Ginsenoside Rg1 Delays Chronological Aging in a Yeast Model via CDC19- and SDH2-Mediated Cellular Metabolism
Abstract
1. Introduction
2. Materials and Methods
2.1. Drugs
2.2. Yeast Strains and Growth Conditions
2.3. Growth
2.4. Colony-Forming Unit Assay
2.5. Scanning Electron Microscopy (SEM)
2.6. Apoptosis Assay
2.7. Stress Tests
2.8. Measurement of Antioxidant Biomarkers
2.9. Reverse Transcription–Quantitative PCR (RT–qPCR)
2.10. Establishment of Proteomics Database
2.11. Mitochondrial Oxidative Phosphorylation (OXPHOS) and Glycolysis Assays
2.12. Construction of CDC19-Overexpression and SDH2-Deletion Strains
2.13. Determination of Core Enzymes in Glycolysis
2.14. ATP Content
2.15. Mitochondrial Membrane Potential (MMP) Analysis
2.16. Statistical Analysis
3. Results
3.1. Rg1 Delays Chronological Aging of S. cerevisiae Cells Better Than Other Ginsenoside Monomers
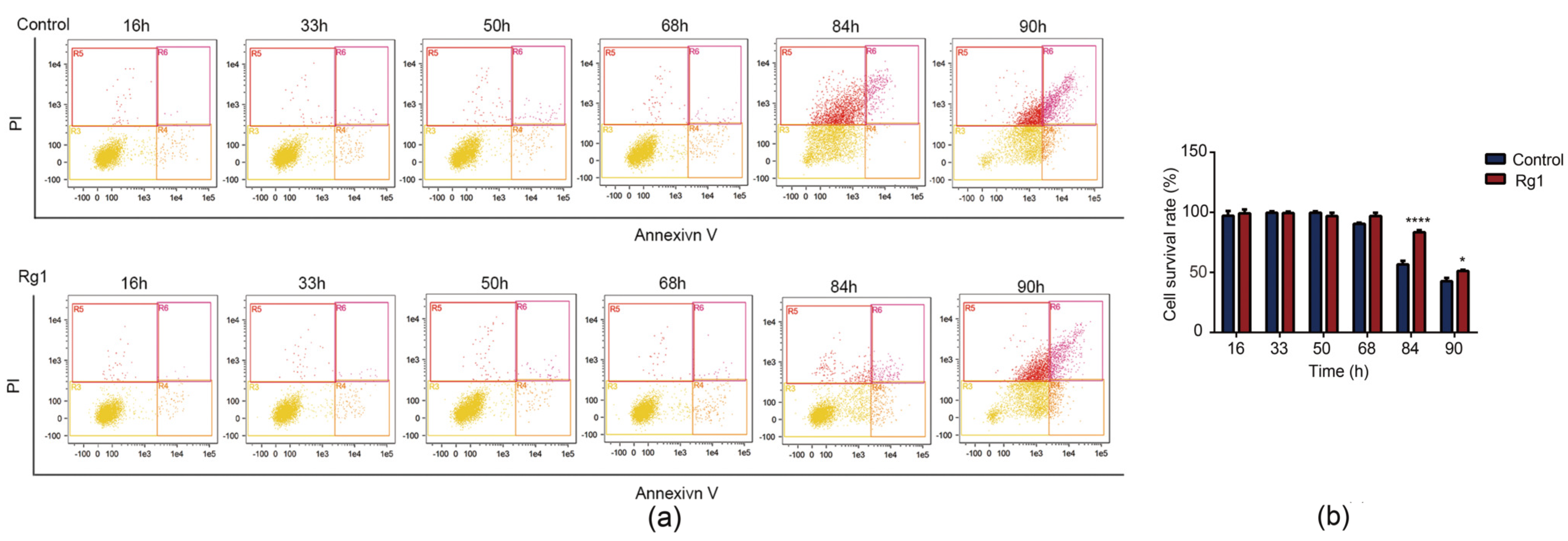
3.2. Rg1 Delays Apoptosis of S. cerevisiae Cells
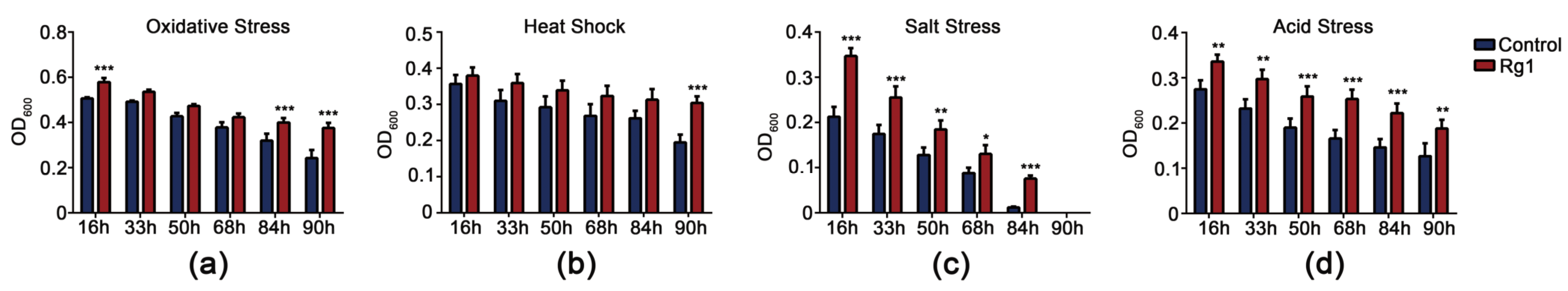
3.3. Rg1 Increased Stress-Resistance of S. Cerevisiae Cells
3.4. Rg1 Enhances the Antioxidant Defense System to Scavenge ROS in S. cerevisiae
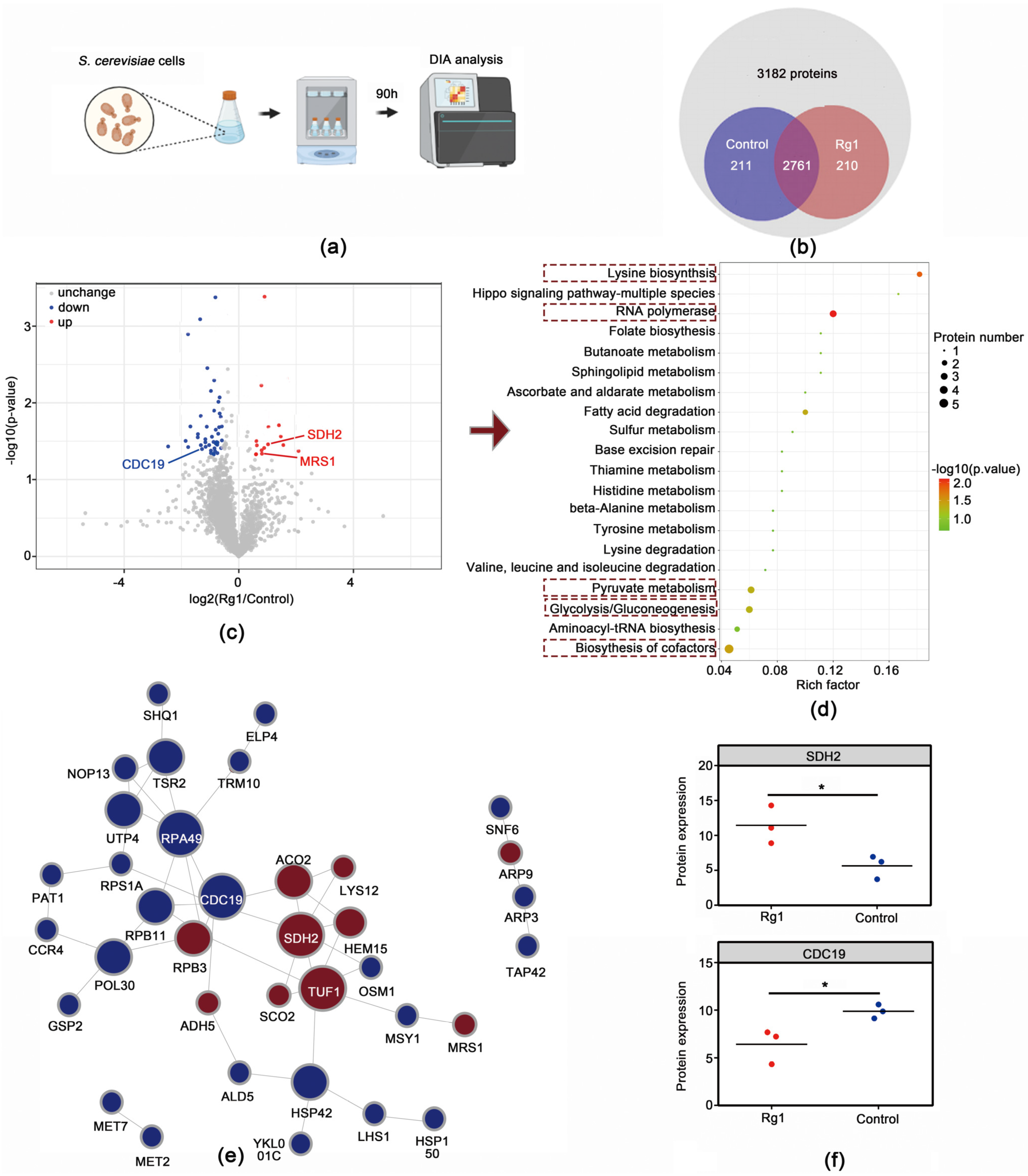
3.5. Rg1 Treatment Altered Proteomic Profiles in Aging S. cerevisiae Cells
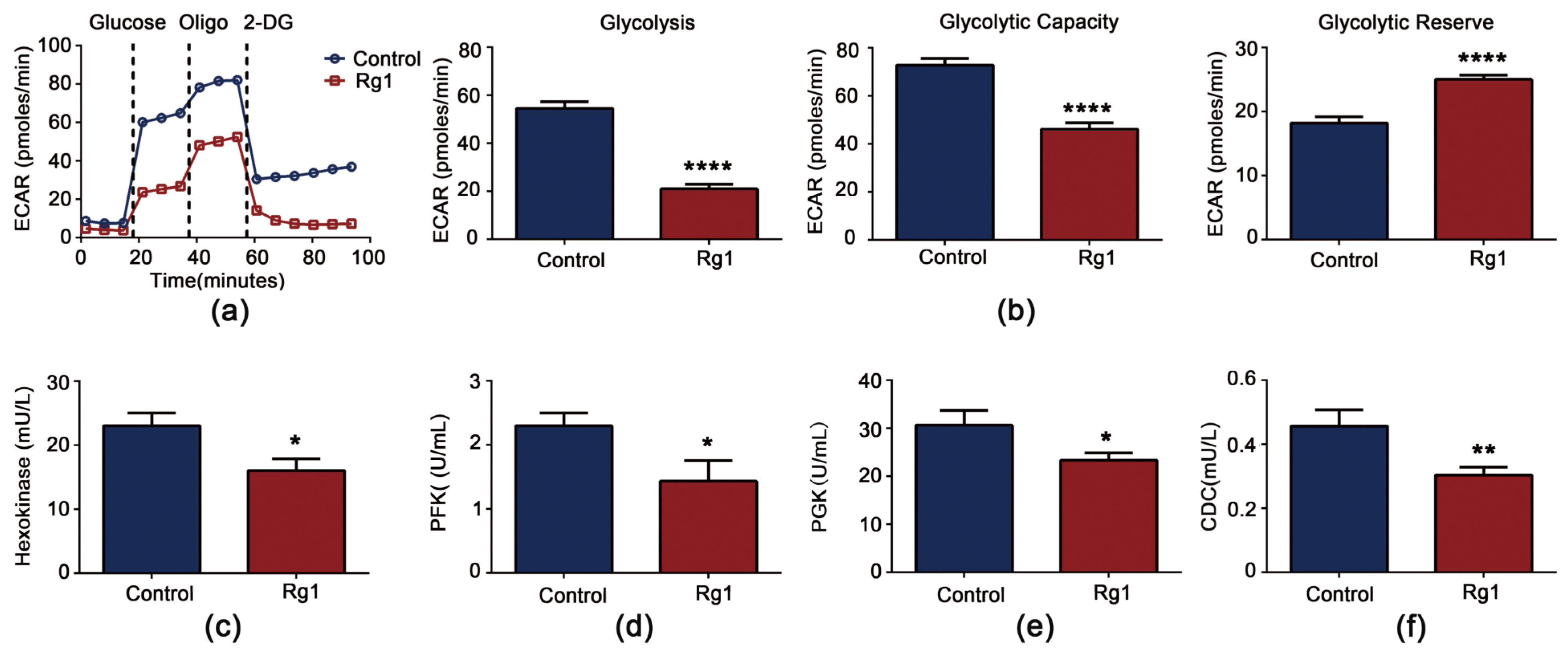
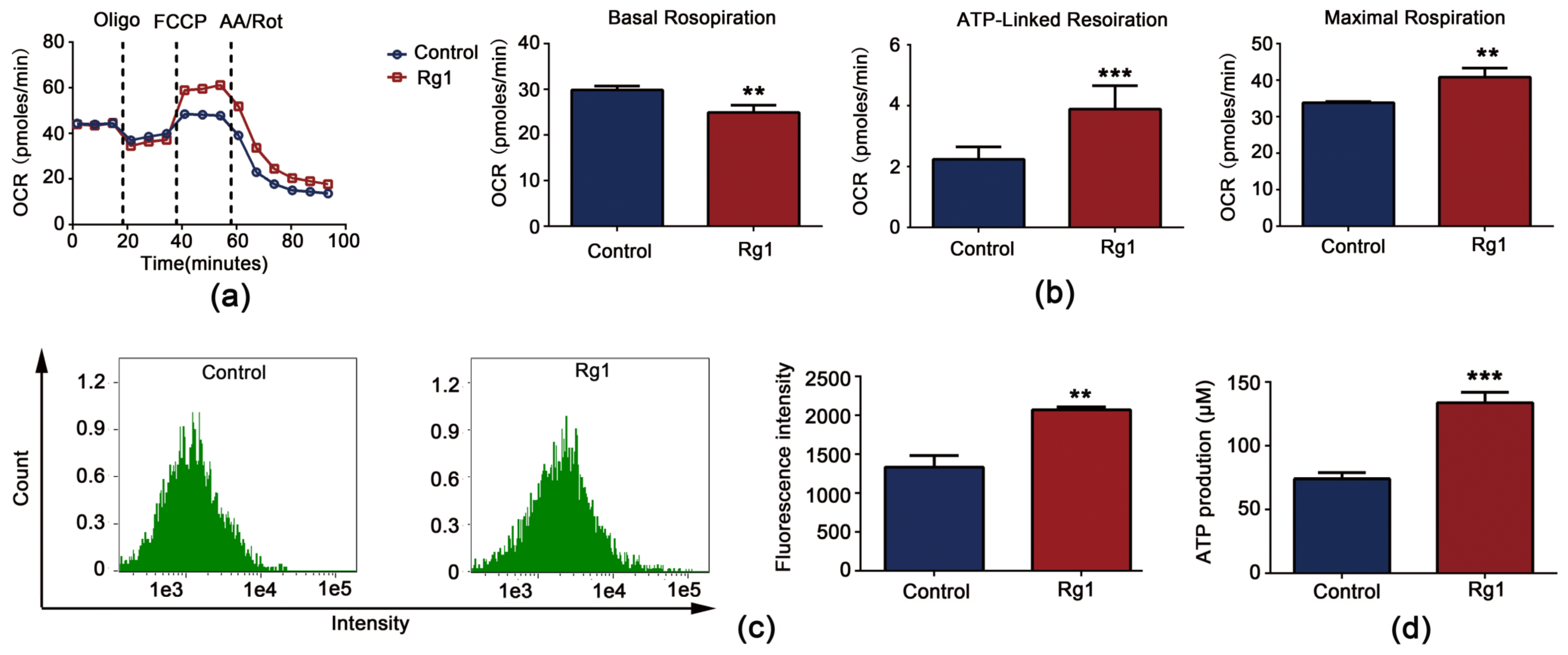
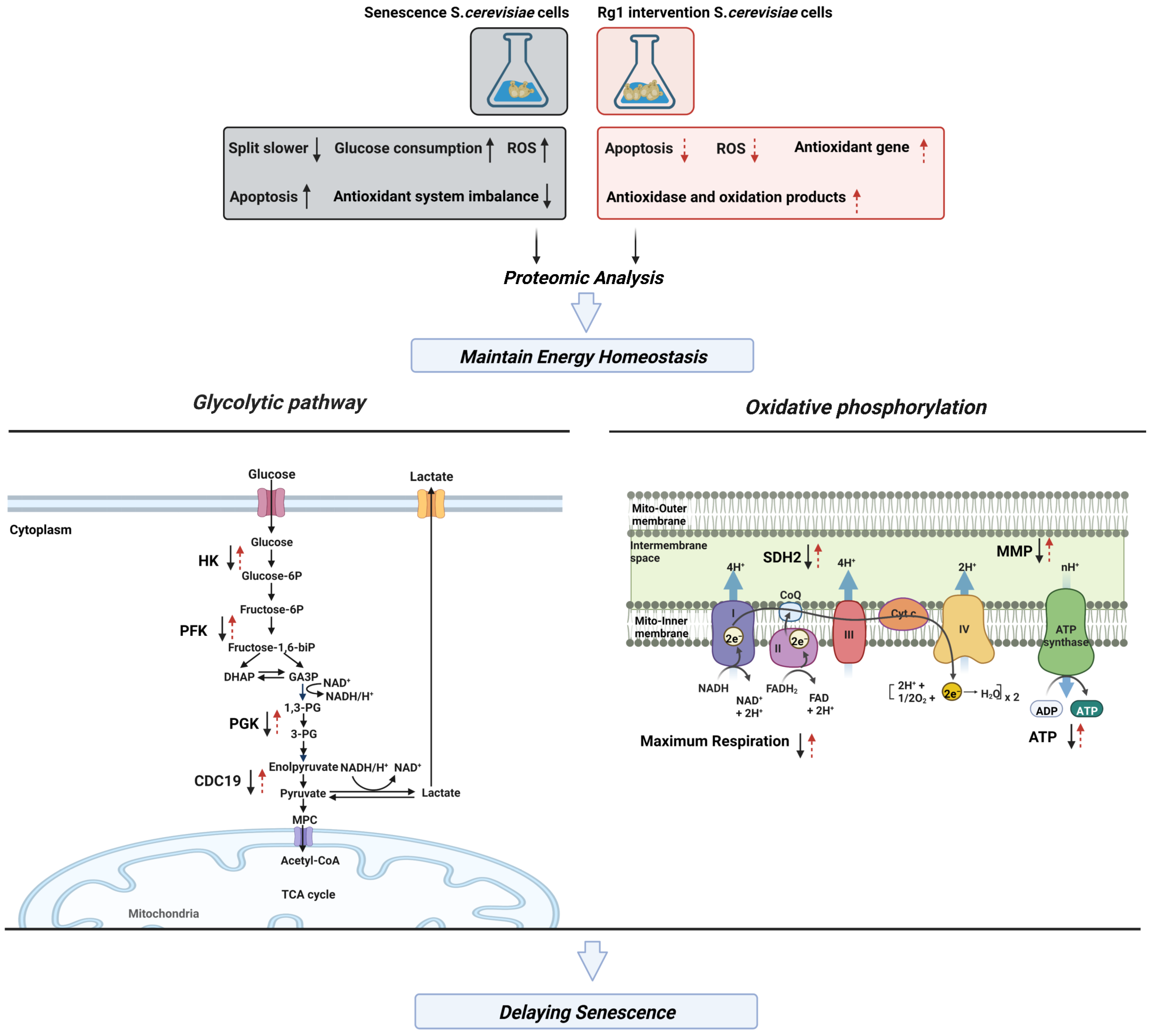
3.6. Rg1 Downregulates Glycolysis and Upregulates OXPHOS to Promote Longevity of S. cerevisiae Cells
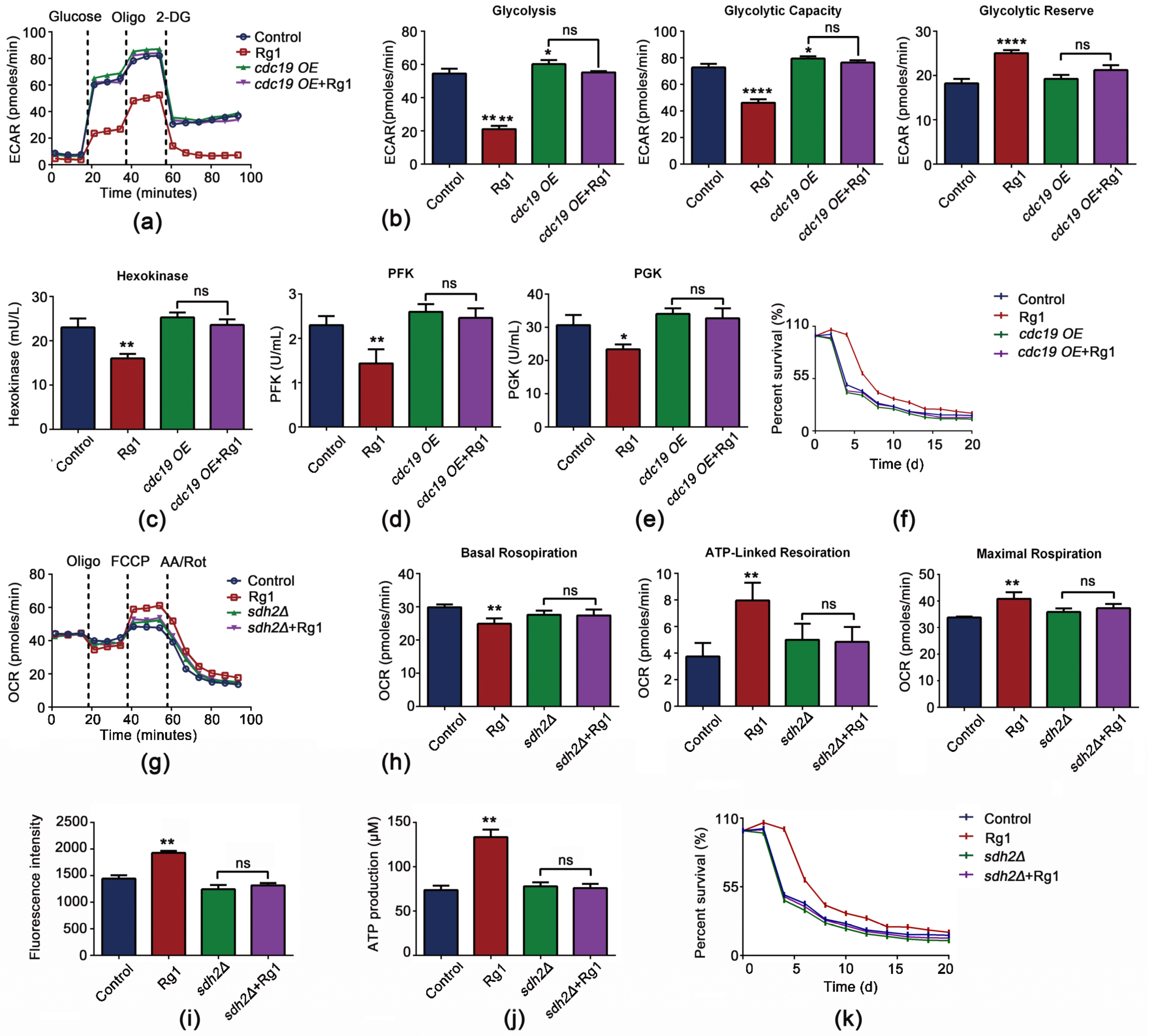
3.7. CDC19 and SDH2 Are Required for Rg1 to Regulate Glycolysis and OXPHOS to Delay Aging
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wagner, K.H.; Cameron-Smith, D.; Wessner, B.; Franzke, B. Biomarkers of Aging: From Function to Molecular Biology. Nutrients 2016, 8, 338. [Google Scholar] [CrossRef]
- Oliveira, Z.B.; Oliveira, A.R.; Miola, V.F.B.; Guissoni, L.M.; Spilla, C.S.G.; Barbalho, S.M. Panax ginseng and aging related disorders: A systematic review. Exp. Gerontol. 2022, 161, 111731. [Google Scholar]
- Zhao, Q.; Liu, Y.; Zhang, S.; Zhao, Y.; Wang, C.; Li, K.; Jin, Z.; Qiao, J.; Liu, M. Studies on the Regulation and Molecular Mechanism of Panax Ginseng Saponins on Senescence and Related Behaviors of Drosophila melanogaster. Front. Aging Neurosci. 2022, 14, 870326. [Google Scholar] [CrossRef]
- Wang, H.; Zhang, S.; Zhai, L.; Sun, L.; Zhao, D.; Wang, Z.; Li, X. Ginsenoside extract from ginseng extends lifespan and health span in Caenorhabditis elegans. Food Funct. 2021, 12, 6793–6808. [Google Scholar] [CrossRef]
- Li, Y.; Wang, L.; Wang, P.; Fan, C.; Zhang, P.; Shen, J.; Yu, S.Y. Ginsenoside-Rg1 Rescues Stress-Induced Depression-Like Behaviors via Suppression of Oxidative Stress and Neural Inflammation in Rats. Oxidative Med. Cell. Longev. 2020, 2020, 2325391. [Google Scholar] [CrossRef]
- Chen, C.; Zhang, H.; Xu, H.; Zheng, Y.; Wu, T.; Lian, Y. Ginsenoside Rb1 ameliorates cisplatin-induced learning and memory impairments. J. Ginseng Res. 2019, 43, 499–507. [Google Scholar] [CrossRef]
- Li, X.; Chu, S.; Lin, M.; Gao, Y.; Liu, Y.; Yang, S.; Zhou, X.; Zhang, Y.; Hu, Y.; Wang, H.; et al. Anticancer property of ginsenoside Rh2 from ginseng. Eur. J. Med. Chem. 2020, 203, 112627. [Google Scholar] [CrossRef]
- Zhao, T.; Wang, X.; Liu, Q.; Yang, T.; Qu, H.; Zhou, H. Ginsenoside Rd Promotes Cardiac Repair After Myocardial Infarction by Modulating Monocytes/Macrophages Subsets Conversion. Drug Des. Dev. Ther. 2022, 16, 2767–2782. [Google Scholar] [CrossRef]
- Brown, C.J.; Kaufman, T.; Trinidad, J.C.; Clemmer, D.E. Proteome changes in the aging Drosophila melanogaster head. Int. J. Mass Spectrom. 2018, 425, 36–46. [Google Scholar] [CrossRef]
- Chen, C.; Yang, C.; Wang, J.; Huang, X.; Yu, H.; Li, S.; Li, S.; Zhang, Z.; Liu, J.; Yang, X.; et al. Melatonin ameliorates cognitive deficits through improving mitophagy in a mouse model of Alzheimer’s disease. J. Pineal Res. 2021, 71, e12774. [Google Scholar] [CrossRef]
- Tanaka, T.; Biancotto, A.; Moaddel, R.; Moore, A.Z.; Gonzalez-Freire, M.; Aon, M.A.; Candia, J.; Zhang, P.; Cheung, F.; Fantoni, G.; et al. Plasma proteomic signature of age in healthy humans. Aging Cell 2018, 17, e12799. [Google Scholar] [CrossRef]
- Ximerakis, M.; Lipnick, S.L.; Innes, B.T.; Simmons, S.K.; Adiconis, X.; Dionne, D.; Mayweather, B.A.; Nguyen, L.; Niziolek, Z.; Ozek, C.; et al. Single-cell transcriptomic profiling of the aging mouse brain. Nat. Neurosci. 2019, 22, 1696–1708. [Google Scholar] [CrossRef]
- Mo, C.; Zhang, W.; Liu, L.; Wang, L.; Xiao, H. High throughput screening technology and the small molecules modulating aging related signals. Comb. Chem. High Throughput Screen. 2012, 15, 242–252. [Google Scholar] [CrossRef]
- Vanderwaeren, L.; Dok, R.; Voordeckers, K.; Nuyts, S.; Verstrepen, K.J. Saccharomyces cerevisiae as a Model System for Eukaryotic Cell Biology, from Cell Cycle Control to DNA Damage Response. Int. J. Mol. Sci. 2022, 19, 11665. [Google Scholar] [CrossRef]
- Foury, F. Human genetic diseases: A cross-talk between man and yeast. Gene 1997, 1, 1–10. [Google Scholar] [CrossRef]
- Subramaniyan, S.; Alugoju, P.; Sj, S.; Veerabhadrappa, B.; Dyavaiah, M. Magnolol protects Saccharomyces cerevisiae antioxidant-deficient mutants from oxidative stress and extends yeast chronological life span. FEMS Microbiol. Lett. 2019, 366, fnz065. [Google Scholar] [CrossRef]
- Roth, R.; Madhani, H.D.; Garcia, J.F. Total RNA Isolation and Quantification of Specific RNAs in Fission Yeast. Methods Mol. Biol. 2018, 1721, 63–72. [Google Scholar]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Hu, M.; Crawford, S.A.; Henstridge, D.C.; Ng, I.H.; Boey, E.J.; Xu, Y.; Febbraio, M.A.; Jans, D.A.; Bogoyevitch, M.A. p32 protein levels are integral to mitochondrial and endoplasmic reticulum morphology, cell metabolism and survival. Biochem. J. 2013, 453, 381–391. [Google Scholar] [CrossRef]
- Wach, A.; Brachat, A.; Pöhlmann, R.; Philippsen, P. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast 1994, 10, 1793–1808. [Google Scholar] [CrossRef]
- Giaever, G.; Chu, A.M.; Ni, L.; Connelly, C.; Riles, L.; Véronneau, S.; Dow, S.; Lucau-Danila, A.; Anderson, K.; André, B.; et al. Functional profiling of the Saccharomyces cerevisiae genome. Nature 2002, 418, 387–391. [Google Scholar] [CrossRef]
- Winzeler, E.A.; Shoemaker, D.D.; Astromoff, A.; Liang, H.; Anderson, K.; Andre, B.; Bangham, R.; Benito, R.; Boeke, J.D.; Bussey, H.; et al. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 1999, 285, 901–906. [Google Scholar] [CrossRef]
- Zimmermann, A.; Hofer, S.; Pendl, T.; Kainz, K.; Madeo, F.; Carmona-Gutierrez, D. Yeast as a tool to identify anti-aging compounds. FEMS Yeast Res. 2018, 18, foy020. [Google Scholar] [CrossRef]
- Liochev, S.I. Reactive oxygen species and the free radical theory of aging. Free Radic. Biol. Med. 2013, 60, 1–4. [Google Scholar] [CrossRef]
- Curtis, R.; Geesaman, B.J.; DiStefano, P.S. Ageing and metabolism: Drug discovery opportunities. Nat. Rev. Drug Discov. 2005, 7, 569–580. [Google Scholar] [CrossRef]
- Eisenberg, T.; Schroeder, S.; Andryushkova, A.; Pendl, T.; Küttner, V.; Bhukel, A.; Mariño, G.; Pietrocola, F.; Harger, A.; Zimmermann, A.; et al. Nucleocytosolic depletion of the energy metabolite acetyl-coenzyme a stimulates autophagy and prolongs lifespan. Cell Metab. 2014, 19, 431–444. [Google Scholar] [CrossRef]
- Xu, P.; Wang, M.; Song, W.M.; Wang, Q.; Yuan, G.C.; Sudmant, P.H.; Zare, H.; Tu, Z.; Orr, M.E.; Zhang, B. The landscape of human tissue and cell type specific expression and co-regulation of senescence genes. Mol. Neurodegener. 2022, 1, 5. [Google Scholar] [CrossRef]
- Jo, S.L.; Yang, H.; Lee, S.R.; Heo, J.H.; Lee, H.W.; Hong, E.J. Curcumae Radix Decreases Neurodegenerative Markers through Glycolysis Decrease and TCA Cycle Activation. Nutrients 2022, 14, 1587. [Google Scholar] [CrossRef]
- Oliveira, A.V.; Vilaça, R.; Santos, C.N.; Costa, V.; Menezes, R. Exploring the power of yeast to model aging and age-related neurodegenerative disorders. Biogerontology 2017, 18, 3–34. [Google Scholar] [CrossRef]
- Khawaja, A.A.; Belak, Z.R.; Eskiw, C.H.; Harkness, T.A.A. High-Throughput Rapid Yeast Chronological Lifespan Assay. Methods Mol. Biol. 2021, 2196, 229–233. [Google Scholar]
- Meunier, J.R.; Choder, M. Saccharomyces cerevisiae colony growth and ageing: Biphasic growth accompanied by changes in gene expression. Yeast 1999, 15, 1159–1169. [Google Scholar] [CrossRef]
- Yang, S.; Dong, Y.; Liu, Y.; Yan, X.; Sun, G.; Jia, G.; Li, X.; Liu, H.; Su, H.; Li, Y. Application of lipidomics strategy to explore aging-related biomarkers and potential anti-aging mechanisms of ginseng. Biogerontology 2021, 22, 589–602. [Google Scholar] [CrossRef]
- Wang, Z.L.; Chen, L.B.; Qiu, Z.; Chen, X.B.; Liu, Y.; Li, J.; Wang, L.; Wang, Y.P. Ginsenoside Rg1 ameliorates testicular senescence changes in D-gal-induced aging mice via anti-inflammatory and antioxidative mechanisms. Mol. Med. Rep. 2018, 17, 6269–6276. [Google Scholar]
- Madhi, I.; Kim, J.H.; Shin, J.E.; Kim, Y. Ginsenoside Re exhibits neuroprotective effects by inhibiting neuroinflammation via CAMK/MAPK/NF-κB signaling in microglia. Mol. Med. Rep. 2021, 24, 698. [Google Scholar] [CrossRef]
- Bonawitz, N.D.; Chatenay-Lapointe, M.; Pan, Y.; Shadel, G.S. Reduced TOR signaling extends chronological life span via increased respiration and upregulation of mitochondrial gene expression. Cell Metab. 2007, 5, 265–277. [Google Scholar] [CrossRef]
- Sunthonkun, P.; Palajai, R.; Somboon, P.; Suan, C.L.; Ungsurangsri, M.; Soontorngun, N. Life-span extension by pigmented rice bran in the model yeast Saccharomyces cerevisiae. Sci. Rep. 2019, 9, 18061. [Google Scholar] [CrossRef]
- Hohmann, M.S.; Habiel, D.M.; Coelho, A.L.; Verri, W.A., Jr.; Hogaboam, C.M. Quercetin Enhances Ligand-induced Apoptosis in Senescent Idiopathic Pulmonary Fibrosis Fibroblasts and Reduces Lung Fibrosis In Vivo. Am. J. Respir. Cell Mol. Biol. 2019, 60, 28–40. [Google Scholar] [CrossRef]
- Sudharshan, S.J.; Narayanan, A.K.; Princilly, J.; Dyavaiah, M.; Nagegowda, D.A. Saccharomyces cerevisiae Betulinic acid mitigates oxidative stress-mediated apoptosis and enhances longevity in the yeast model. Free. Radic. Res. 2023, accepted. [Google Scholar]
- Westcott, W.L. Resistance training is medicine: Effects of strength training on health. Curr. Sport. Med. Rep. 2012, 11, 209–216. [Google Scholar] [CrossRef]
- Kudryavtseva, A.V.; Krasnov, G.S.; Dmitriev, A.A.; Alekseev, B.Y.; Kardymon, O.L.; Sadritdinova, A.F.; Fedorova, M.S.; Pokrovsky, A.V.; Melnikova, N.V.; Kaprin, A.D.; et al. Mitochondrial dysfunction and oxidative stress in aging and cancer. Oncotarget 2016, 7, 44879–44905. [Google Scholar] [CrossRef]
- Lee, Y.; Min, K.; Son, H.; Park, A.R.; Kim, J.C.; Choi, G.J.; Lee, Y.W. ELP3 is involved in sexual and asexual development, virulence, and the oxidative stress response in Fusarium graminearum. Mol. Plant Microbe Interact. 2014, 27, 1344–1355. [Google Scholar] [CrossRef] [PubMed]
- Su, L.J.; Zhang, J.H.; Gomez, H.; Murugan, R.; Hong, X.; Xu, D.; Jiang, F.; Peng, Z.Y. Reactive Oxygen Species-Induced Lipid Peroxidation in Apoptosis, Autophagy, and Ferroptosis. Oxidative Med. Cell. Longev. 2019, 2019, 5080843. [Google Scholar] [CrossRef] [PubMed]
- Shi, W.; Tan, C.; Liu, C.; Chen, D. Mitochondrial fission mediated by Drp1-Fis1 pathway and neurodegenerative diseases. Rev. Neurosci. 2022, accepted. [Google Scholar] [CrossRef]
- Hadj, A.I.Y.; Smaoui, K.; Chaabène, I.; Mabrouk, I.; Djemal, L.; Elleuch, H.; Allouche, M.; Mokdad-Gargouri, R.; Gargouri, A. Human p53 induces cell death and downregulates thioredoxin expression in Saccharomyces cerevisiae. FEMS Yeast Res. 2008, 8, 1254–1262. [Google Scholar] [CrossRef]
- Roichman, A.; Elhanati, S.; Aon, M.A.; Abramovich, I.; Di Francesco, A.; Shahar, Y.; Avivi, M.Y.; Shurgi, M.; Rubinstein, A.; Wiesner, Y.; et al. Restoration of energy homeostasis by SIRT6 extends healthy lifespan. Nat. Commun. 2021, 12, 3208. [Google Scholar] [CrossRef] [PubMed]
- Raffaghello, L.; Longo, V. Metabolic Alterations at the Crossroad of Aging and Oncogenesis. Int. Rev. Cell Mol. Biol. 2017, 332, 1–42. [Google Scholar] [PubMed]
- Al Zaid Siddiquee, K.; Arauzo-Bravo, M.J.; Shimizu, K. Metabolic flux analysis of pykF gene knockout Escherichia coli based on 13C-labeling experiments together with measurements of enzyme activities and intracellular metabolite concentrations. Appl. Microbiol. Biotechnol. 2004, 63, 407–417. [Google Scholar] [CrossRef] [PubMed]
- Sakamuri, S.; Sure, V.N.; Kolli, L.; Liu, N.; Evans, W.R.; Sperling, J.A.; Busija, D.W.; Wang, X.; Lindsey, S.H.; Murfee, W.L.; et al. Glycolytic and Oxidative Phosphorylation Defects Precede the Development of Senescence in Primary Human Brain Microvascular Endothelial Cells. GeroScience 2022, 44, 1975–1994. [Google Scholar] [CrossRef]
- Cho, S.J.; Moon, J.S.; Lee, C.M.; Choi, A.M.; Stout-Delgado, H.W. Glucose Transporter 1-Dependent Glycolysis Is Increased during Aging-Related Lung Fibrosis, and Phloretin Inhibits Lung Fibrosis. Am. J. Respir. Cell Mol. Biol. 2017, 56, 521–531. [Google Scholar] [CrossRef]
- Müller, M.; Ahumada-Castro, U.; Sanhueza, M.; Gonzalez-Billault, C.; Court, F.A.; Cárdenas, C. Mitochondria and Calcium Regulation as Basis of Neurodegeneration Associated with Aging. Front. Neurosci. 2018, 12, 470. [Google Scholar] [CrossRef]
- Jackson, J.; Wischhof, L.; Scifo, E.; Pellizzer, A.; Wang, Y.; Piazzesi, A.; Gentile, D.; Siddig, S.; Stork, M.; Hopkins, C.E.; et al. SGPL1 stimulates VPS39 recruitment to the mitochondria in MICU1 deficient cells. Mol. Metab. 2022, 61, 101503. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Martinez, A.; Calleja, M.; Peralta, S.; Matsushima, Y.; Hernandez-Sierra, R.; Whitworth, A.J.; Kaguni, L.S.; Garesse, R. Modeling pathogenic mutations of human twinkle in Drosophila suggests an apoptosis role in response to mitochondrial defects. PLoS ONE 2012, 7, e43954. [Google Scholar] [CrossRef] [PubMed]
- Hariton, F.; Xue, M.; Rabbani, N.; Fowler, M.; Thornalley, P.J. Sulforaphane Delays Fibroblast Senescence by Curbing Cellular Glucose Uptake, Increased Glycolysis, and Oxidative Damage. Oxidative Med. Cell. Longev. 2018, 2018, 5642148. [Google Scholar] [CrossRef] [PubMed]
- Pastor, M.M.; Proft, M.; Pascual-Ahuir, A. Mitochondrial function is an inducible determinant of osmotic stress adaptation in yeast. J. Biol. Chem. 2009, 44, 30307–30317. [Google Scholar] [CrossRef] [PubMed]



| Gene | GenBank Accession | Primer Sequence (5′–3′) |
|---|---|---|
| Tubulin | NM-001179929 | F: CCAAGGGCTATTTACGTGGA |
| R: GGTGTAATGGCCTCTTGCAT | ||
| SOD2 | NM-001179138 | F: GCATTACACCAAGCACCATCA |
| R: CCAGGTTTTCCCAGAATAGACA | ||
| CTT1 | NM-001181217 | F: TCATCACCCATACGCTTCT |
| R: GGACATTTGTAACCCACATTCT | ||
| GSH1 | NM-001181534 | F: GCTGTTCGTGCTTACAAGTGAC |
| R: ATGCCTCCAAATCCGTTCT | ||
| YAP1 | NM-001182362 | F: GACGGCGTGGGGAAGAAGGC |
| R: CCGACATCCAGGCGGCGTTT | ||
| GLR1 | NM-001183905 | F: TATTTGGATGGGCTAGATTC |
| R: TTCAACATTACCGTCCTTATT | ||
| PFK | NM-001181369 | F: GATGCTTCTGGGTTCCGTAT |
| R: ACCTTGACTTTGAGCCTTGT | ||
| CYC1 | NM-001181706 | F: GGTTCTGCTAAGAAAGGTGCTA |
| R: CCTTCAGCTTGACCAGAGTG | ||
| CDC19 | NM-001178183 | F: AGAAGAACCTCCATCATT |
| R: AGACTTGTGGTATTCGTA | ||
| PGK | NM-001178725 | F: TGTCTTGGCTTCTCACTTGG |
| R: TTCGTTTCTTTCACCGTTTG | ||
| NDI1 | NM-001182483 | F: ATCATTATCTGCCGTTAGCCA |
| R: CAAATGTGTTAGGTTCCGCA | ||
| FIS1 | NM-001179415 | F: AGTCCCGTAGACGAGAATGC |
| R: CCACCTGCTTGTTATTACGCT | ||
| TPI | NM-001180358 | F: AACTTTCTTTGTCGGTGGTA |
| R: TTCCTTAATGGATTGTTTGG | ||
| HXK | NM-001181119 | F: AAAACCACAAGCCAGAAAGG |
| R: GGGAAATCCATAACCCAACC |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, S.; Qiao, J.; Jiang, C.; Pan, D.; Yu, S.; Chen, J.; Liu, S.; Zhang, P.; Zhao, D.; Liu, M. Ginsenoside Rg1 Delays Chronological Aging in a Yeast Model via CDC19- and SDH2-Mediated Cellular Metabolism. Antioxidants 2023, 12, 296. https://doi.org/10.3390/antiox12020296
Wang S, Qiao J, Jiang C, Pan D, Yu S, Chen J, Liu S, Zhang P, Zhao D, Liu M. Ginsenoside Rg1 Delays Chronological Aging in a Yeast Model via CDC19- and SDH2-Mediated Cellular Metabolism. Antioxidants. 2023; 12(2):296. https://doi.org/10.3390/antiox12020296
Chicago/Turabian StyleWang, Siming, Juhui Qiao, Chunyan Jiang, Daian Pan, Shiting Yu, Jingjing Chen, Shichao Liu, Peiguang Zhang, Daqing Zhao, and Meichen Liu. 2023. "Ginsenoside Rg1 Delays Chronological Aging in a Yeast Model via CDC19- and SDH2-Mediated Cellular Metabolism" Antioxidants 12, no. 2: 296. https://doi.org/10.3390/antiox12020296
APA StyleWang, S., Qiao, J., Jiang, C., Pan, D., Yu, S., Chen, J., Liu, S., Zhang, P., Zhao, D., & Liu, M. (2023). Ginsenoside Rg1 Delays Chronological Aging in a Yeast Model via CDC19- and SDH2-Mediated Cellular Metabolism. Antioxidants, 12(2), 296. https://doi.org/10.3390/antiox12020296







