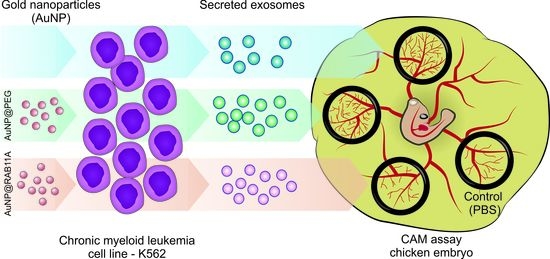
Exploring RAB11A Pathway to Hinder Chronic Myeloid Leukemia-Induced Angiogenesis In Vivo
Abstract
:1. Introduction
2. Materials and Methods
2.1. Preparation and Characterization of Gold Nanoconjugates
2.2. Cell Cultures Maintenance
2.3. RAB11A Silencing in K562 Cell Line
2.4. Cell Viability
2.5. Evaluation of RAB11A Expression in K562 Cell Line by RT-qPCR
2.6. Western-Blot for Evaluation of Rab11a Protein Expression in K562 Cell Line
2.7. Exosomes Isolation
2.8. ELISA for Exosomes Characterization
2.9. Ex-OVO Angiogenesis Assays
2.10. Statistical Analysis
3. Results
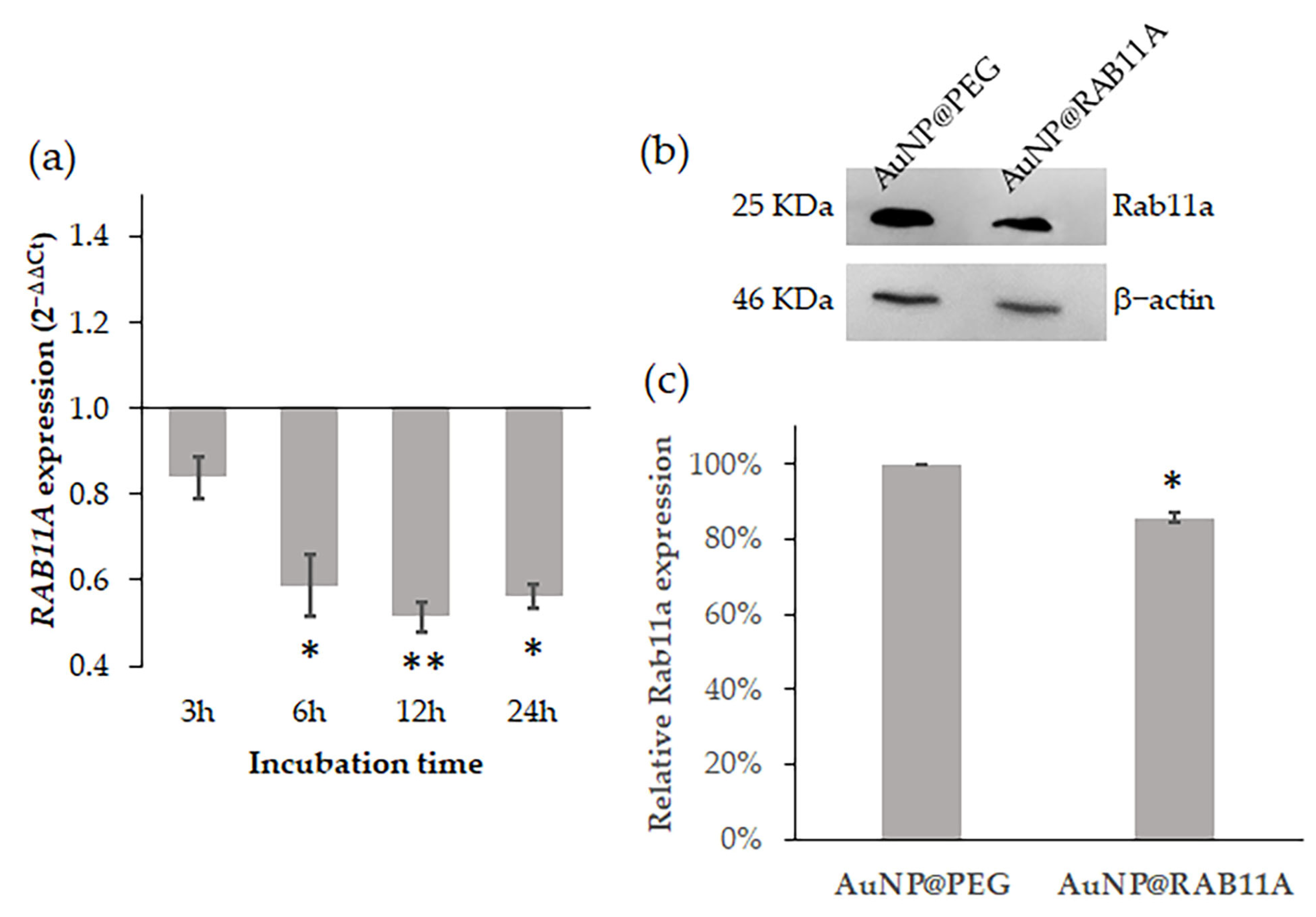
3.1. RAB11A mRNA Silencing with AuNP@RAB11A
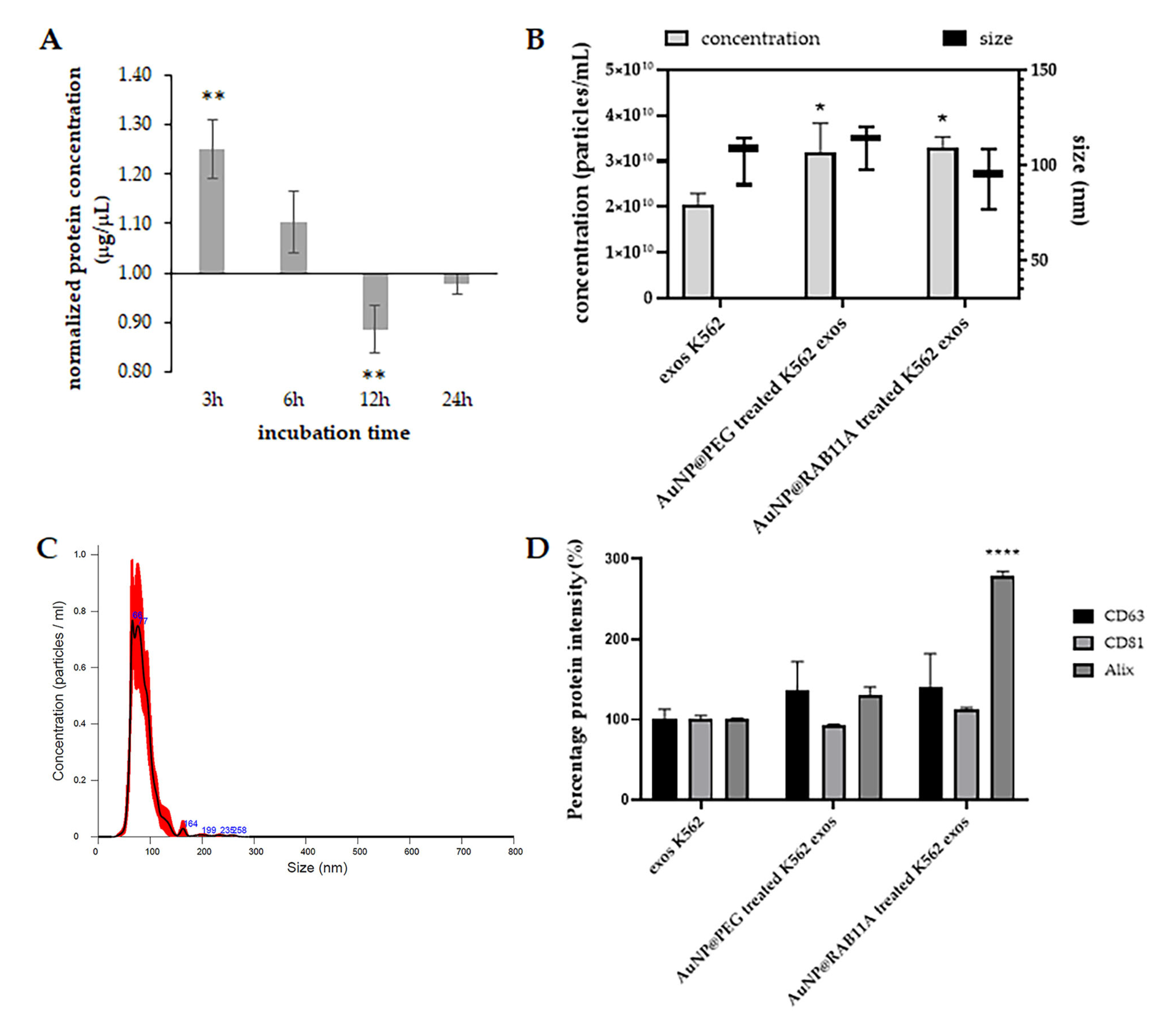
3.2. Exosomes Secreted by K562 Treated with AuNP@RAB11A Are Smaller and Present Different Protein Content than K562 Exosomes Counterparts
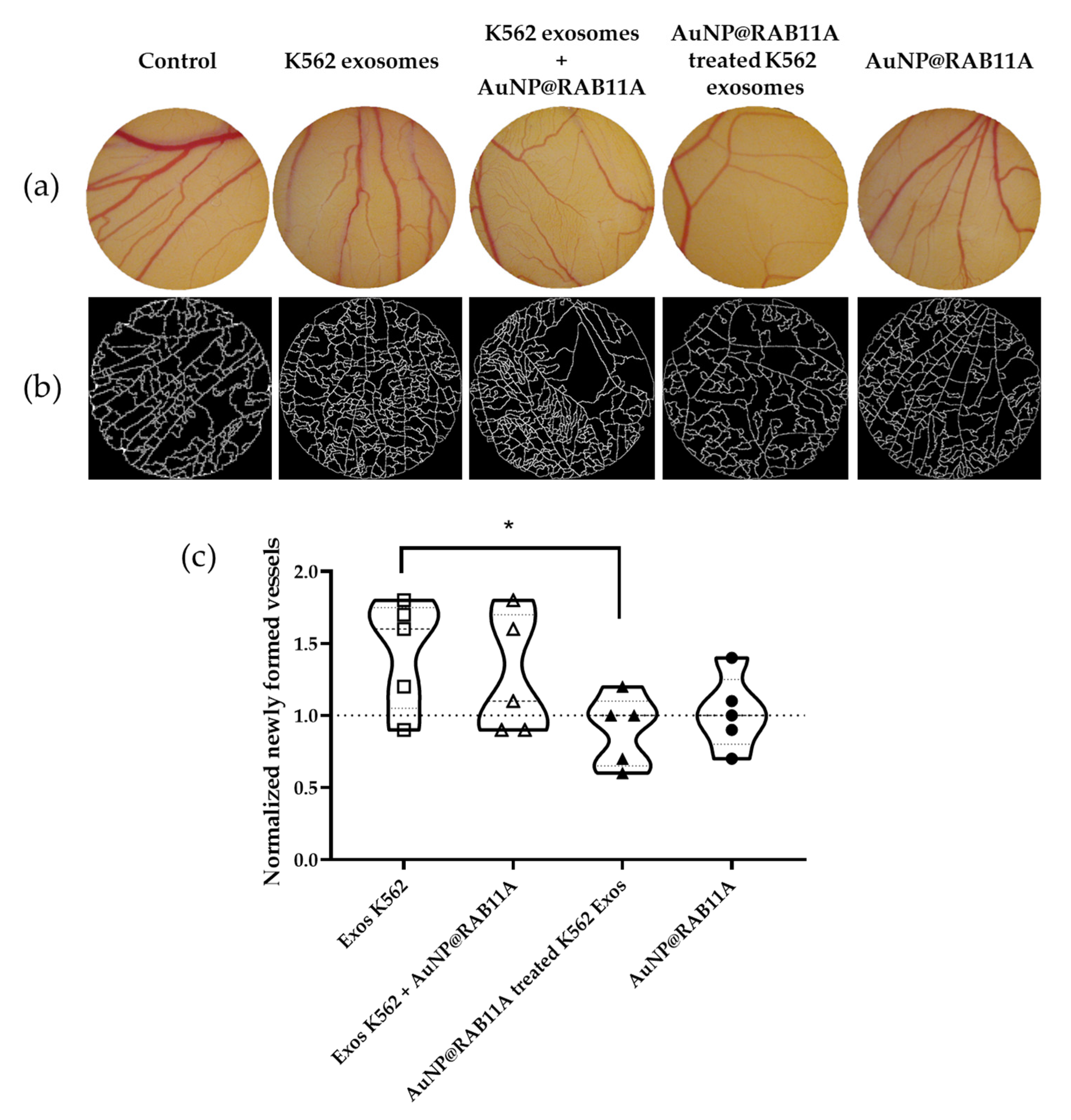
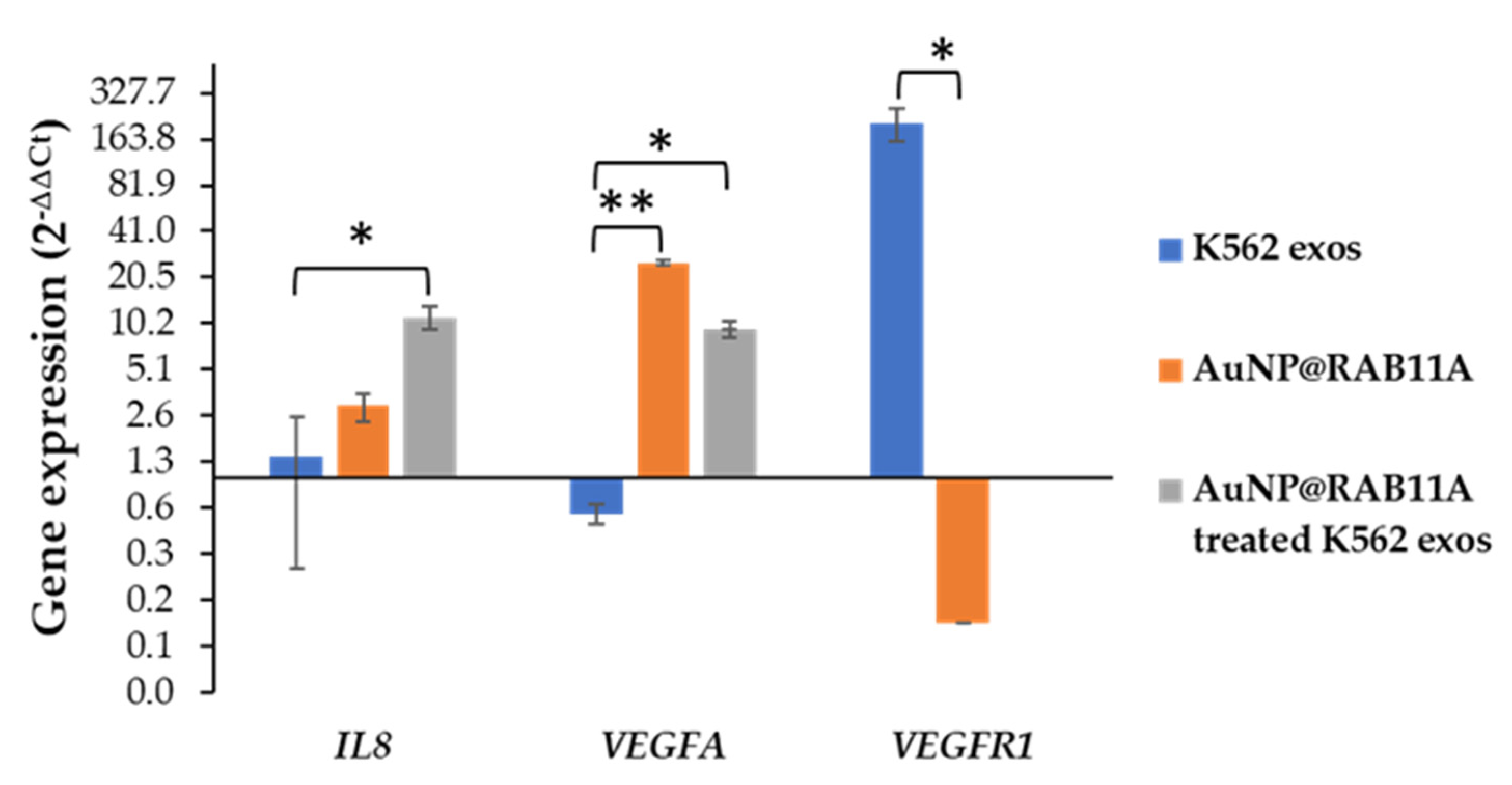
3.3. The Pro-Angiogenic Potential of Exosomes Secreted by K562 Treated with AuNP@RAB11A Is Lower than the Pro-Angiogenic Potential of K562 Exosomes Counterparts
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Shtivelman, E.; Lifshitz, B.; Gale, R.P.; Canaani, E. Fused transcript of abl and bcr genes in chronic myelogenous leukaemia. Nature 1985, 315, 550–554. [Google Scholar] [CrossRef] [PubMed]
- Rinke, J.; Hochhaus, A.; Ernst, T. CML-Not only BCR-ABL1 matters. Best Pract. Res. Clin. Haematol. 2020, 33, 101194. [Google Scholar] [CrossRef] [PubMed]
- Ng, J.J.; Ong, S.T. Therapy Resistance and Disease Progression in CML: Mechanistic Links and Therapeutic Strategies. Curr. Hematol. Malign Rep. 2022, 17, 181–197. [Google Scholar] [CrossRef]
- Abdulmawjood, B.; Costa, B.; Roma-Rodrigues, C.; Baptista, P.V.; Fernandes, A.R. Genetic Biomarkers in Chronic Myeloid Leukemia: What Have We Learned So Far? Int. J. Mol. Sci. 2021, 22, 12516. [Google Scholar] [CrossRef]
- Hochhaus, A.; Baccarani, M.; Silver, R.T.; Schiffer, C.; Apperley, J.F.; Cervantes, F.; Clark, R.E.; Cortes, J.E.; Deininger, M.W.; Guilhot, F.; et al. European LeukemiaNet 2020 recommendations for treating chronic myeloid leukemia. Leukemia 2020, 34, 966–984. [Google Scholar] [CrossRef] [Green Version]
- Korkolopoulou, P.; Viniou, N.; Kavantzas, N.; Patsouris, E.; Thymara, I.; Pavlopoulos, P.M.; Terpos, E.; Stamatopoulos, K.; Plata, E.; Anargyrou, K.; et al. Clinicopathologic correlations of bone marrow angiogenesis in chronic myeloid leukemia: A morphometric study. Leukemia 2003, 17, 89–97. [Google Scholar] [CrossRef] [Green Version]
- Cojbasic, I.; Macukanovic-Golubovic, L.; Mihailovic, D.; Vucic, M.; Cojbasic, Z. The significance of angiogenesis for predicting optimal therapeutic response in chronic myeloid leukaemia patients. Pol. J. Pathol. 2017, 68, 241–251. [Google Scholar] [CrossRef]
- Schmidt, T.; Carmeliet, P. Angiogenesis: A Target in Solid Tumors, Also in Leukemia? Hematol. Am. Soc. Hematol. Educ. Program Book 2011, 2011, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Deng, M.; Zha, J.; Zhao, H.; Jia, X.; Shi, Y.; Li, Z.; Fu, G.; Yu, L.; Fang, Z.; Xu, B. Apatinib exhibits cytotoxicity toward leukemia cells by targeting VEGFR2-mediated prosurvival signaling and angiogenesis. Exp. Cell Res. 2020, 390, 111934. [Google Scholar] [CrossRef] [PubMed]
- Kargozar, S.; Baino, F.; Hamzehlou, S.; Hamblin, M.R.; Mozafari, M. Nanotechnology for angiogenesis: Opportunities and challenges. Chem. Soc. Rev. 2020, 49, 5008–5057. [Google Scholar] [CrossRef] [PubMed]
- Mashouri, L.; Yousefi, H.; Aref, A.R.; Ahadi, A.M.; Molaei, F.; Alahari, S.K. Exosomes: Composition, biogenesis, and mechanisms in cancer metastasis and drug resistance. Mol. Cancer 2019, 18, 75. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Corrado, C.; Saieva, L.; Raimondo, S.; Santoro, A.; De Leo, G.; Alessandro, R. Chronic myelogenous leukaemia exosomes modulate bone marrow microenvironment through activation of epidermal growth factor receptor. J. Cell. Mol. Med. 2016, 20, 1829–1839. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Raimondo, S.; Saieva, L.; Corrado, C.; Fontana, S.; Flugy, A.; Rizzo, A.; De Leo, G.; Alessandro, R. Chronic myeloid leukemia-derived exosomes promote tumor growth through an autocrine mechanism. Cell Commun. Signal. 2015, 13, 8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Taverna, S.; Amodeo, V.; Saieva, L.; Russo, A.; Giallombardo, M.; De Leo, G.; Alessandro, R. Exosomal shuttling of miR-126 in endothelial cells modulates adhesive and migratory abilities of chronic myelogenous leukemia cells. Mol. Cancer 2014, 13, 169. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mineo, M.; Garfield, S.H.; Taverna, S.; Flugy, A.; De Leo, G.; Alessandro, R.; Kohn, E.C. Exosomes released by K562 chronic myeloid leukemia cells promote angiogenesis in a Src-dependent fashion. Angiogenesis 2012, 15, 33–45. [Google Scholar] [CrossRef] [Green Version]
- Taverna, S.; Flugy, A.; Saieva, L.; Kohn, E.C.; Santoro, A.; Meraviglia, S.; De Leo, G.; Alessandro, R. Role of exosomes released by chronic myelogenous leukemia cells in angiogenesis. Int. J. Cancer 2012, 130, 2033–2043. [Google Scholar] [CrossRef] [Green Version]
- Roma-Rodrigues, C.; Fernandes, A.R.; Baptista, P.V. Counteracting the effect of leukemia exosomes by antiangiogenic gold nanoparticles. Int. J. Nanomed. 2019, 14, 6843–6854. [Google Scholar] [CrossRef] [Green Version]
- Welz, T.; Wellbourne-Wood, J.; Kerkhoff, E. Orchestration of cell surface proteins by Rab11. Trends Cell Biol. 2014, 24, 407–415. [Google Scholar] [CrossRef]
- Zhao, L.; Yu, Q.; Gao, C.; Xiang, J.; Zheng, B.; Feng, Y.; Li, R.; Zhang, W.; Hong, X.; Zhan, Y.Y.; et al. Studies of the Efficacy of Low-Dose Apatinib Monotherapy as Third-Line Treatment in Patients with Metastatic Colorectal Cancer and Apatinib’s Novel Anticancer Effect by Inhibiting Tumor-Derived Exosome Secretion. Cancers 2022, 14, 2492. [Google Scholar] [CrossRef]
- Ballmer-Hofer, K.; Andersson, A.E.; Ratcliffe, L.E.; Berger, P. Neuropilin-1 promotes VEGFR-2 trafficking through Rab11 vesicles thereby specifying signal output. Blood 2011, 118, 816–826. [Google Scholar] [CrossRef] [Green Version]
- Jung, J.-J.; Tiwari, A.; Inamdar, S.M.; Thomas, C.P.; Goel, A.; Choudhury, A. Secretion of Soluble Vascular Endothelial Growth Factor Receptor 1 (sVEGFR1/sFlt1) Requires Arf1, Arf6, and Rab11 GTPases. PLoS ONE 2012, 7, e44572. [Google Scholar] [CrossRef]
- Kong, L.; Xiao, F.; Wang, L.; Li, M.; Wang, D.; Feng, Z.; Huang, L.; Wei, Y.; Li, H.; Liu, F.; et al. Intermedin promotes vessel fusion by inducing VE-cadherin accumulation at potential fusion sites and to achieve a dynamic balance between VE-cadherin-complex dissociation/reconstitution. MedComm 2020, 1, 84–102. [Google Scholar] [CrossRef] [PubMed]
- Gelfand, M.V.; Hagan, N.; Tata, A.; Oh, W.J.; Lacoste, B.; Kang, K.T.; Kopycinska, J.; Bischoff, J.; Wang, J.H.; Gu, C. Neuropilin-1 functions as a VEGFR2 co-receptor to guide developmental angiogenesis independent of ligand binding. Elife 2014, 3, e03720. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Marlin, M.C. Rab family of GTPases. Methods Mol. Biol. 2015, 1298, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Benwell, C.J.; Taylor, J.; Robinson, S.D. Endothelial neuropilin-2 influences angiogenesis by regulating actin pattern development and α5-integrin-p-FAK complex recruitment to assembling adhesion sites. FASEB J. 2021, 35, e21679. [Google Scholar] [CrossRef]
- Blanc, L.; Vidal, M. New insights into the function of Rab GTPases in the context of exosomal secretion. Small GTPases 2018, 9, 95–106. [Google Scholar] [CrossRef] [Green Version]
- Savina, A.; Vidal, M.; Colombo, M.I. The exosome pathway in K562 cells is regulated by Rab11. J. Cell Sci. 2002, 115, 2505–2515. [Google Scholar] [CrossRef]
- Savina, A.; Fader, C.M.; Damiani, M.T.; Colombo, M.I. Rab11 promotes docking and fusion of multivesicular bodies in a calcium-dependent manner. Traffic 2005, 6, 131–143. [Google Scholar] [CrossRef]
- Pasparakis, G. Recent developments in the use of gold and silver nanoparticles in biomedicine. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnology 2022, 14, e1817. [Google Scholar] [CrossRef]
- Roma-Rodrigues, C.; Rivas-García, L.; Baptista, P.V.; Fernandes, A.R. Gene therapy in cancer treatment: Why go nano? Pharmaceutics 2020, 12, 233. [Google Scholar] [CrossRef] [Green Version]
- Yang, Y.; Zheng, X.; Chen, L.; Gong, X.; Yang, H.; Duan, X.; Zhu, Y. Multifunctional gold nanoparticles in cancer diagnosis and treatment. Int. J. Nanomed. 2022, 17, 2041–2067. [Google Scholar] [CrossRef] [PubMed]
- Zhao, R.; Xiang, J.; Wang, B.; Chen, L.; Tan, S. Recent Advances in the Development of Noble Metal NPs for Cancer Therapy. Bioinorg. Chem. Appl. 2022, 2022, 2444516. [Google Scholar] [CrossRef]
- Cifuentes-Rius, A.; Deepagan, V.G.; Xie, J.; Voelcker, N.H. Bright future of gold nanoclusters in theranostics. ACS Appl. Mater. Interfaces 2021, 13, 49581–49588. [Google Scholar] [CrossRef] [PubMed]
- Roma-Rodrigues, C.; Raposo, L.R.; Valente, R.; Fernandes, A.R.; Baptista, P.V. Combined cancer therapeutics—Tackling the complexity of the tumor microenvironment. WIREs Nanomed. Nanobiotechnol. 2021, 13, e1704. [Google Scholar] [CrossRef]
- Ahmad, F.; Salem-Bekhit, M.M.; Khan, F.; Alshehri, S.; Khan, A.; Ghoneim, M.M.; Wu, H.F.; Taha, E.I.; Elbagory, I. Unique properties of surface-functionalized nanoparticles for bio-application: Functionalization mechanisms and importance in application. Nanomaterials 2022, 12, 1333. [Google Scholar] [CrossRef] [PubMed]
- Villar-Alvarez, E.; Leal, B.H.; Cambón, A.; Pardo, A.; Martínez-Gonzalez, R.; Fernández-Vega, J.; Al-Qadi, S.; Mosquera, V.X.; Bouzas, A.; Barbosa, S.; et al. Triggered RNAi therapy using metal inorganic nanovectors. Mol. Pharm. 2019, 16, 3374–3385. [Google Scholar] [CrossRef]
- Barbosa, S.; Topete, A.; Alatorre-Meda, M.; Villar-Alvarez, E.M.; Pardo, A.; Alvarez-Lorenzo, C.; Concheiro, A.; Taboada, P.; Mosquera, V. Targeted combinatorial therapy using gold nanostars as theranostic platforms. J. Phys. Chem. C 2014, 118, 26313–26323. [Google Scholar] [CrossRef]
- Villar-Alvarez, E.; Cambón, A.; Pardo, A.; Arellano, L.; Marcos, A.V.; Pelaz, B.; del Pino, P.; Bouzas Mosquera, A.; Mosquera, V.X.; Almodlej, A.; et al. Combination of light-driven co-delivery of chemodrugs and plasmonic-induced heat for cancer therapeutics using hybrid protein nanocapsules. J. Nanobiotechnol. 2019, 17, 106. [Google Scholar] [CrossRef] [Green Version]
- Vinhas, R.; Mendes, R.; Fernandes, A.R.; Baptista, P.V. Nanoparticles—Emerging Potential for Managing Leukemia and Lymphoma. Front. Bioeng. Biotechnol. 2017, 5, 79. [Google Scholar] [CrossRef] [Green Version]
- Schneider-Stock, R.; Ribatti, D. The CAM Assay as an Alternative In Vivo Model for Drug Testing. Handb. Exp. Pharm. 2020. [Google Scholar] [CrossRef]
- Roma-Rodrigues, C.; Heuer-Jungemann, A.; Fernandes, A.R.; Kanaras, A.G.; Baptista, P.V. Peptide-coated gold nanoparticles for modulation of angiogenesis in vivo. Int. J. Nanomed. 2016, 11, 2633–2639. [Google Scholar] [CrossRef] [Green Version]
- Pedrosa, P.; Heuer-Jungemann, A.; Kanaras, A.G.; Fernandes, A.R.; Baptista, P.V. Potentiating angiogenesis arrest in vivo via laser irradiation of peptide functionalised gold nanoparticles. J Nanobiotechnol. 2017, 15, 85. [Google Scholar] [CrossRef] [Green Version]
- Lipinski, A.R.; Heymann, J.; Meissner, C.; Karlas, A.; Brinkmann, V.; Meyer, T.F.; Heuer, D. Rab6 and Rab11 regulate Chlamydia trachomatis development and golgin-84-dependent Golgi fragmentation. PLoS Pathog. 2009, 5, e1000615. [Google Scholar] [CrossRef]
- Oliveira, H.; Roma-Rodrigues, C.; Santos, A.; Veigas, B.; Bras, N.; Faria, A.; Calhau, C.; de Freitas, V.; Baptista, P.V.; Mateus, N.; et al. GLUT1 and GLUT3 involvement in anthocyanin gastric transport- Nanobased targeted approach. Sci. Rep. 2019, 9, 789. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vinhas, R.; Fernandes, A.R.; Baptista, P.V. Gold Nanoparticles for BCR-ABL1 Gene Silencing: Improving Tyrosine Kinase Inhibitor Efficacy in Chronic Myeloid Leukemia. Mol. Ther. Nucleic Acids 2017, 7, 408–416. [Google Scholar] [CrossRef] [Green Version]
- Strober, W. Trypan blue exclusion test of cell viability. Curr. Protoc. Immunol. 2001. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Roberts, T.C.; Hart, J.R.; Kaikkonen, M.U.; Weinberg, M.S.; Vogt, P.K.; Morris, K.V. Quantification of nascent transcription by bromouridine immunocapture nuclear run-on RT-qPCR. Nat. Protoc. 2015, 10, 1198–1211. [Google Scholar] [CrossRef] [Green Version]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef] [Green Version]
- Longatti, A.; Schindler, C.; Collinson, A.; Jenkinson, L.; Matthews, C.; Fitzpatrick, L.; Blundy, M.; Minter, R.; Vaughan, T.; Shaw, M.; et al. High affinity single-chain variable fragments are specific and versatile targeting motifs for extracellular vesicles. Nanoscale 2018, 10, 14230–14244. [Google Scholar] [CrossRef] [Green Version]
- Conde, J.; Rosa, G.; Baptista, P.V. Gold-Nanobeacons as a Theranostic System for the Detection and Inhibition of Specific Genes. Nat. Protoc. Exch. 2013. Available online: https://www.nature.com/protocolexchange/protocols/2881 (accessed on 1 February 2022).
- Conde, J.; Rosa, J.; de la Fuente, J.M.; Baptista, P.V. Gold-nanobeacons for simultaneous gene specific silencing and intracellular tracking of the silencing events. Biomaterials 2013, 34, 2516–2523. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.-H.; Lee, C.-W.; Chiou, A.; Wei, P.-K. Size-dependent endocytosis of gold nanoparticles studied by three-dimensional mapping of plasmonic scattering images. J. Nanobiotechnol. 2010, 8, 33. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ng, C.T.; Tang, F.M.; Li, J.J.; Ong, C.; Yung, L.L.; Bay, B.H. Clathrin-mediated endocytosis of gold nanoparticles in vitro. Anat. Rec. 2015, 298, 418–427. [Google Scholar] [CrossRef]
- O’Sullivan, M.J.; Lindsay, A.J. The Endosomal Recycling Pathway-At the Crossroads of the Cell. Int. J. Mol. Sci. 2020, 21, 6074. [Google Scholar] [CrossRef] [PubMed]
- Lai, F.; Stubbs, L.; Artzt, K. Molecular analysis of mouse Rab11b: A new type of mammalian YPT/Rab protein. Genomics 1994, 22, 610–616. [Google Scholar] [CrossRef]
- Yan, Z.; Wang, Z.G.; Segev, N.; Hu, S.; Minshall, R.D.; Dull, R.O.; Zhang, M.; Malik, A.B.; Hu, G. Rab11a Mediates Vascular Endothelial-Cadherin Recycling and Controls Endothelial Barrier Function. Arter. Thromb. Vasc. Biol. 2016, 36, 339–349. [Google Scholar] [CrossRef] [Green Version]
- Seerapu, H.R.; Borthakur, S.; Kong, N.; Agrawal, S.; Drazba, J.; Vasanji, A.; Fantin, A.; Ruhrberg, C.; Buck, M.; Horowitz, A. The cytoplasmic domain of neuropilin-1 regulates focal adhesion turnover. FEBS Lett. 2013, 587, 3392–3399. [Google Scholar] [CrossRef] [Green Version]
- Dong, W.; Wu, X. Overexpression of Rab11-FIP2 in colorectal cancer cells promotes tumor migration and angiogenesis through increasing secretion of PAI-1. Cancer Cell Int. 2018, 18, 35. [Google Scholar] [CrossRef] [Green Version]
- Majidpoor, J.; Mortezaee, K. Steps in metastasis: An updated review. Med. Oncol. 2021, 38, 3. [Google Scholar] [CrossRef]
- Yates, A.D.; Achuthan, P.; Akanni, W.; Allen, J.; Allen, J.; Alvarez-Jarreta, J.; Amode, M.R.; Armean, I.M.; Azov, A.G.; Bennett, R.; et al. Ensembl 2020. Nucleic Acids Res. 2020, 48, D682–D688. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Neiva, K.; Lingen, M.W.; Ellis, L.M.; Nor, J.E. VEGF-dependent tumor angiogenesis requires inverse and reciprocal regulation of VEGFR1 and VEGFR2. Cell Death Differ. 2010, 17, 499–512. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sekiguchi, K.; Ito, Y.; Hattori, K.; Inoue, T.; Hosono, K.; Honda, M.; Numao, A.; Amano, H.; Shibuya, M.; Unno, N.; et al. VEGF Receptor 1-Expressing Macrophages Recruited from Bone Marrow Enhances Angiogenesis in Endometrial Tissues. Sci. Rep. 2019, 9, 7037. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chelouche-Lev, D.; Miller, C.P.; Tellez, C.; Ruiz, M.; Bar-Eli, M.; Price, J.E. Different signalling pathways regulate VEGF and IL-8 expression in breast cancer: Implications for therapy. Eur. J. Cancer 2004, 40, 2509–2518. [Google Scholar] [CrossRef] [PubMed]
- Yuan, A.; Yu, C.-J.; Luh, K.-T.; Kuo, S.-H.; Lee, Y.-C.; Yang, P.-C. Aberrant p53 Expression Correlates With Expression of Vascular Endothelial Growth Factor mRNA and Interleukin-8 mRNA and Neoangiogenesis in Non–Small-Cell Lung Cancer. J. Clin. Oncol. 2002, 20, 900–910. [Google Scholar] [CrossRef]
- Samaras, V.; Piperi, C.; Levidou, G.; Zisakis, A.; Kavantzas, N.; Themistocleous, M.S.; Boviatsis, E.I.; Barbatis, C.; Lea, R.W.; Kalofoutis, A.; et al. Analysis of interleukin (IL)-8 expression in human astrocytomas: Associations with IL-6, cyclooxygenase-2, vascular endothelial growth factor, and microvessel morphometry. Hum. Immunol. 2009, 70, 391–397. [Google Scholar] [CrossRef]
- Bancroft, C.C.; Chen, Z.; Yeh, J.; Sunwoo, J.B.; Yeh, N.T.; Jackson, S.; Jackson, C.; Van Waes, C. Effects of pharmacologic antagonists of epidermal growth factor receptor, PI3K and MEK signal kinases on NF-κB and AP-1 activation and IL-8 and VEGF expression in human head and neck squamous cell carcinoma lines. Int. J. Cancer 2002, 99, 538–548. [Google Scholar] [CrossRef]
- Ostenfeld, M.S.; Jeppesen, D.K.; Laurberg, J.R.; Boysen, A.T.; Bramsen, J.B.; Primdal-Bengtson, B.; Hendrix, A.; Lamy, P.; Dagnaes-Hansen, F.; Rasmussen, M.H.; et al. Cellular disposal of miR23b by RAB27-dependent exosome release is linked to acquisition of metastatic properties. Cancer Res. 2014, 74, 5758–5771. [Google Scholar] [CrossRef] [Green Version]
- Li, W.; Mu, D.; Tian, F.; Hu, Y.; Jiang, T.; Han, Y.; Chen, J.; Han, G.; Li, X. Exosomes derived from Rab27a-overexpressing tumor cells elicit efficient induction of antitumor immunity. Mol. Med. Rep. 2013, 8, 1876–1882. [Google Scholar] [CrossRef] [Green Version]
- Roma-Rodrigues, C.; Pereira, F.; Alves de Matos, A.P.; Fernandes, M.; Baptista, P.V.; Fernandes, A.R. Smuggling gold nanoparticles across cell types—A new role for exosomes in gene silencing. Nanomedicine 2017, 13, 1389–1398. [Google Scholar] [CrossRef]
- Clotilde Théry, C.; Witwer, K.W.; Aikawa, E.; Alcaraz, M.J.; Anderson, J.D.; Andriantsitohaina, R.; Antoniou, A.; Arab, T.; Archer, F.; Atkin-Smith, G.K.; et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J. Extracell. Vesicles 2018, 7, 1535750. [Google Scholar] [CrossRef] [PubMed] [Green Version]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Roma-Rodrigues, C.; Fernandes, A.R.; Baptista, P.V. Exploring RAB11A Pathway to Hinder Chronic Myeloid Leukemia-Induced Angiogenesis In Vivo. Pharmaceutics 2023, 15, 742. https://doi.org/10.3390/pharmaceutics15030742
Roma-Rodrigues C, Fernandes AR, Baptista PV. Exploring RAB11A Pathway to Hinder Chronic Myeloid Leukemia-Induced Angiogenesis In Vivo. Pharmaceutics. 2023; 15(3):742. https://doi.org/10.3390/pharmaceutics15030742
Chicago/Turabian StyleRoma-Rodrigues, Catarina, Alexandra R. Fernandes, and Pedro V. Baptista. 2023. "Exploring RAB11A Pathway to Hinder Chronic Myeloid Leukemia-Induced Angiogenesis In Vivo" Pharmaceutics 15, no. 3: 742. https://doi.org/10.3390/pharmaceutics15030742
APA StyleRoma-Rodrigues, C., Fernandes, A. R., & Baptista, P. V. (2023). Exploring RAB11A Pathway to Hinder Chronic Myeloid Leukemia-Induced Angiogenesis In Vivo. Pharmaceutics, 15(3), 742. https://doi.org/10.3390/pharmaceutics15030742