACTCAAT GTTTTCTTGT GGTC-3′ |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jabasini, M.; Ewis, A.; Sato, Y.; Nakahori, Y.; Baba, Y. Anomalous Separation of Small Y-Chromosomal DNA Fragments on Microchip Electrophoresis. Sci. Pharm. 2016, 84, 507-513. https://doi.org/10.3390/scipharm84030507
Jabasini M, Ewis A, Sato Y, Nakahori Y, Baba Y. Anomalous Separation of Small Y-Chromosomal DNA Fragments on Microchip Electrophoresis. Scientia Pharmaceutica. 2016; 84(3):507-513. https://doi.org/10.3390/scipharm84030507
Chicago/Turabian StyleJabasini, Mohammad, Ashraf Ewis, Youichi Sato, Yutaka Nakahori, and Yoshinobu Baba. 2016. "Anomalous Separation of Small Y-Chromosomal DNA Fragments on Microchip Electrophoresis" Scientia Pharmaceutica 84, no. 3: 507-513. https://doi.org/10.3390/scipharm84030507
APA StyleJabasini, M., Ewis, A., Sato, Y., Nakahori, Y., & Baba, Y. (2016). Anomalous Separation of Small Y-Chromosomal DNA Fragments on Microchip Electrophoresis. Scientia Pharmaceutica, 84(3), 507-513. https://doi.org/10.3390/scipharm84030507





