Abstract
MYCN Proto-Oncogene, BHLH Transcription Factor (MYCN) has been one of the most studied genes in neuroblastoma. It is known for its oncogenetic mechanisms, as well as its role in the prognosis of the disease and it is considered one of the prominent targets for neuroblastoma therapy. In the present work, we attempted to review the literature, on the relation between MYCN and neuroblastoma from all possible mechanistic sites. We have searched the literature for the role of MYCN in neuroblastoma based on the following topics: the references of MYCN in the literature, the gene’s anatomy, along with its transcripts, the protein’s anatomy, the epigenetic mechanisms regulating MYCN expression and function, as well as MYCN amplification. MYCN plays a significant role in neuroblastoma biology. Its functions and properties range from the forming of G-quadraplexes, to the interaction with miRNAs, as well as the regulation of gene methylation and histone acetylation and deacetylation. Although MYCN is one of the most primary genes studied in neuroblastoma, there is still a lot to be learned. Our knowledge on the exact mechanisms of MYCN amplification, etiology and potential interventions is still limited. The knowledge on the molecular mechanisms of MYCN in neuroblastoma, could have potential prognostic and therapeutic advantages.
1. Introduction
1.1. A Brief “Ensemble” to MYCN
MYCN is one of the most studied genes with respect to neuroblastoma. This gene was identified back in 1983 by Kohl et al. (1983) [1] and Schwab et al. (1983) [2], which was initially found to be amplified in neuroblastoma cases, “homologous to v-myc but different from MYC in human neuroblastoma” [3]. Later on, MYCN was found to manifest high expression levels, as a result of gene amplification, in neuroblastoma cell lines, metastatic neuroblastoma, retinoblastoma and lung tumors [4]. The first report on MYCN (as previously was named as myc, n-myc), was by Schwab and Bishop (1988), where they have reported that besides its role in human tumors, it participated in various biological processes, including senescence [5], resistance to therapy [6] and most interestingly it was found that “circular extra-chromosomal DNA molecules could transport amplified MYCN proto-oncogenes in human neuroblastomas” [7].
MYCN has been extensively studied both in fetal development, as well as for its role in human neoplasms. MYCN products are nuclear phosphoproteins with a short half-life. It has been shown that in terms of its physiological cellular function, it plays a role in neural development of the sympathetic system. Especially, the expression of MYCN increases significantly during the fetal stages. It has been shown, in multiple reports, that MYCN overexpression is directly associated with poor prognosis of neuroblastoma and it is thought that there is a strong correlation between gene overexpression and the evolutionary course of tumors [8]. On the other hand, MYCN has received less attention with respect to pediatric brain tumors, while recent reports have highlighted its role in the disease [9,10,11]. Apart from its role in neuroblastoma, MYCN has been studied in some extent for other pediatric brain neoplasms such as medulloblastoma [12,13], astrocytoma [14], glioblastoma [15,16], and others. One of the main questions in MYCN biology is the link between its overexpression and amplification. The gene’s overexpression does not always correlate to the gene’s copy numbers and this is a topic under continuing investigation.
The MYCN oncogene is one of the most important genetic biomarkers for the diagnosis, prognosis and treatment of neuroblastoma. MYCN overexpression is associated with poor prognosis and rapid tumor growth [9,17,18]. However, it does not function as a unique f actor rather it is reported to participate in a network of other factors, procuring neuroblastoma’s pathology. Examples of other genes linked to MYCN biology is the ID2 gene, which is also linked to MYCN overexpression and the fast development of the disease as studies have demonstrated the ability of MYCN to act as a transcription factor for ID2 [19]. Similar case is the NF2 transcription factor, whose methylation is linked to neuroblastoma progression [20]. One significant player in tumor progression, TP73 tumor suppressor gene, could not be left out of tumor progression. MYCN overexpression and TP53 downregulation is considered (and actually very recently) as a twin-target for neuroblastoma therapy [21], but also is linked to advanced stages of the disease.
Beside the aforementioned gene aberrations, chromosomal aberrations are also known to play a significant role in neuroblastoma progression. For example, deletion of the 1p36 region is also associated with poor disease prognosis, and the region has not yet been associated with a tumor suppressor gene [22,23]. Further studies, lead to the conclusion that the chromosomal region 1p36 contains some tumor suppressor genes which, however, have not yet been detected. Another negative prognostic indicator for the poor progression of the disease is the unbalanced enhancement of the chromosomal region 17q. The expression of TRKA, TRKB and TRKC receptors (neurotrophic tyrosine kinase receptors Types 1, 2, 3) has been associated with a good prognosis of the disease, while the gene appears to be suppressed in cases where we have MYCN amplification [24]. Other chromosomal aberrations are also known to play a significant role in neuroblastoma progression such as 17q and 22p [25]. These chromosomal regions are known to code for tumor suppressor genes, which when deleted lead to disease progression.
1.2. The Frequency of MYCN in the Literature
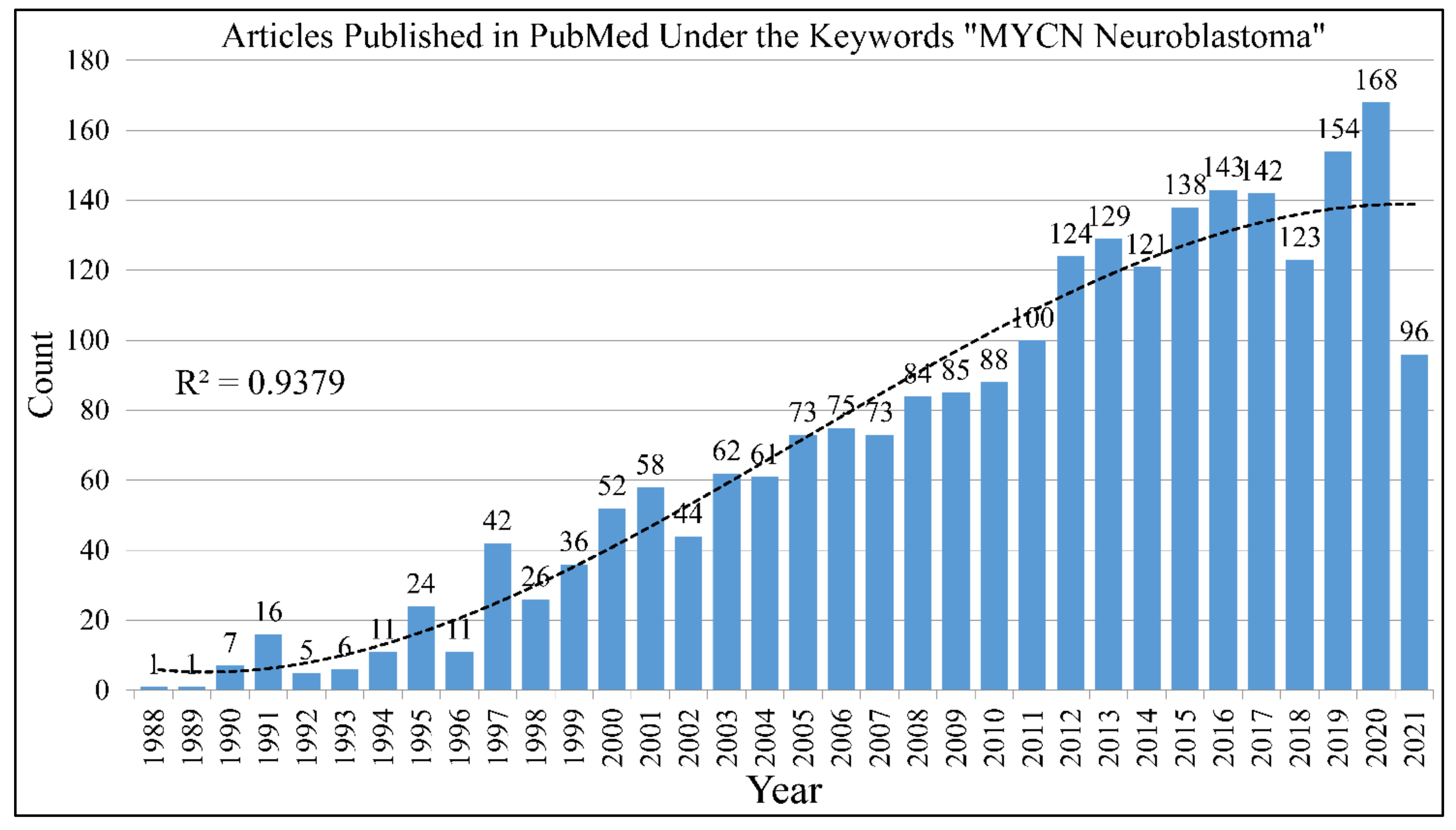
The search with the keywords “MYCN Neuroblastoma” in PubMed returns 2166 results until June 12th 2021 (Figure 1). The gene, is one of the most studied genes for a specific disease. Interest on the MYCN gene and neuroblastoma biology remained constant throughout the years. In its almost 40 year history since its first reports, a lot is still to be learned on its role in neuroblastoma. The study of MYCN for other types of tumors returns much less results, as for example the keywords “MYCN brain tumor” returns 237 hits until 12 June 2021.
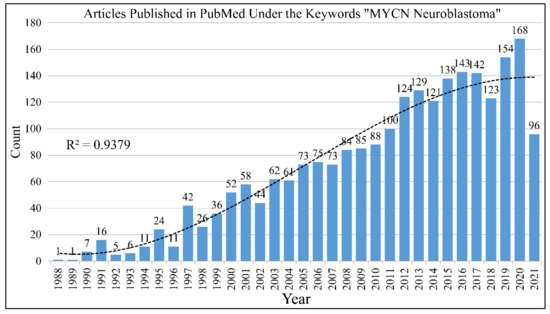
Figure 1.
The literature concerning the role of MYCN in neuroblastoma. A total of 2166 citations come up in the PubMed database with the keywords “MYCN” and “neuroblastoma”.
1.3. A Brief Reference (and History) to Neuroblastoma
Neuroblastoma was discovered in 1910 by Wright, JH (1910), where in his work he referred to the tumor as “neurocytoma” or “neuroblastoma”. In particular, he referred to neuroblastoma as “The essential cells of the tumor are considered to be more or less undifferentiated nerve cells or neurocytes or neuroblasts, and hence the names neurocytoma and neuroblastoma” [26]. Knowledge on the nature and biology of neuroblastoma increased drastically since that time. Prognosis and therapy have greatly improves although in many cases neuroblastoma is still fatal.
Neuroblastomas afford the most common extracranial solid tumors of childhood [27]. They commonly occur during the first five years of life and may arise during infancy. Potential localizations include the sympathetic nervous system and occasionally the brain, but they are also common in the abdomen; most cases arise in either the adrenal medulla or the peritoneal sympathetic ganglia. Ganglion cell tumors, neuroblastoma and ganglioneuroma, are more common mainly in childhood, but their clinical manifestation vary.
These neoplasms occur most commonly during the first five years of life and may arise during infancy. Most neuroblastomas are sporadic, although familial cases occur as well. Neuroblastomas may occur anywhere in the sympathetic nervous system and occasionally within the brain, but they are also common in the abdomen; most cases arise in either the adrenal medulla or the peritoneal sympathetic ganglia. The adrenal medulla derives from the neural crest. It contains two major cell types chromaffin and ganglion cells. Chromaffin cells (pheochromocytes) are the predominant medullary cells. They are postganglionic sympathetic neurons that have lost their axons and dendrites. They synthesize and release their catecholamines (epinephrine or norepinephrine) in response to neural stimulation (especially stress) mediated by preganglionic sympathetic neurons. The few parasympathetic ganglion cells present exhibit typical morphological characteristics of autonomic ganglion cells.
Under normal circumstances, as aforementioned, chromaffin cells produce two types of catecholamines in response to pregaglionic sympathetic stimulation (e.g., stress). Both elevate blood glucose by stimulating hepatic glycogenolysis. They also increase blood flow to the heart. Epinephrine increases the heart rate and dilates blood vessels to the organs needed to escape stress, such as cardiac and skeletal muscles. It dilates bronchioles and constricts the vessels in organs (e.g., skin, digestive tract, kidneys) that are not essentials in reacting to stress. Norepinephrine also constricts blood vessels in non-essential organs increasing peripheral resistance and as a result it increases blood pressure and blood flow to the heart, brain and skeletal muscles.
Under abnormal function, hypersecreting chromaffin cell tumors (pheochromocytomas) cause a sustained stress response (especially hypertension) even in the absence of stress. While neuroblastoma is a highly malignant tumor of early life (related by maturation to ganglioneuroma), the other principal tumor of the sympathetic nervous system, pheochromocytoma, is usually a benign tumor of adults, unrelated to either neuroblastoma or ganglioneuroma [28]. Ganglion cell tumors, neuroblastoma and ganglioneuroma, are more common, mainly in childhood, but their clinical manifestation vary.
1.3.1. Clinical Characteristics of Neuroblastoma
Clinical features are extremely variable and reflect the widespread distribution of neural crest tissue. Metastases are common at diagnosis, approximately half of newly diagnosed patients have distant metastases, and often cause the symptoms that lead to the diagnosis of neuroblastoma. These symptoms are divided into specific and nonspecific. Remote effects are occasionally seen. There are three most widely used neuroblastoma staging systems: Evans, Pediatric Oncology Group (POG) and TNM- Union Internationale Contre le Cancer (UICC) [29].
Tumor markers are extremely useful in evaluating children who have neuroblastoma. Concerning urinary markers, catecholamines, are elaborated by most tumors are useful for diagnosis, follow up on response to therapy, and detection of recurrence. On the other hand, serum markers are used, whom elevations are often associated with a poor prognosis. Such markers are ferritin and lactate dehydrogenase. Last but not least, MYCN represents an oncogene marker whose amplification within the tumor cells is also associated with poor prognosis [30]. MYCN oncogene is a member of myc family of oncogenes that encode transcription factors regulating cell growth. The additional copies of this oncogene contribute to the rapid growth of the neuroblastomas that have MYCN amplification and thus poor prognosis [31]. Prognosis depends on the following factors: age, stage, histopathology (degree of differentiation of tumor cells and their pattern of growth) and tumor markers [32].
Therapy includes surgery, aggressive multi-agent chemotherapy treatment, especially if the patient has poor prognostic features such as MYCN amplification and autologous bone marrow transplantation according to the stage diagnosed. In addition, spontaneous regression without any therapy is common in a particular stage [33].
1.3.2. Oncogenetic Mechanisms in Neuroblastoma
Cell cycle is divided in four phases, designated G1, S (when DNA synthesis occurs), G2 and M (mitosis, when the cell actually divides). Cells that have ceased dividing (permanently or temporarily) are said to be in a resting phase called G0. The ordered progression from one phase to the next is coordinated by a complex set of proteins, and the cellular signals that control cell division do so by modulating the activities of those proteins which include cyclins, CDKs (cyclin-dependent kinases), CDKIs (CDK inhibitors) [34]. Progression through G1, is also marked by the accumulation of other transcription factors, which presumably act on other genes required for DNA replication. Several of these factors are regulated by association with other proteins.
The ultimate stage of the signal transduction pathway is regulation of DNA transcription in the nucleus. Components of the signal transduction factors that regulate the activity of specific genes whose proteins products influence cellular growth and proliferation. Genes that encode these transcription factors include Myc, Fos and Jun. Mutations may occur in any of these steps involved in regulation of cell growth and differentiation. Accumulation of such mutations within a cell lineage may result in progressive deregulation of growth eventually producing a tumor cell. Proto-oncogenes encode products that control cell growth and differentiation. When mutated, they may become oncogenes, which can cause cancer. Most oncogenes act as dominant gain-of-function mutations that lead to deregulation of cell cycle control [35]. In contrast to tumor suppressor genes, most oncogenes do not exhibit germline mutations that cause inherited cancer syndromes [36]. Instead, somatic mutations are observed that lead to sporadic cancers.
Retroviruses are a type of RNA virus that is capable of using reverse transcriptase to transcribe RNA into DNA. In this way, the RNA genome of the retrovirus is converted to DNA, which can be inserted into a chromosome of a host cell. In an earlier cycle of infection, a retrovirus may have incorporated in a mutant oncogene from the genome of the host. When the retrovirus invades a new cell, it can transfer the oncogene in the genome of the new host, thus transforming the cell. Transforming retroviruses have also identified the nuclear transcription factor genes Myc, Jun and Fos, as other molecular components capable of initiating cell transformation. The transcription factor gene associated with neuroblastoma is the oncogene MYCN that encodes a DNA-binding protein and is located on the 2p24 chromosome [37]. A recent study showed that in neuroblastoma the human endogenous retroviruses was abundant, indicating a possible role in the ontogenesis of the disease [38]. Interestingly, another recent report has shown that enterovirii responsible for other conditions, are implicated in the nerve tissue and possibly in tumorigenesis, including neuroblastoma [39]. The role of viral genome in neuroblastoma is still under investigation and consists of an active research field.
Myc protein represents a specific example of HLH (helix–loop–helix) proteins which consist of short alpha helix connected by a loop to a longer alpha helix. The loop allows dimerization of two HLH proteins to occur and form a Y-shaped dimer. Dimerization may occur between two of the same proteins (homodimers) or two different proteins (heterodimers) [40].
Differences in the level of expression of encoded proteins can, among others, cause proto-oncogenes to become oncogenic. In childhood neuroblastoma, the amount of the MYCN gene is increased from a diploid number to many dozens per cell. MYCN amplification is associated with poor prognosis especially in older children than infants and is a significant factor in the aggressiveness of the tumor [41].
An older DNA microarray analysis suggested there is a link between DNA methylation and MYCN gene expression, i.e., there is evidence DNA methylation may directly control MYCN oncoprotein levels [42]. Epigenetic silencing of potential tumor suppressor genes as an alternative mechanism in the absence of genetic mutations has not yet been studied systematically in neuroblastomas and needs further investigation.
In contrast to MYCN gene amplification, the degree of expression of the MYCN gene in the tumor does not predict prognosis [43,44], but it has been previously shown that the MYCN product (MYCN) is a nuclear phosphoprotein, which can transcriptionally activate many genes, either directly (e.g., ID2) or indirectly [45]. In childhood neuroblastoma, the amount of the MYCN gene is increased from a diploid number to many dozens per cell. MYCN amplification has been associated with poor prognosis especially in older children when compared to infants, while it affords a significant factor for tumor aggressiveness [41].
1.3.3. Prognosis of Neuroblastoma
Since the advent of MYCN discovery, it is directly linked to the disease’s prognosis. Up-to-date, several factors have been added to the criteria for neuroblastoma’s prognosis. A recent study highlighted that exportin-T overexpression, is an independent poor prognosis factor in MYCN amplified neuroblastoma [46]. Further on, the most recent neuroblastoma risk classification system indicated that the presence of image defined risk factors (IDRF), MYCN amplification and age are important prognostic factors related to poor prognosis [47]. Another prognostic factor recently mentioned was the overexpression of c-myc, which was confirmed to be linked to poor prognosis independently of MYCN amplification [48]. Further on, in MYCN non-amplified neuroblastoma, the DST gene was linked to good prognosis, with patients overexpressing DST manifesting higher survival rates [49]. In cases of metastatic neuroblastoma (those with bone marrow metastasis) aberrations in chromosome 10, along with MYCN amplification [22]. Finally, an interesting recent study indicated the role of the microenvironment in neuroblastoma prognosis. In particular, it has been reported that low expression of IL5 and NKT in the tumor neuroblastoma, was linked to poor outcome in MYCN non-amplified tumors [50].
1.4. Scope of the Present Work
In the present work, we review the literature for the significance of MYCN, in neuroblastoma. We present old and new knowledge on the prognostic, diagnostic and therapeutic properties of MYCN in neuroblastoma.
2. The MYCN Gene
The Myc genes consist of a family of regulator genes and proto-oncogenes whose products are transcription factors. The Myc family consists of “three related human genes: c-myc (MYC), l-myc (MYCL), and n-myc (MYCN)”. MYC was the first gene of this family to be discovered. Its name was derived from the v-myc gene, which was the first gene to be discovered in this family. The term c-myc, was given due to the homology of c-myc to the v-myc gene, which is of viral origin. The v-myc gene is an oncogene present in the avian myelocytomatosis virus, as well as a human oncogene upregulated in various tumors. Following the discovery of c-myc other members of this family were discovered and named n-Myc and l-Myc [51].
2.1. The “Anatomy” of MYCN
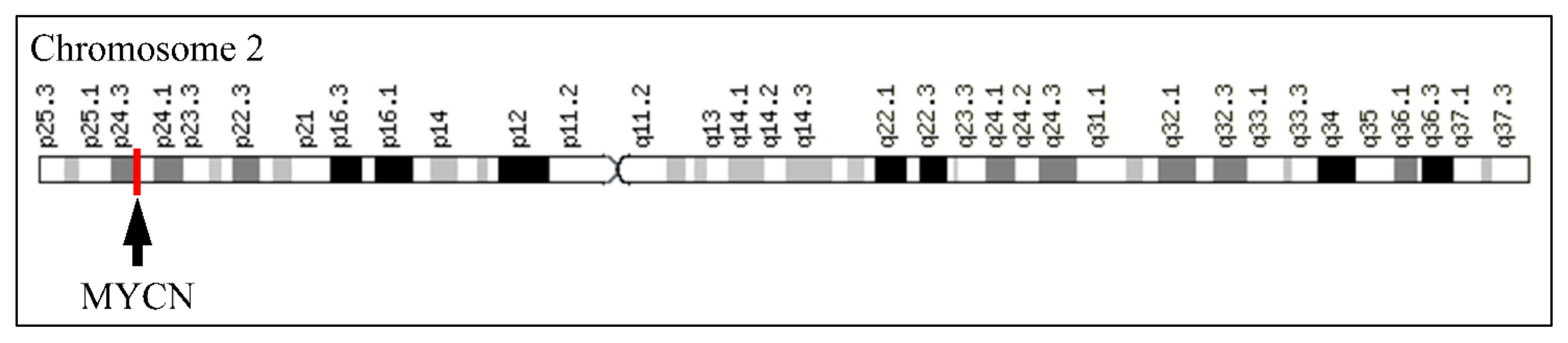
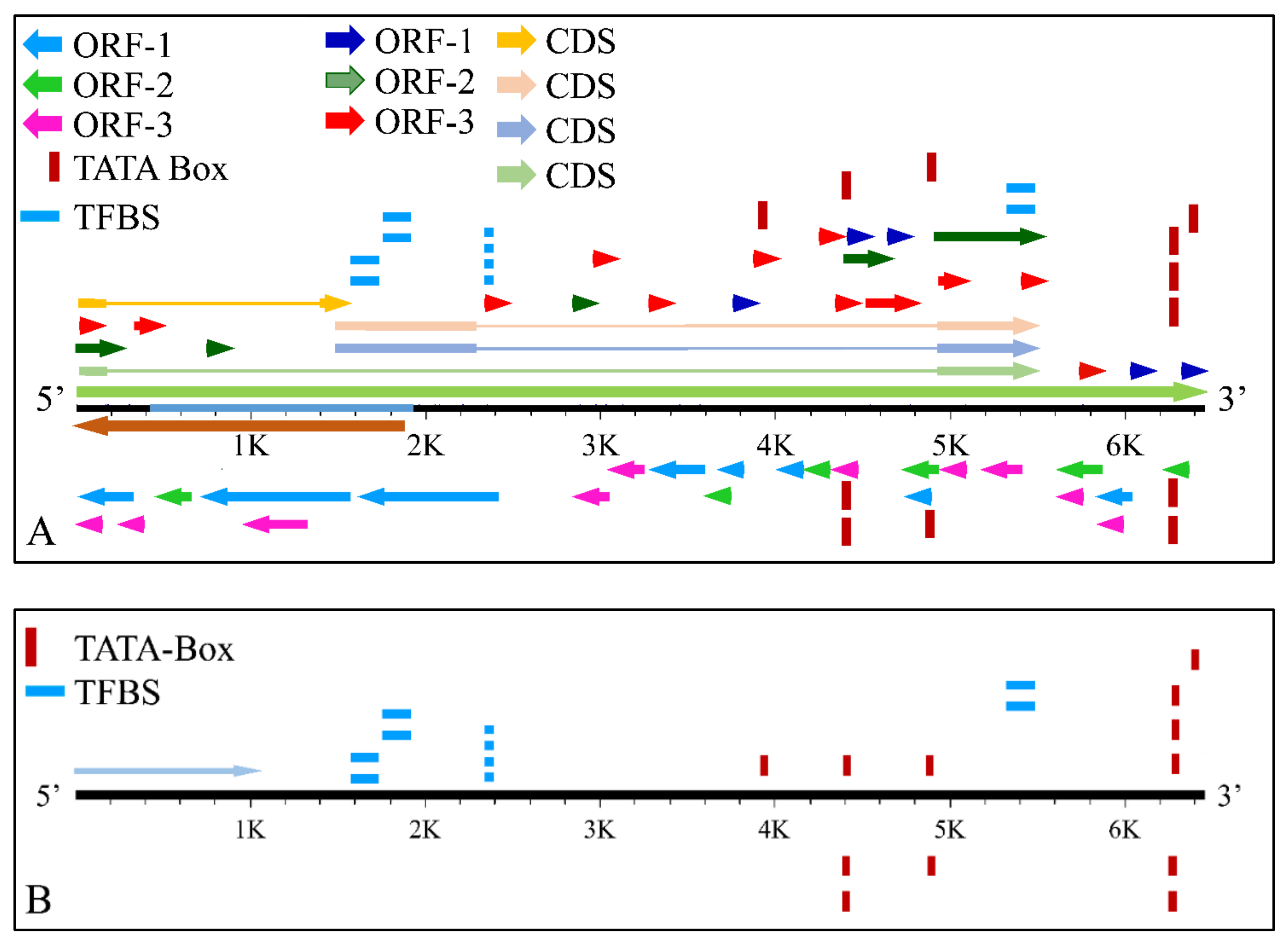
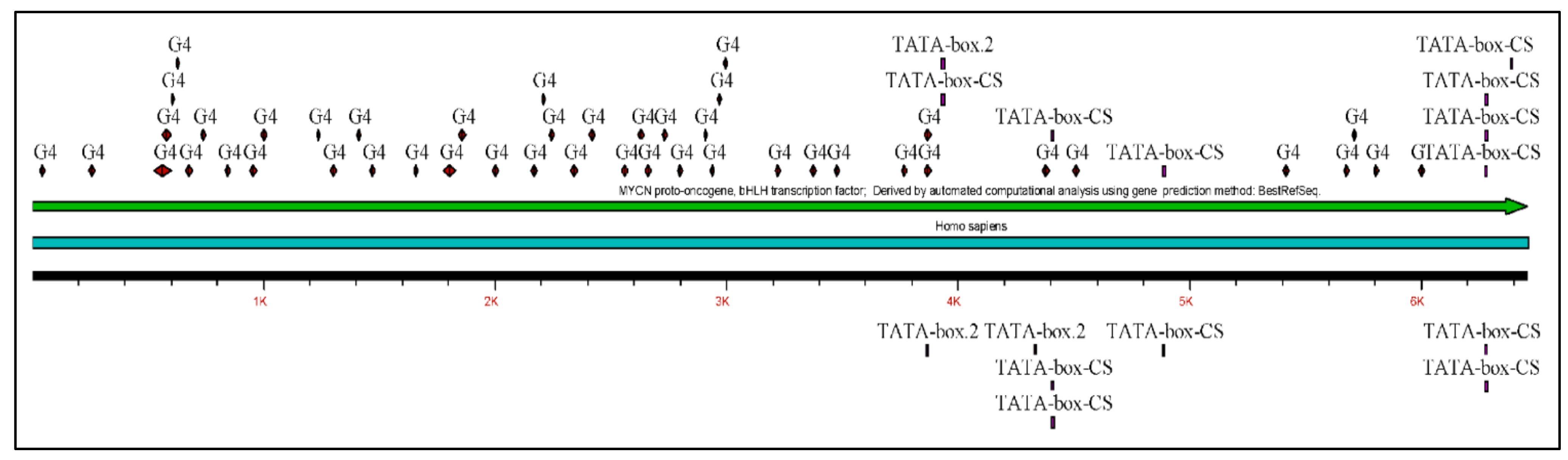
The MYCN, is constructed by 6455 nucleotides and is located on Chromosome 2 with exact location 2p24.3 and has three exons (Figure 2). It can be accessed through the gene ID 4613 (https://www.ncbi.nlm.nih.gov/gene/4613, accessed on 18 June 2021) in the NCBI, Gene database with official name is “MYCN proto-oncogene, bHLH transcription factor”. The complete anatomy of the gene is presented in Figure 3A and Table 1. The gene has twelve TATA-Box sites, which are located between nucleotides 3930 and 6382 (Figure 3B and Table 2).
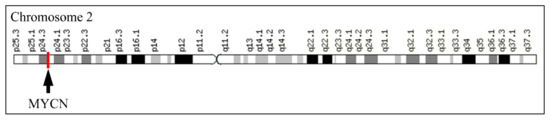
Figure 2.
The location of MYCN on Chromosome 2.
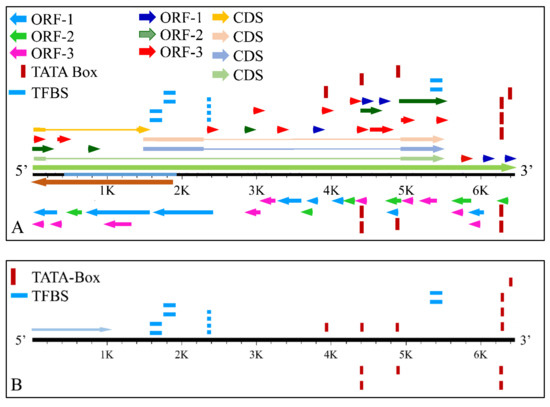
Figure 3.
The analysis of the MYCN gene with its complete anatomy (A) and the TATA boxes (B) (ORF: open reading frame, TFBS: transcription factor binding site, CDS: gene coding region).

Table 1.
The analysis of the MYCN gene (table has been reproduced from the NCBI provided data https://www.ncbi.nlm.nih.gov/gene/4613, accessed on 20 June 2021.).

Table 2.
TATA-Boxes on the MYCN gene.
2.1.1. Transcription Factor Binding Sites
Gene regulation is further controlled by transcription factors, on the so-called transcription factor binding sites (TFBS). TFBS can be predicted either experimentally or bioinformatically. In the present work we have attempted to identify possible TFBS on the MYCN gene using the Webgestalt (http://www.webgestalt.org/, accessed on 20 June 2021) web-based tool [52,53,54,55]. Hence, predicted TFBS were the NF1, E2F, AP4 and FREAC2. Further on, two regions on the gene’s product were identified as potential interacting sites with AURKA and FBXW7 [56], and one site was predicted to be a Leucine-zipper region.
2.1.2. Gene Ontology (GO) Annotation
The gene ontology annotation for the gene reports as functions: “enables DNA binding” [57], “enables DNA-binding transcription activator activity” [58], “RNA polymerase II-specific” [59], “enables DNA-binding transcription factor activity” [60], “enables DNA-binding transcription factor activity, RNA polymerase II-specific” [60], “enables RNA polymerase II cis-regulatory region sequence-specific DNA binding” [58], “enables kinase binding” [56], “enables protein binding” [56,57,61,62,63,64,65,66,67,68,69] and “enables protein dimerization activity” [70]. In addition, the gene’s biological processes are: “involved in branching morphogenesis of an epithelial tube”, “involved in cartilage condensation”, “involved in embryonic digit morphogenesis”, “involved in embryonic skeletal system morphogenesis”, “involved in lung development”, “involved in negative regulation of astrocyte differentiation”, “involved in negative regulation of gene expression” [71], “involved in negative regulation of reactive oxygen species metabolic process”, “involved in positive regulation of cell death”, “involved in positive regulation of gene expression” [72], “involved in positive regulation of mesenchymal cell proliferation”, “involved in positive regulation of production of miRNAs involved in gene silencing by miRNA” [71], “involved in positive regulation of transcription by RNA polymerase II” [58], “involved in positive regulation of transcription, DNA-templated” [73], “involved in regulation of inner ear auditory receptor cell differentiation”, “involved in regulation of transcription by RNA polymerase II” [60] and “involved in regulation of transcription by RNA polymerase II” [59].
2.1.3. G-Quadraplexes in MYCN
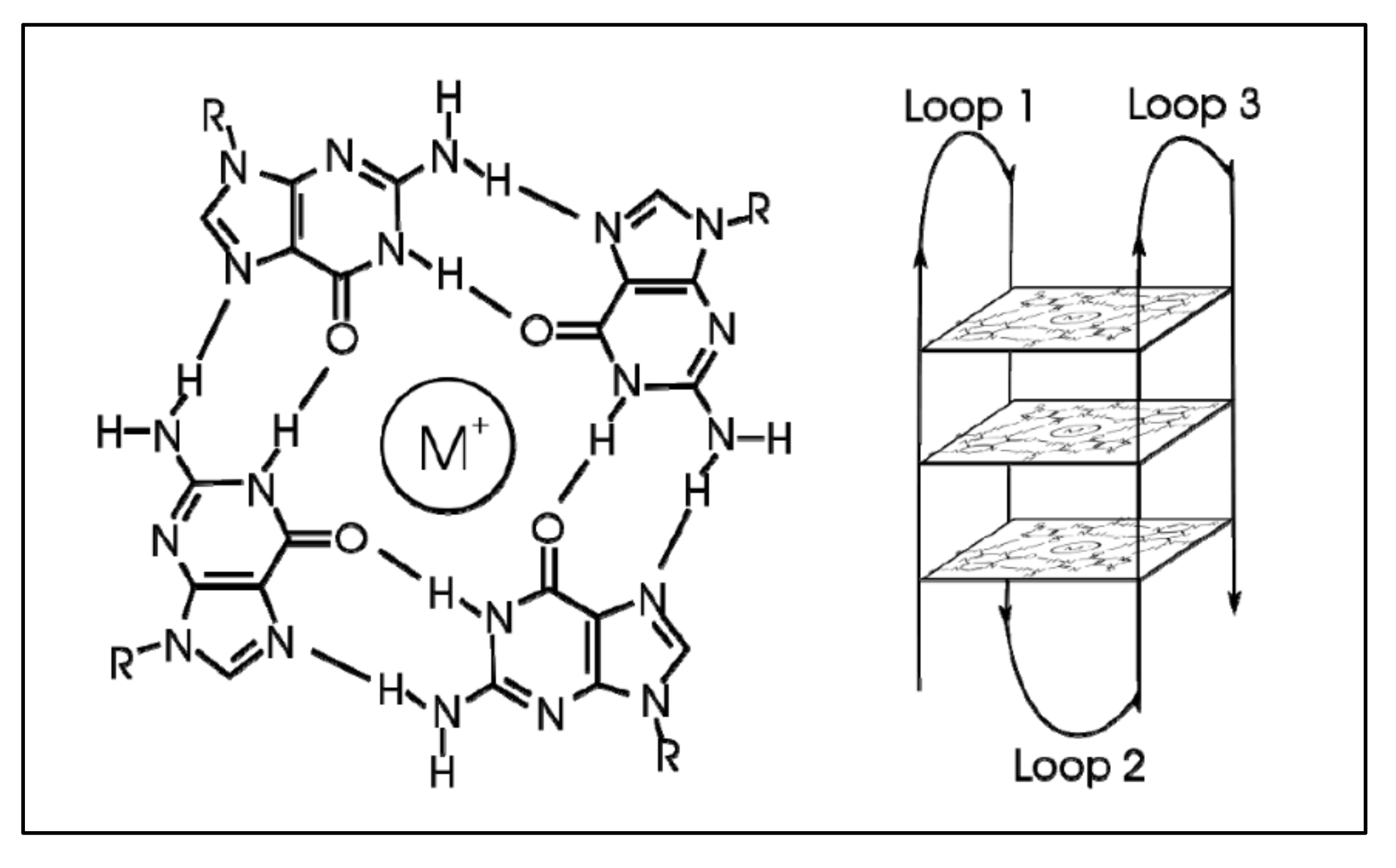
G-quadraplexes were first discovered in the early 1960s, where four-stranded DNA structures were discovered in nucleotide sequences rich in guanines [74]. G-quadruplex are tertiary DNA structures, which are formed in nucleic acid polymers that are rich in guanine [75] (Figure 4). G-quadraplexes can take different shapes, where each one contains “guanine-tetrads”, forming single- [76], double- [77], and quadruple-strands [78]. G-quadraplexes usually form in the telomeric regions, yet they form on oncogene sites [79]. The mechanism through which such a quadruplex forms is known to be through the Hoogsteen hydrogen bonding, creating a square planar or also known as “guanine-tetrad”. When multiple tetrads assemble or “stack” form the G-quadraplex. There are several variations of G-quadraplexes. These different forms can be divided into the intramolecular and intermolecular complexes. The intramolecular complexes are further divided to the parallel, anti-parallel, hybrid and higher order complexes, while the intermolecular complexes are further divided to bimolecular, trimolecular and tetramolecular structures [80]. The quadraplex is stabilized by the presence of a cation, which in most cases is potassium, yet other cations can also participate in quadruplex stabilization. G-quadraplexes can be predicted computationally and even experimentally through crystallization studies.
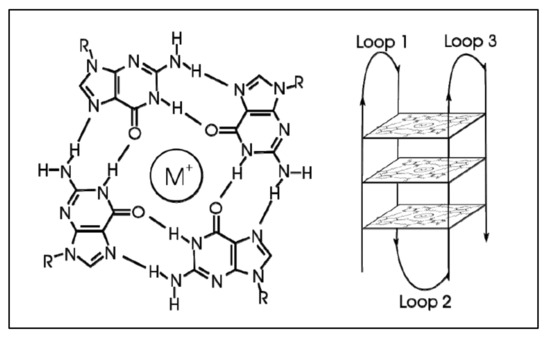
Figure 4.
G-Quadraplex. A tetrad forms with the addition of an ion (left), while multiple tetrads form the quadraplex (https://en.wikipedia.org/wiki/G-quadruplex#cite_note-ReferenceC-4, accessed on 20 June 2021).
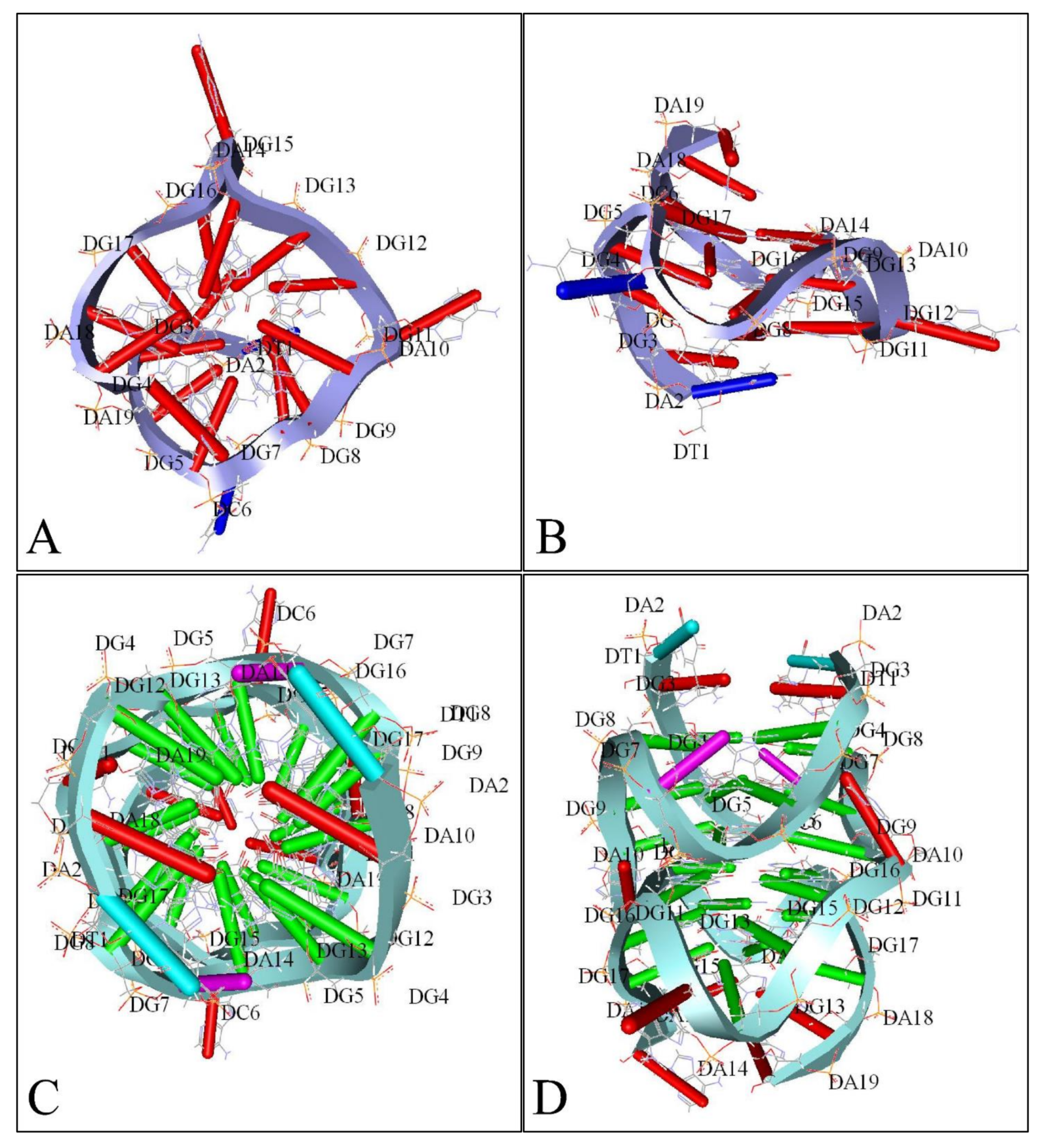
Similarly, several studies (but not many) have investigated the presence of G-quadraplexes in the MYCN gene. We have also investigated the possibility of the presence of G-quadraplexes on the MYCN gene, using the QGRS Mapper [81]. In Figure 5 we provide an overview of the possible sites, where G-quadraplexes can form based on the nucleotide sequence as well as in Figure 6, we have designed a map of G-Quadraplexes on the gene itself. Interestingly, further studies have discovered the 3D structure of MYCN G-quadraplexes. One recent study, has reported on the potential G-quadraplex structures of MYCN (Figure 7), where they have investigated the nucleotide sequences 5′-T-A-G-G-G-C-G-G-G-A-G-G-G-A-G-G-G-A-A-3′ (Figure 7A,B) as well as the nucleotide sequence 5′-T-A-G-G-G-C-G-G-G-A-G-G-G-A-G-G-G-A-A-T-A-G-G-G-C-G-G-G-A-G-G-G-A-G-G-G-A-A-3′ (Figure 7C,D).
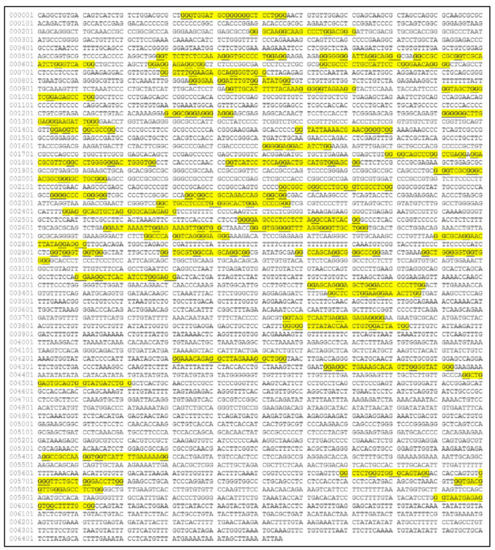
Figure 5.
The predicted G-quadraplex sequences on the MYCN nucleotide sequence (image obtained from the QGRS Mapper web-tool, https://bioinformatics.ramapo.edu/QGRS/index.php, accessed on 21 June 2021. Map was reconstructed using the gene ID 4613 from the NCBI site).
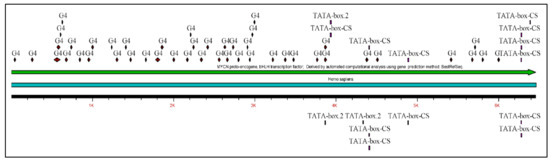
Figure 6.
The reconstructed MYCN map based on the previous predictions from Figure 5 (legend: G4: G-Quadraplex position).
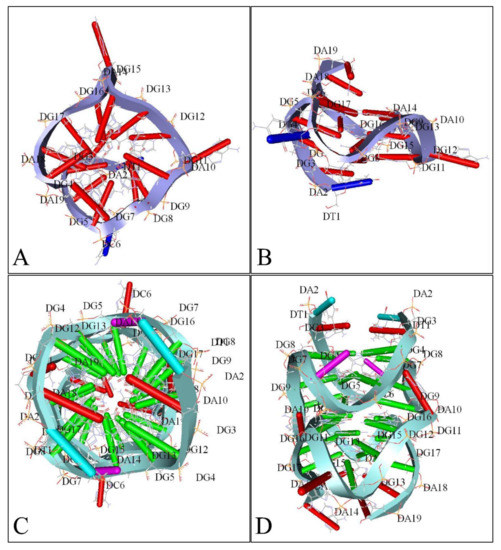
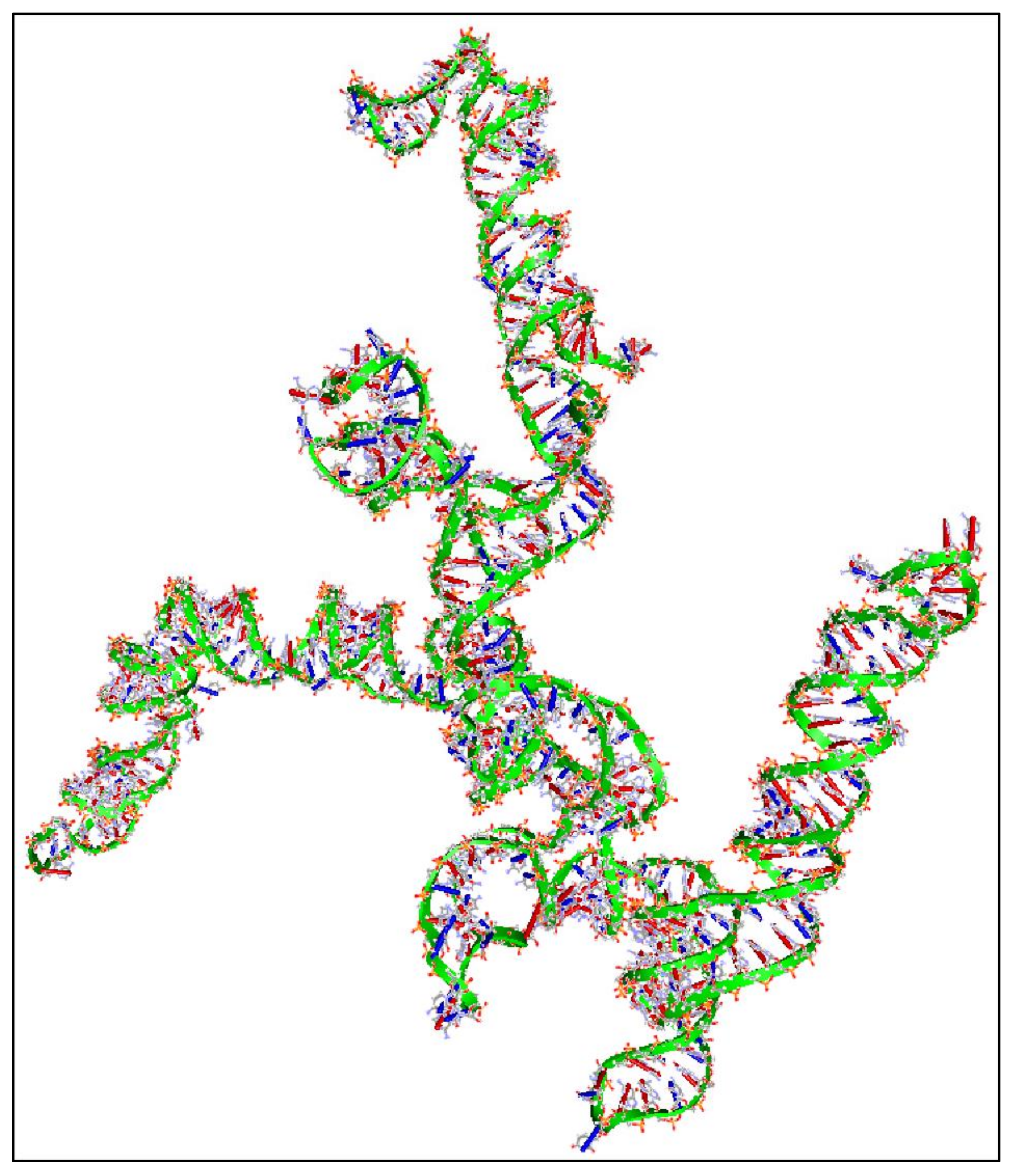
Figure 7.
A 3D reconstruction of MYCN G-Quadraplexes. The transversal (A) and lateral (B) views of the 5′-T-A-G-G-G-C-G-G-G-A-G-G-G-A-G-G-G-A-A-3′nucleotide sequence’s G-quadraplex are presented as depicted from the deposited structure 2LEE (https://www.rcsb.org/structure/2LEE, accessed on 21 June 2021) in the Protein Data Bank (PDB) database (https://www.rcsb.org/, accessed on 21 June 2021) [82]. Similarly, The transversal (C) and lateral (D) views of the 5′-T-A-G-G-G-C-G-G-G-A-G-G-G-A-G-G-G-A-A-T-A-G-G-G-C-G-G-G-A-G-G-G-A-G-G-G-A-A-3′ nucleotide sequence’s G-quadraplex are presented as depicted from the deposited structure 2LED (https://www.rcsb.org/structure/2LED, accessed on 21 June 2021) in the PDB [82] (available structures have been reproduced with the Accelrys Discovery Studio software v.2.5).
The presence of G-quadraplexes in the MYCN gene, plays a functional role, where it has been shown that they are very stable at the promoter site, regulating the gene’s expression [83]. The importance of MYCN G-quadraplexes, with respect to its prognostic, diagnostic and therapeutic use is still under investigation. However, some studies have highlighted that chemotherapeutics are able to recognize G-quadraplexes as for example, the alkaloids tetrandrine and isotetrandrine were able to bind to MYCN G-quadraplex [84]. From the same study, it was shown that tetrandrine had a “high possibility of binding to the MYCN G-quadraplexes” through hydrogen bonding, while isotetrandrine did not manifest the same affinity. However, it appeared that G-quadraplexes could be possible attractive sites for tumor therapy [84]. Similarly, another study showed that Enniatin B, “a well-known antibacterial, antihelmintic, antifungal, herbicidal, and insecticidal compound” [85], manifested high binding affinity to MYCN G-quadraplex implying possible therapeutic effects [86].
2.2. The “Anatomy” of MYCN’s Transcripts
The MYCN gene transcribes to four known transcripts namely the MYCN proto-oncogene, bHLH transcription factor (MYCN), Transcript Variant 1, mRNA (NM_001293228.2, accessed on 20 June 2021), the MYCN proto-oncogene, bHLH transcription factor (MYCN), Transcript Variant 3, mRNA (NM_001293231.2, accessed on 20 June 2021), the MYCN proto-oncogene, bHLH transcription factor (MYCN), Transcript Variant 2, mRNA (NM_001293233.2, accessed on 20 June 2021) and the MYCN proto-oncogene, bHLH transcription factor (MYCN), Transcript Variant 2, mRNA (NM_005378.6, accessed on 20 June 2021).
2.2.1. MYCN Proto-Oncogene, bHLH Transcription Factor (MYCN), Transcript Variant 1, mRNA
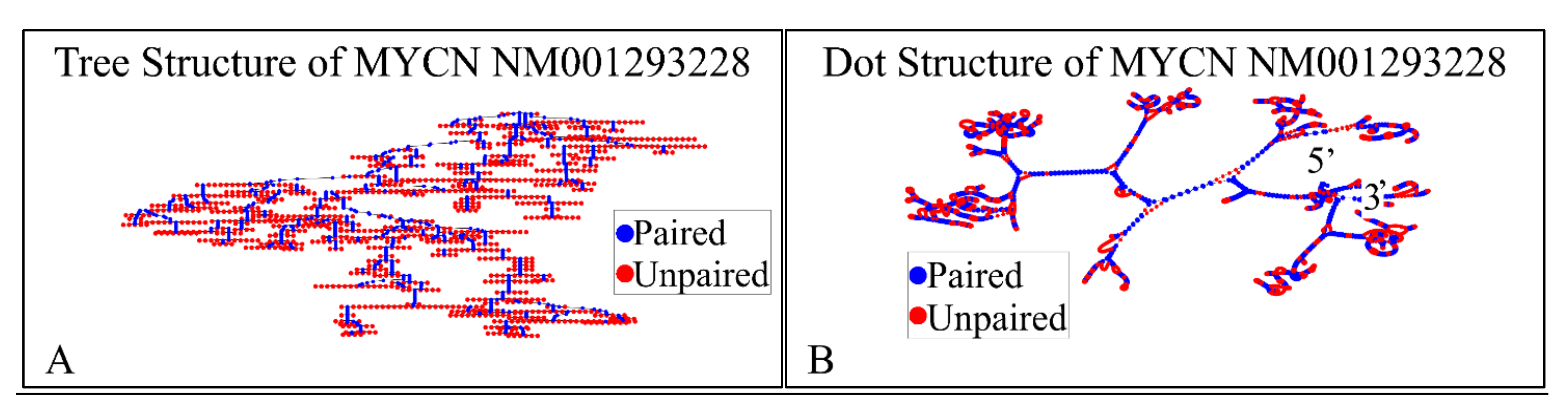
The NM001293228 mRNA, is a 2923 nucleotide long transcript. It consists of three exons, where the first ranges from Nucleotide 1 to 504, the second from Nucleotide 505 to 1411 and the third from Nucleotide 1412 to 2923. The gene coding region is from Nucleotide 622 to 2016. We have also calculated the secondary structure of the transcript, which is shown in Figure 8. There is not known how the secondary structure of this transcript participated in neuroblastoma progression and ontogenesis.
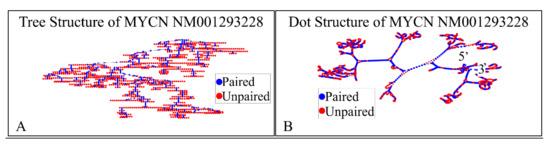
Figure 8.
The predicted secondary structure of the MYCN, NM001293228 transcript. In particular, the tree structure (A) and the dot structure (B) of the transcript are presented (predictions were performed with the Matlab® (The Mathworks, Inc., Natick, MA, USA), using the RNAfold method).
In addition, we have calculated the 3D structure of the first exon of this specific transcript (Nucleotides 1–504). The reason, why we have tested the first 500 nucleotides, was due to the fact that it is computationally very challenging to create 3D predictions of large molecules. Just for reference, the secondary structure prediction of the ~3000 long nucleotide, needed approximately 15 h of computation in i7 8-core computer with 24 GB memory capacity and two 1GB CUDA graphics cards. This points out, not only the difficulty of calculating and predicting the 3D structures of RNAs, but also, and even more, the prediction of their functional properties. Out of pure curiosity, we wanted to examine and visualize how a 3D mRNA would look, and in addition, if we could find something about its functional properties (Figure 9). There are no known properties for the functional role of the 3D MYCN’s RNA structure, which makes it an interesting topic for future research.
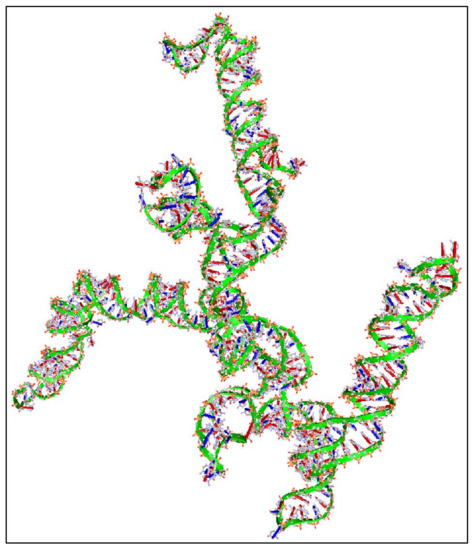
Figure 9.
The predicted 3D structure of the MYCN, NM001293228 transcript and in particular, of the first exon (Nucleotides 1–504) (3D structure was predicted using the RNAcomposer web-tool [87,88] and it was then visualized using the Accelrys Discovery Studio).
2.2.2. MYCN Proto-Oncogene, bHLH Transcription Factor (MYCN), Transcript Variant 3, mRNA
The NM001293231 mRNA, is a 1706 nucleotide long transcript. It consists of two exons, where the first ranges from Nucleotides 1 to 194 and the second from Nucleotides 195 to 1706, while the gene coding region is from Nucleotide 38 to 799. Similarly, we have also calculated the secondary structure of the transcript, which is shown in Figure 10. As in the case of NM001293228, there are no known relations of the secondary structure of this transcript to neuroblastoma progression and ontogenesis or to other molecular functions. Since this transcript was smaller, we were able to model the 3D structures of exon 1 (Figure 11A) as well as part of the CDS region of the transcript (Figure 11B). Similarly to the NM001293228 transcript, the prediction of the secondary structure required around 10 h with the same hardware configuration as aforementioned. There are no known properties for the functional role of the 3D MYCN’s RNA structure, which makes it an interesting topic for future research.
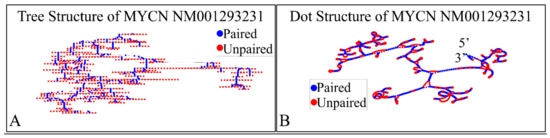
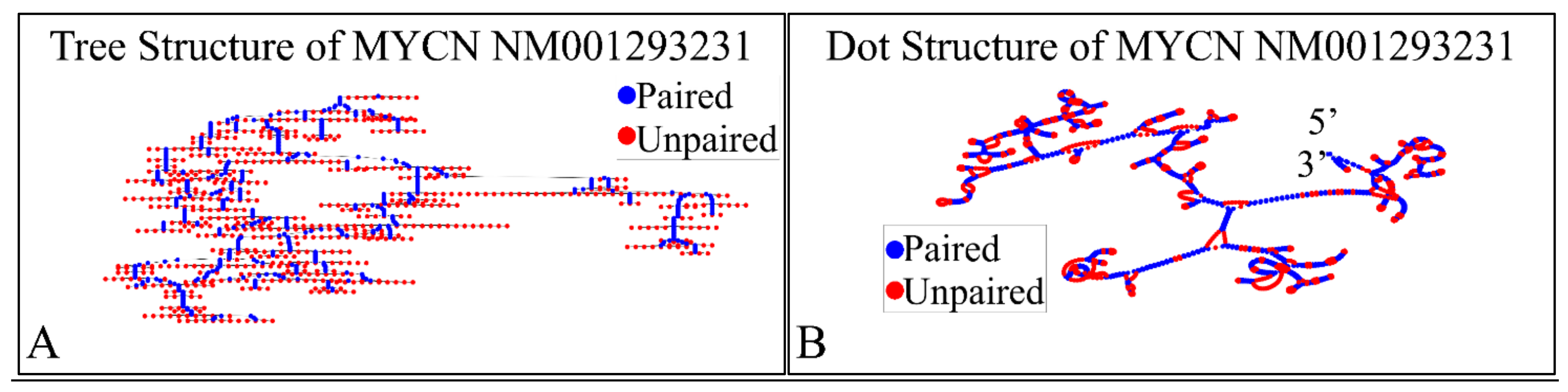
Figure 10.
The predicted secondary structure of the MYCN, NM001293231 transcript. In particular, the tree structure (A) and the dot structure (B) of the transcript are presented (predictions were performed with the Matlab® (The Mathworks, Inc. Natick, MA, USA), using the RNAfold method).
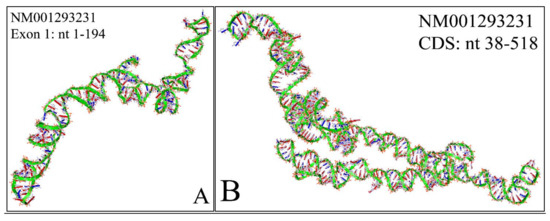
Figure 11.
The predicted 3D structure of the MYCN, NM001293231 transcript and in particular, of the first exon (Nucleotides 1–194) (A) and the CDS (Coding DNA Sequence) sequence (nucleotides 38-518) (B) (3D structure was predicted using the RNAcomposer web-tool [87,88] and it was then visualized using the Accelrys Discovery Studio).
2.2.3. MYCN Proto-Oncogene, bHLH Transcription Factor (MYCN), Transcript Variant 2, mRNA
The NM001293233 mRNA, is a 2613 nucleotide long transcript. It consists of three exons, where the first ranges from Nucleotide 1 to 194, the second from Nucleotide 195 to 1101 and the third from Nucleotide 1102 to 2613. The gene coding region is from nucleotide 38 to 376. Similarly, we have also calculated the secondary structure of the transcript, which is shown in Figure 12. As in the case of the previous transcripts, there are no known relations of the secondary structure of this transcript to neuroblastoma progression and ontogenesis or to other molecular functions. In the case of this transcript the CDS region is 339 nucleotides long and we were able to predict the complete 3D structure of it (Figure 13). There are no known properties for the functional role of the 3D MYCN’s RNA structure, which makes it an interesting topic for future research. In addition, this variant lacks the Segment 1b in the 5′ region, compared to Variant 1 [89]. This variant has two ORFs, the MYCNOT, which is translated from the upstream ORF. The MYCNOT has been found to be a long non-coding RNA embedded within the MYCN gene [90]. Further on, this variant (along with the following NM005378 variant) are suspected to participate in the G-Quadraplex structures, formed by the MYCN gene, and thus probably regulating its functions in cell physiology. A recent report has highlighted that this transcript variant probably is responsible for the MYCN mRNA expression levels, as in several cases MYCN amplification, is not correlated to high mRNA levels [91]. However, up-to-date its function is still not well-defined, but there is a hint of its role in anti-apoptosis, which is in agreement to the overall role of MYCN in neuroblastoma.
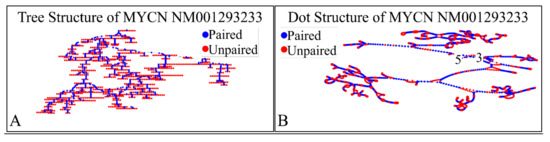
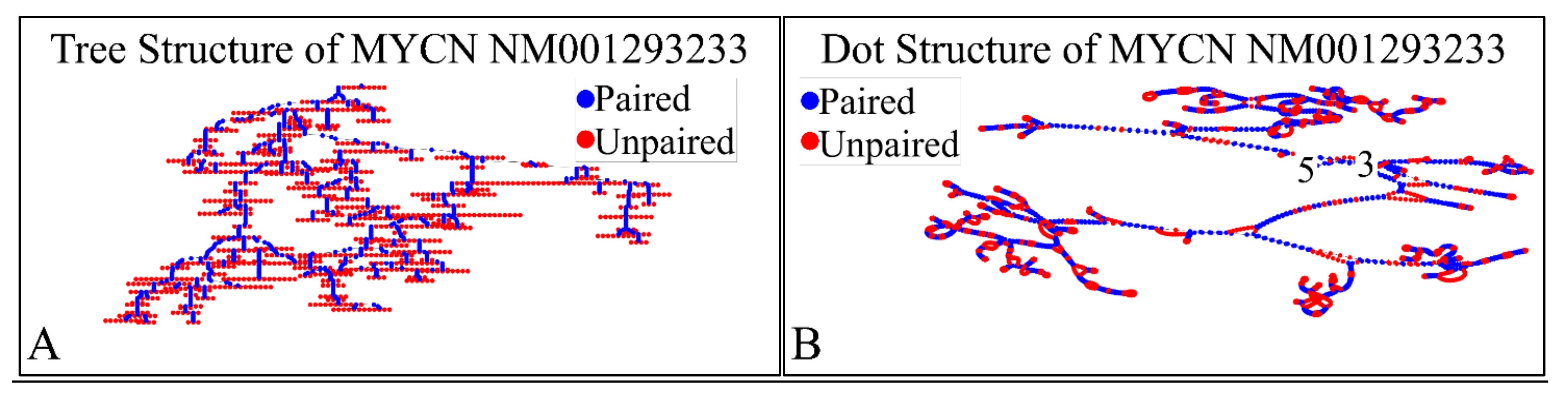
Figure 12.
The predicted secondary structure of the MYCN, NM001293233 transcript. In particular, the tree structure (A) and the dot structure (B) of the transcript are presented (predictions were performed with the Matlab® (The Mathworks, Inc. Natick, MA, USA), using the RNAfold method).
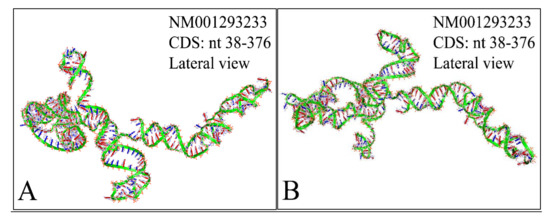
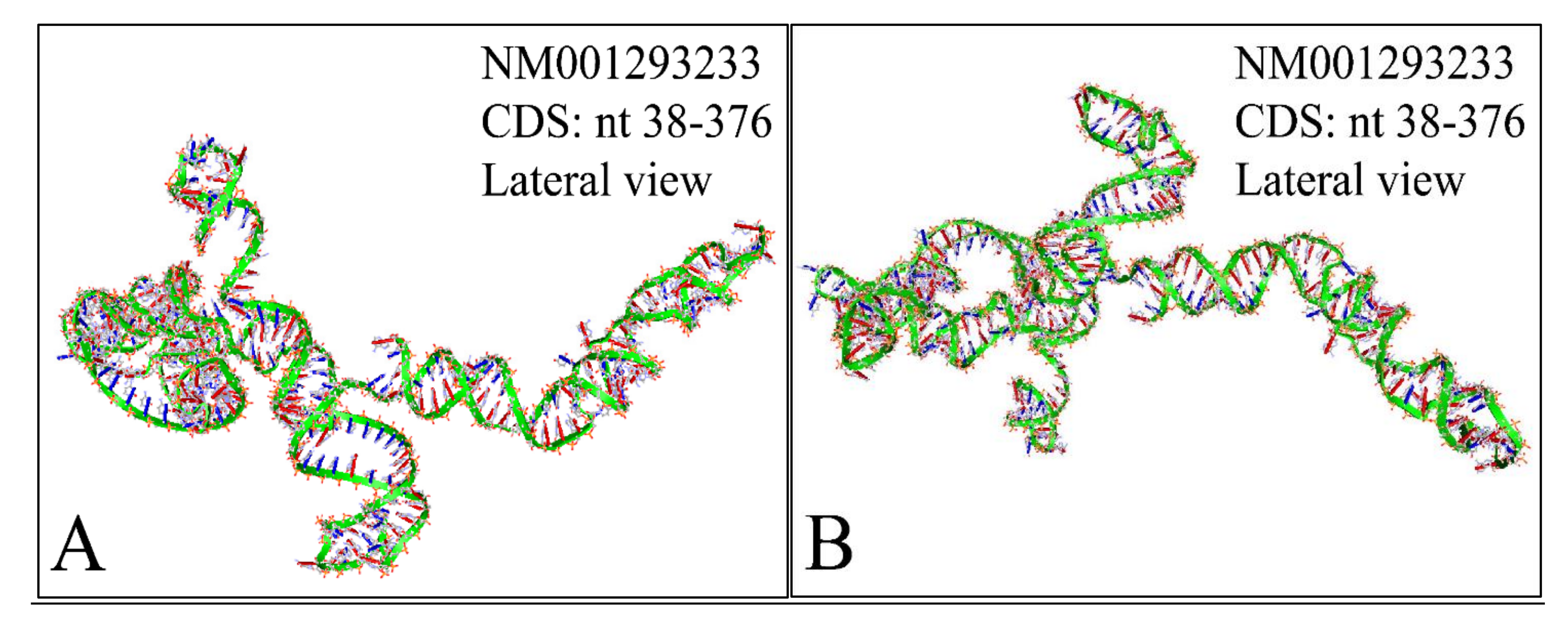
Figure 13.
The predicted 3D structure of the MYCN, NM001293233 CDS region (nucleotides 38–376). The proximal (A) and the distal (B) lateral views are presented (3D structure was predicted using the RNAcomposer web-tool [87,88] and it was then visualized using the Accelrys Discovery Studio).
2.2.4. MYCN Proto-Oncogene, bHLH Transcription Factor (MYCN), Transcript Variant 2, mRNA
The NM005378 mRNA, is a 2613 nucleotide long transcript. It is similar to the NM001293233 transcript and consists of three exons, where the first ranges from Nucleotide 1 to 194, the second from nucleotide 195 to 1101 and the third from Nucleotide 1102 to 2613. The gene coding region is from nucleotide 312 to 1706. As in the case of the NM001293233 transcript, this also lacks segment 1b in the 5′ region. It includes two ORFs, where the one variant is translated from the downstream ORF, encoding the same isoform 1. The function of this transcript is unknown, and it is still under investigation. Overall, the transcription regulation of MYCN is considered a very complex phenomenon, since the MYCN transcripts have a very short half-life, making them difficult to study [92].
2.3. The “Anatomy” of MYCN’s Protein
The MYCN product (MYCN) is a nuclear phosphoprotein, which can activate transcriptionally many genes, either directly (e.g., ID2) or indirectly [45]. A recent DNA microarray analysis suggested there is a link between DNA methylation and MYCN gene expression i.e., there is evidence DNA methylation may directly control MYCN oncoprotein levels [42]. Epigenetic silencing of potential tumor suppressor genes as an alternative mechanism in the absence of genetic mutations has not yet been studied systematically in neuroblastomas and needs further investigation.
Each of the aforementioned transcripts is registered to provide variants of the MYCN protein. In particular, the NM001293228 produces the NP_001280157.1 N-myc proto-oncogene protein isoform 1, the NM001293231 transcript produces the NP_001280160.1 N-myc proto-oncogene Protein Isoform 2, the NM001293233 transcript produces the NP_001280162.1 N-myc proto-oncogene Protein Isoform 3 and the NM005378 transcript produces the NP_005369.2 N-myc proto-oncogene Protein Isoform 1. There is one reference to the solved structure of the MYCN protein (Figure 14) [73]. This is a member of the MYC family and encodes a protein with a basic helix–loop–helix (bHLH) domain. This protein is located in the nucleus and must dimerize with another bHLH protein in order to bind DNA. Multiple alternatively spliced transcript variants encoding different isoforms have been found for this gene. The NP001280157 variant represents the full-length transcript. “Its Exon 1 includes Segments 1a and 1b (also known as Exon 1a and Exon 1b) encoding Isoform 1” (Provided by https://www.ncbi.nlm.nih.gov/protein/NP_001280157.1, accessed on 5 July 2021) [89]. The NP001280160 variant (transcript variant 3) “lacks Segment 1b and Exon 2, which results in an upstream AUG start codon, as compared to Variant 1. The resulting isoform (Isoform 2) has a shorter and distinct N-terminus, compared to Isoform 1” (provided by https://www.ncbi.nlm.nih.gov/protein/NP_001280160.1, accessed on 5 July 2021). On the other hand, the NP001280162 variant (Transcript Variant 2) lacks Segment 1b in the 5′ region, compared to Variant 1. “This variant includes two open reading frames; the isoform (3, also known as MYCNOT [89]) represented by NM001293233is translated from the upstream open reading frame. The Isoform 3 has an identical N-terminus to that of the Isoform 2, and the function of the Isoform 3 is currently unknown” (provided by https://www.ncbi.nlm.nih.gov/protein/NP_001280162.1, accessed on 5 July 2021). Finally, the NP005369 isoform is derived from the Transcript Variant 2 and lacks Segment 1b in the 5′ region, compared to Variant 1. “This variant includes two open reading frames; Isoform 1 represented by the NM005378 transcript, is translated from the downstream open reading frame. This transcript and Variant 1 encode the same Isoform 1” (provided by https://www.ncbi.nlm.nih.gov/protein/NP_005369.2, accessed on 5 July 2021).
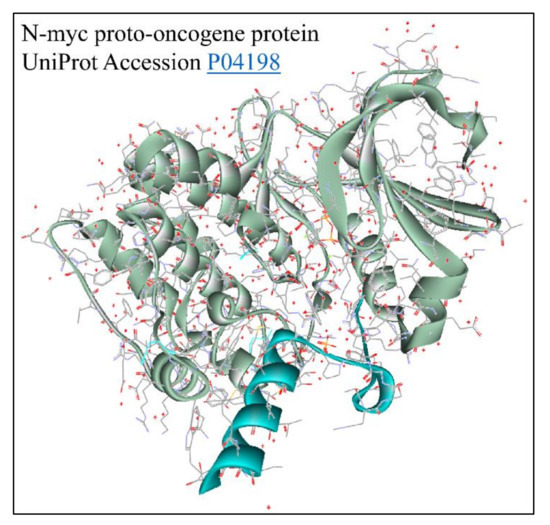
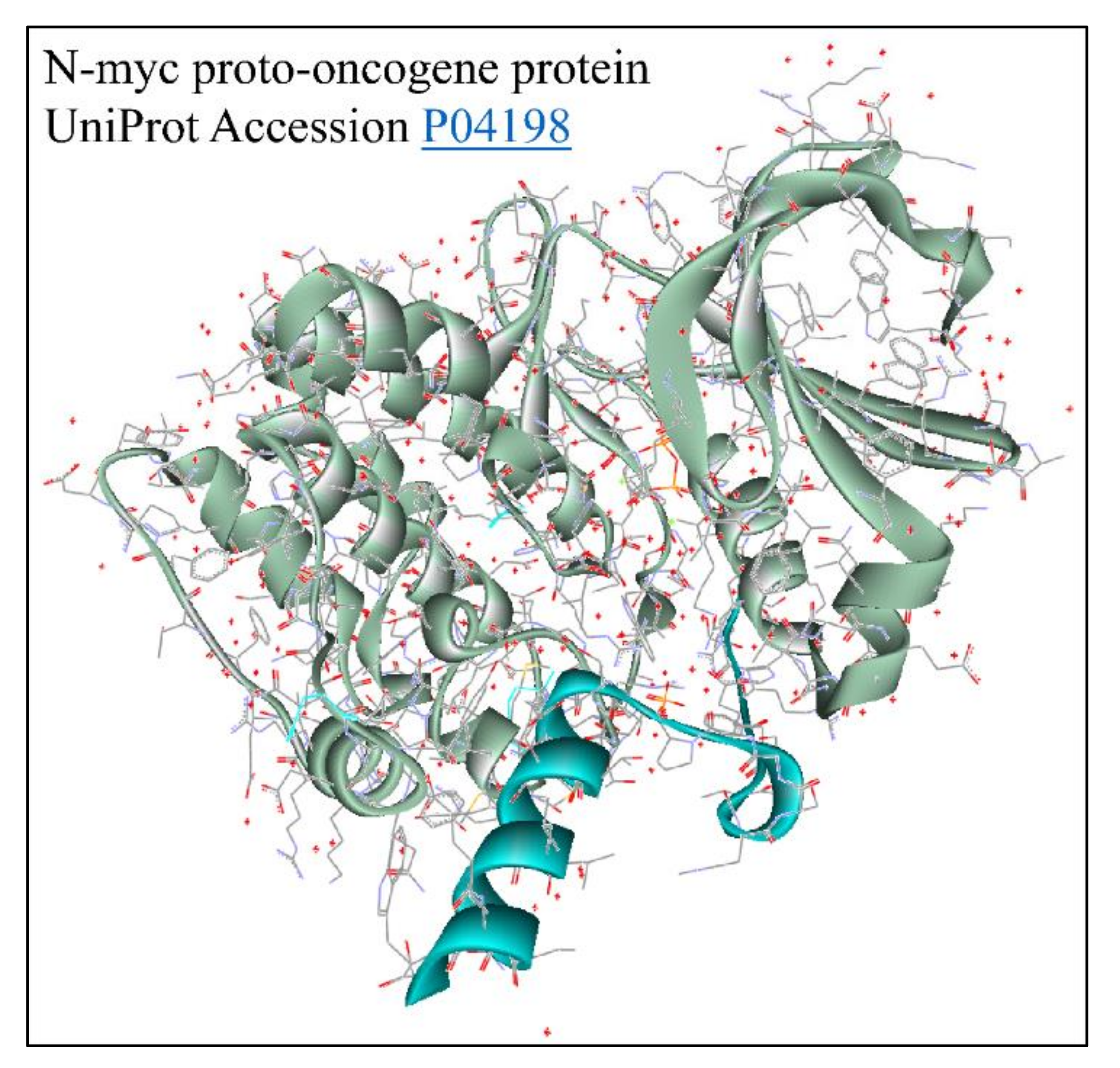
Figure 14.
The predicted 3D structure of the MYCN protein with UniProt accession Nr. P04198 [73] (protein was visualized with Accelrys Discovery Studio).
2.4. The Cellular Functions of MYCN
One of the first discovered functions of MYCN, was its role in cell cycle regulation. As the cell cycle is a tightly regulated process, MYCN has been found to play a significant role in its regulation. It has been reported that MYCN amplification is linked to the failure of cells to arrest in the G1 phase [93]. This is facilitated by the inhibition of PI3K, which downregulates MYCN protein levels, lowering proliferation and leading to a reduction in S- and M phase cells [94]. Along with PI3K regulation, reduced MYCN expression is correlated to other cell cycle regulators such as the CDK inhibitor p27, E2 factor (E2F) and inhibitor of differentiation 2 (ID2). P27 is known to be upregulated in case of MYCN inhibition, which in turn reduces CDK levels and thus G1-arrest inhibition [95]. Thus, MYCN amplification is directly linked to the aberrant enhancement of cell cycle.
Further on, MYCN has been found to play a “dual role” in the regulation of apoptosis [96]. Recent reports have indicated that MYCN is implicated in the upregulation of the “pro-apoptotic regulator phorbol-12-myristate-13-acetate-induced protein 1” (NOXA). MYCN does not induce apoptosis directly, yet it sensitizes the cell to be more responsive to cytotoxic agents [97]. Therefore, it is known that MYCN inactivation, leads to tumor regression through proliferation arrest and the induction of apoptosis [96]. Interestingly, MYCN is involved in the control of chromatic acetylation levels in the cell, which has been exploited as a therapeutic target since the HDAC inhibitor suberoylanilide hydroxamic acid (SAHA), significantly reduced MYCN levels in MYCN-amplified cells, leading to induction of apoptosis [98].
As the regulation of cell cycle and apoptosis, were expected to be the “theatrical stage” for MYCN, metabolism is the prime highlight. As cancer cells require large amounts of energy, they rely to glycolysis for their energy resources, yet another way of surviving comes from glutamine catabolism [99]. Tumors with amplified MYCN have been shown to manifest increased glutamine transport and glutamate metabolism. This has been exploited as a potential therapeutic intervention, where glutamine deprivation in neuroblastoma led to increased cell death [100].
The role of MYCN in cellular physiology is well-studied and there are numerous reports concerning this topic. Yet, it becomes apparent that there is still much to be learned fort its biology.
3. Detection of the MYCN Gene
Neuroblastoma tumors show remarkable biological heterogeneity. Therefore, to predict the biological behavior of an individual tumor several parameters have been proposed to predict the prognosis of neuroblastoma patients. These include DNA ploidy and deletion of the short arm of Chromosome 1, MYCN gene amplification and TrkA expression and telomerase activity [101]. The use of molecular analysis as a prognostic factor relies on the simplicity, reliability and the rapidity of the chosen procedure. The detection of gene amplification can be carried out using the fluorescence in situ hybridization (FISH) [102] and Southern Blotting (SB) techniques, both of which require a significant amount of high quality DNA and several days in order to obtain the results. Sometimes, the samples obtained by aspiration or biopsy, are small and other times archival paraffin embedded tissue may be used, making it difficult to obtain enough quantity/quality of DNA. In cases where the assay is required in order to determine the appropriate therapeutic regimen the above techniques are unsuitable since they are not rapid.
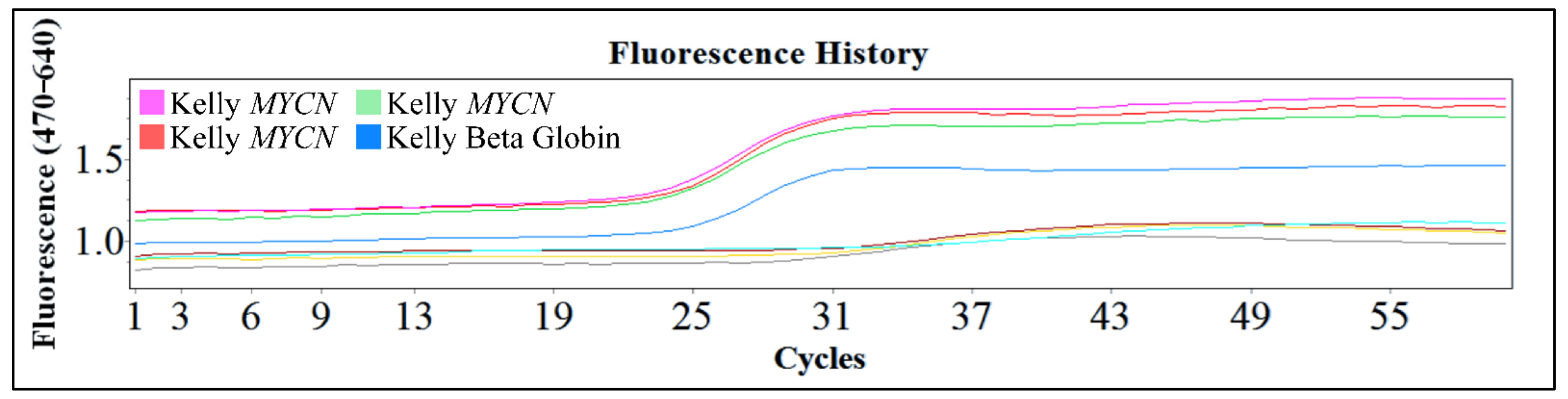
One of the popular methods used is the polymerase chain reaction (PCR) and the reverse transcription PCR (RT-PCR). These are powerful procedures for the amplification of small amounts of DNA or mRNA respectively, for molecular analysis. These procedures are efficient since they require only small amounts of sample, they are rapid and in the case of PCR even partially degraded DNA can be used. The downside of these techniques is the fact that in both cases the results are qualitative. Therefore, the exact gene copy number of MYCN cannot be evaluated with the PCR technique. However, the use of this innovative technology, gives the advantage of examining a large number of samples and quantifying the gene and its expression. In Figure 15, we present an example of Real-Time PCR, from unpublished in-house experimentation with the Kelly cell line. This cell line was an ideal model for MYCN amplification, since it is reported to have a ×100 MYCN amplification [2,4,5].
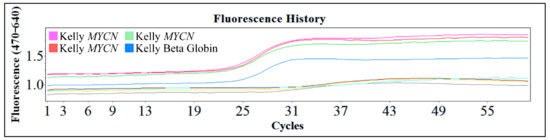
Figure 15.
An example of real-time PCR with the Kelly cell line from in-house unpublished data.
4. Molecular Mechanistics of the MYCN Gene
Neuroblastoma is presented at diagnosis with extreme heterogeneity. This is probably to the fact that neuroblastoma is inherently a complex and heterogeneous tumor from a biological and genetic perspective. A series of genomic lesions have been discovered in neuroblastoma, each depending on the methodological advancements in the course of time. Initially, the first observations concerned the aberrant number of chromosomes, followed by chromosomal aberrations, such as deletions, translocations, fusions and amplifications. Further on, advances in gene expression came to add more information to the knowledge on neuroblastoma’s biology. This kind of knowledge, initially concerning pure research findings, was added to the diagnostic and prognostic tools for the tumor, as well as to the treatment algorithms of neuroblastoma.
4.1. Other Molecular Biomarkers Besides MYCN
Ploidy is characteristic of all tumors. In the case of neuroblastoma, tumors are separated into two main categories; the semi-diploid (45% of tumors) and quasi-triploid tumors (55% of tumors). Ploidy has a prognostic significance in patients younger than two years of age. It has been reported that triploid tumors are usually presented with loss of whole chromosomes, whereas diploid or almost-diploid tumors manifest a major impairment of genome stability leading to chromosomal rearrangements, such as unbalanced permutations. In general, it is reported that triploid tumors have a better prognosis [103,104].
Aside from MYCN aberrations, there are several other chromosomal abnormalities present in neuroblastoma. A much known chromosomal aberration is the 1p deletion, which is often associated with MYCN amplification, but it is also frequent without it. It affects the 1p36 region and it is found in the 23–35% of all neuroblastic tumor cases. It is associated with patient’s unfavorable prognosis, where in some cases it remains an independent prognostic indicator, especially in localized, non-metastatic tumors. It is possible that neuroblastoma tumor suppressor oncogenes are located in the 1p36 region and their deletion favors oncogenesis. Such genes include the CHD5 and KIF1B genes, as previously reported [105,106,107,108].
Another chromosomal aberration includes the 11q deletion and in particular deletion of the 11q23 region. This type of chromosomal aberration, is found in the 26–44% of neuroblatoma patients and is inversely related to MYCN amplification. This finding divides neuroblastoma patients to two large categories; those with MYCN amplification and those with 11q23, indicating that these two aberrations are mutually exclusive. This aberration, is known to have prognostic value, especially for low- and intermediate-risk neuroblastoma. In this region, the CADM1 gene is located, another tumor suppressor gene, whose deletion is probably linked to neuroblastoma progression [106,107,109,110].
Similarly to the previous deletions, neuroblastoma manifests a deletion in the 3p region, which is known to co-exist with the 11q23 deletion. Both deletions are known, up-to-date, to be present without a MYCN amplification or a 1p36 deletion. In addition, this deletion is more frequent in patients of older diagnostic age, indicating that it is probable a distant event in the process of oncogenesis. Similarly to the aforementioned aberrations, this region hosts the RASSF1A gene, whose methylation is also known to be linked to tumor progression [107].
A frequent chromosomal aberration of neuroblastoma is the duplication of the 17q region. It occurs in the 80% of all neuroblastoma cases and is often the result of unbalanced shifts on chromosomes 1p or 11q. Genes located in that region include the NME1, NME2 and PPM1D genes (91). In the previous aberrations, it was hypothesized that tumor progression was caused due to the deletions of tumor suppressor genes, while in the present case tumor progression is probably connected to the amplification of oncogenes.
Gene mutations in neuroblastoma have been largely studied and the candidate genes are countless. However, an interesting case is presented with the mutations of the ALK gene. These mutations are found to be present in 15% of all sporadic neuroblastoma cases. The ALKF1174L mutation has been shown to enhance the oncogenic activity of the amplified MYCN oncogene. The ALK gene codes for a tyrosine kinase and is found not only in neuroblastoma but also in lung malignancies and lymphomas. Mutations of the ALK gene and the signal transduction pathways involving it, probably induce oncogenesis and are thought as possible therapeutic targets [111,112,113,114,115,116,117,118]. In addition to MYCN mutations analysis in neuroblastoma, gene expression studies have been used in order to classify the tumor. For example, the expression footprint of six neuroblastoma-related genes (ALK, BIRC5, CCND1, MYCN, NTRK1 and PHOX2B) have been used to differentiate neuroblastoma molecular subtypes [119].
4.2. Epigenetic Regulation of the MYCN Gene
Besides the aforementioned chromosomal aberrations, much is known on the epigenetic regulation of MYCN in neuroblastoma. Epigenetic regulation of MYCN, includes its interaction with miRNA, as well as the methylation status of the gene. It is known that MYCN is targeted by miRNAs, suppressing MYCN protein expression [107,117,120]. In this section we will deal with the current knowledge on the interactions between MYCN and miRNAs, as well as its epigenetic regulation through methylation.
4.2.1. MYCN and miRNAs
MiRNAs are small single-stranded non-coding RNA molecules (of about 22 nucleotide long) found in all organisms and function as RNA silencing molecules, regulating gene expression post-transcriptionally [121]. MiRNAs bind to their RNA-targets via the nucleotide complementarity principle [122]. The result from the interaction between miRNAs and mRNAs is the silencing of the second, through three main processes; The first concerns mRNA cleavage, the second concerns the destabilization of the mRNA through shortening of its poly(A) tail, and finally the third concerns the inhibition of translation in the ribosomes [123]. Similarly, the MYCN gene is known to be regulated by miRNAs [124]. A search for possible miRNA targets of MYCN revealed a total of 234 miRNAs, using the miRDB database [125,126,127,128], which we present in the Supplementary Materials Table S1. Since the MYCN gene is known to be amplified in neuroblastoma it is expected that most miRNAs, would be negative regulators of the gene. We have searched each miRNA separately for its known roles in neuroblastoma and with respect to MYCN. The details of the predicted miRNAs were searched with the miRBase web-tool [129,130,131,132,133,134].
The majority of miRNAs are reported to be down-regulated in neuroblastoma and in particular, they are found to directly suppress MYCN expression (Table 3). Therefore, in all neuroblastoma cases those miRNAs were found to be down-regulated. Some interesting observations were remarked for several miRNAs, which were found to be up-regulated in neuroblastoma and with a direct link to MYCN. In particular, the miR-17~92 cluster, which includes the miR-17, miR-18a, miR-19a, miR-20a, miR-19b-1 and miR-92a-1 members [135] (Table 3). Those miRNAs, are reported to be up-regulated in neuroblastoma and in particular their over-expression is facilitated by MYCN. It is possible that those miRNAs are enhanced by the gene by an indirect mechanism that attempts to promote tumor progression.

Table 3.
MYCN-related miRNAs. Out of 234 miRNAs targeting MYCN, twenty were studied for their role in neuroblastoma.
4.2.2. MYCN Methylation and Methylation-Related Mechanisms
Gene methylation is known to play an extremely important role in all parts of cellular and tissue physiology. Similarly, several studies have been conducted for the role of methylation in neuroblastoma and MYCN. Up to date, there are not much data on the methylation status of MYCN in neuroblastoma. Yet, there are several reports concerning the role of methylation of other genes in neuroblastoma, in both tumors with amplified MYCN or not. One of the main findings on neuroblastoma is that the tumor is characterized by the extensive methylation of many tumor suppressor genes [159]. This is the case in almost all neuroblastoma cases irrespectively of MYCN amplification. Interestingly, a recent report has highlighted that miRNAs also have been found methylated in neuroblastoma cell lines, hinting towards their role in tumor progression [160]. Some of these miRNAs included LET7G, MIR124-2, MIR1490, MIR15599, MIR23B, MIR24-1, MIR27B, MIR34C, MIR34B2 and MIR196A-1 [160] (Table 4). In almost all cases gene promoter methylation was mainly related to tumor progression and poor prognosis (Table 4). We have found two exceptions to this rule. The first concerned the NR4A3 gene and actually the Nr4a3 gene in a mouse model (Table 4). A recent study has highlighted that cells with MYCN amplification were found to have a hypo-methylation pattern of the NR4A3 gene’s Exon 3, while those tumors with a methylated Exon 3, manifested potentially better prognosis [161]. In addition, three different studies indicated another exception, concerning the CD44 gene (Table 4). In both MYCN amplified and non-amplified neuroblastoma, CD44 was found to play a dual role that is when un-methylated was manifesting high expression levels, redirecting to tumor suppression, while when hyper-methylated it was related to tumor progression [162,163,164].

Table 4.
MYCN-related methylation in neuroblastoma.
4.2.3. Other Epigenetic and Post-Translational Mechanisms in Neuroblastoma
Up to date there are not many studies concerning the role of other epigenetic (e.g., acetylation) or post-translational mechanisms in neuroblastoma. The existent studies have investigated this phenomenon through the application of inhibitors and by observing the outcome. For example, a recent study has found that the inhibitor SF1126, which inhibits BRD4 bromodomain binding to acetylated lysine residues with histone H3 as well as PI3K activity in a MYCN amplified neuroblastoma cell line, manifested tumor inhibitory effects and was proposed as a potential therapeutic agent [194].
In another interesting study, it has been proposed that a known anti-depressant, fluoxetine, was able to induce apoptosis in neuroblastoma cells. Interestingly, fluoxetine was able to induce apoptosis through two mechanistic pathways; the caspase cascade and the probably the hyper-acetylation of histone H3 and H4, upregulation of p300 histone acetyltransferase (HAT), as well as the downregulation of histone deacetylases (HDAC) [195]. However, this was observed in MYCN amplified cells, while it was not apparent in MYCN non-amplified or MYCN knock-down cells [195]. The results from this study, indicated that MYCN is actively participating in other post-transcriptional and post-translational modifications. The case of fluoxetine, is of great interest since it is a substance that is used for the treatment of different conditions. In the same context, another recent work indicated the use of anti-diabetic drugs, such as metformin and phenformin, in the treatment of neuroblastoma. The treatment of neuroblastoma cells, both with or without MYCN amplification, with metformin and phenformin resulted to increased apoptosis rates and inhibition of cell proliferation. One the possible mechanisms through which these drugs act is the augmentation of H3 acetylation [196].
Another study, indicated that the drug romidepsin, a selective histone deacetylase inhibitor (HDAC1/2), was able to induce apoptosis to neuroblastoma cells. In this study, it appeared that neuroblastoma progression is probably linked to the extensive histone acetylation and its inhibition was able to destabilize tumor progression [197]. Further on, it has been reported that MYCN and HDAC genes are functioning in cooperation in neuroblastoma. At the same time another gene the Grainyhead-like 1 (GRHL1), which is known to be critical in Drosophila neural development, was found to be one of the main targets of HDAC inhibitor treatment in neuroblastoma [198]. It was found that an increase in the histone H4 “pan-acetylation” in association with its promoter was followed by extensive transcriptional activation [198]. The HDAC3 and MYCN genes are physically co-localized to the GRHL1 promoter and they are probably able to repress its transcription. In cases of high GRHL1 expression, in neuroblastoma, tumor progression was suppressed, as well as patient survival was more favorable. Thus, it appeared that neuroblastoma with or without MYCN amplification, manifested low proliferation rates and sensitivity to therapy if GRHL1 was present [198].
4.3. MYCN Gene Amplification: “The Oldest Trick in the Book”
MYCN is known to regulate the proliferation, growth, differentiation and survival of cells of the developing nervous system [199]. Initial karyotyping techniques, has shown that large chromosomal abnormalities are found in the 1p region of Chromosome 1. This finding became known in the mid-1980s, where it was found that this chromosomal aberration was linked to ectopic MYCN amplification, which is located on the short arm of chromosome 2p24 [200]. It became apparent that the presence of MYCN amplification affected patients with advanced neuroblastoma and manifested an unfavorable prognosis. Numerous studies have shown that MYCN overexpression leads to the development of neuroblastic tumors and it seems that this is achieved by regulating p53 levels, regulating key mechanisms of cell apoptosis [201]. In general, although MYCN controls a large number of genes, only a small number of regulatory genes have been studied. Interestingly, the high expression of MYCN-controlled genes is not only characteristic of MYCN-amplified tumors but is also found in other high-risk neuroblastic tumors that do not show MYCN amplification. However, in addition to MYCN, several other cytogenetic abnormalities have been frequently observed in neuroblastic tumors. Most involve the loss of genomic material.
MYCN amplification, drives one out of six cases of neuroblastoma. The mechanisms through which amplification takes place are still obscure, yet a recent report indicated that gene amplification is related to a sort of “enhancer high jacking” of the MYCN gene [202]. In particular, they have reported that there are two mechanistic approaches for the gene’s amplification. The first includes the co-amplification of the proximal enhancer driven by the noradrenergic core regulatory circuit (CRC), while the second involves the ectopic “enhancer hijacking”, contributing to the loss of local gene regulatory elements [202].
A number of genetic characteristics of neuroblastic tumors have significant prognostic value. These include tumor ploidy, MYCN amplification, and for tumors without MYCN amplification the type of chromosomal lesions that emerge. Tumors with poly-ploidy have a better prognosis as compared to tumors that are diploid. This is especially true for patients without MYCN amplification and of younger age (less than 18 months of age) [203]. On the other hand, triploid tumors are known to have the best prognosis, while tetraploid tumors have a prognosis similar to diploid tumors (inferior) [104]. The most important and most known aberration of neuroblastoma, with an unfavorable prognosis, is MYCN amplification.
One of the first discoveries concerning the molecular mechanistic of neuroblastoma, concerned the discovery of MYCN amplification. Thirty years ago it was realized that an increase in the MYCN copy-number is associated with rapid tumor progression in neuroblastoma [31]. Following that discovery, MYCN amplification emerged and it was discovered its relation to the unfavorable outcome of neuroblastoma [203,204,205]. In the meantime, MYCN amplification became a “standard operating procedure” in terms of neuroblastoma diagnostics, as well as its categorization [206]. MYCN amplification can be detected at any stage of the tumor’s progression, although it is particularly rare in Stage 1 tumors.
The frequency of MYCN amplification has been found to be at 25% of all neuroblastomas and in 40% of high-risk neuroblastomas [96,201]. The definition for MYCN amplification requires the detection of at least four copies more as compared to the normal alleles. Frequently, MYCN amplification is found in neuroblastoma cell lines, which ranges from 20 to 40 times [200]. From a mechanistic point of view, it has been reported that MYCN activity is probably related to NM23-H1 and NM23-H2 genes located in the 17q region [207]. In addition, MYCN expression has been reported to be linked to activation by the TP53 gene and thus regulating the escape from apoptosis of neuroblastic cells [201]. Another important molecular event thought to participate in MYCN amplification is the presence of ALK mutations in neuroblastoma [112]. Interestingly, along with MYCN amplification, ALK has also been reported to be amplified in neuroblastoma [208,209]. In addition, it has been found that in tumors without MYCN amplification a number of other chromosomal aberrations are present. Such examples include the gain or deletion of complete chromosomes such as gain of Chromosome 17, Chromosome 7, Chromosome 2 and deletions of Chromosomes 3, 4, and 14 [210]. Similarly, it is possible to observe the gain or deletion of chromosomal regions, such as the loss of Location 1p of Chromosome 1, 11q on Chromosome 11, 14q on Chromosome 14 and 3p in Chromosome 3. Similarly, most frequent chromosomal gains are the 17q and 2p [210]. The latter, is also known to be present with ALK amplification. The deletion of specific chromosomal regions is known to be present in neuroblastoma progression due to the concurrent deletion of tumor suppressor genes that is these specific regions are the loci of tumor suppressors and their absence favors tumor progression. Such examples are the 1p, 3p and 11q regions. Similarly, the gain of chromosomal regions is tightly related to the amplification of oncogenes, favoring tumor ontogenesis [211,212,213]. An older report has indicated that the presence of complete chromosomal loss or gain was related to more favorable outcome, as compared to the loss or gain of specific chromosomal regions [214,215].
4.4. Therapeutic Interventions with MYCN
One of the first interventions in the treatment of neuroblastoma is chemotherapy, but of equal value comes surgical intervention and removal. Surgery is not always the first choice, as neuroblastoma manifests a diffuse character during its growth, making surgery impossible. Yet, there are several factors that can modify the decision for surgical intervention. All recent reports agree on the fact that disease staging is crucial for the treatment of the disease [47,216]. It has been reported that surgical treatment and its success is linked to the image defined risk factors (IDRF), which include the crossing and extending of the tumor, the engulfment of vessels, the compression of other organs and the encasing of nerves [217]. It would be interesting to find molecular factors that could lead to the decision for surgical intervention, yet to the best of our knowledge, none exist up-to-date.
Since the discovery of MYCN amplification in neuroblastoma, it became apparent that MYCN could pose an attractive therapeutic target. The first report including MYCN as potential target came by Lu et al. (2003) [218]. In the time course of research, it became evident that simultaneous inhibition of multiple targets could prove useful in the treatment of neuroblastoma. For example, a recent study has reported that the simultaneous inhibition of MYCN and mTOR using bromodomain extra-terminal protein inhibitors and temsirolimus manifested significant cell survival suppression [219]. Another study showed that the use of PARP and CHK1 inhibitors synergized to induce death in neuroblastoma cells and in primary cultures of MYCN amplified and MYCN overexpressing cells [220]. In agreement with our previous comments for the role of MYCN in cellular metabolism, a recent study highlighted this approach as a possible therapeutic intervention. In particular, they have reported that the pyrimidine biosynthetic enzyme dihydroorotate dehydrogenase (DHODH) consisted of a potent therapeutic target for MYCN-amplified neuroblastoma. Thus, they have shown that inhibition of DHODH suppressed the proliferation of MYCN-amplified neuroblastoma cells [221]. Up-to-date there are numerous studies concerning the use of inhibitors in neuroblastoma, either directly on MYCN, as well as indirectly through the inhibition of other molecules. We have summarized the most recent inhibitors and molecules in Table 5.

Table 5.
Therapeutic targets in neuroblastoma.
5. Conclusions
It the present study, we have reviewed the literature for the connection between MYCN and neuroblastoma. We have investigated mostly all possible aspects in MYCN biology including, the presence of MYCN G-quadraplexes in the gene’s structure, the “anatomy” of MYCN’s transcripts, as well as the epigenetic regulation in relation to neuroblastoma. MYCN, is one the most well studied genes in neuroblastoma and yet, there is much to be learned on its biology. We have seen that the ontogeny of neuroblastoma is closely related to MYCN biology, where the presence of chromosomal aberrations is tightly linked to the deletion or the gain of tumor suppressor and oncogenes respectively. MYCN, is known to play a significant role in the chromosomal stability of neuroblastoma, as well as it is able to change the methylation and acetylation status of the genome and the genome-regulatory mechanisms. In that sense, several approaches have attempted to exploit the properties of MYCN and its biology, in order to use them as prognostic and more importantly therapeutic targets. It is certain that we still need much more research in order to comprehend the depth of the disease and exploit that knowledge for its therapy.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/diseases9040078/s1, Table S1: predicted miRNAs as potential targets for the MYCN gene. miRNAs were predicted using the miRDB database [125,126,127,128].
Author Contributions
Conceptualization, G.I.L.; investigation, M.B., K.H., A.Z. and G.I.L.; writing—original draft preparation, M.B., K.H. and G.I.L.; writing—review and editing, A.Z. and G.I.L.; visualization, G.I.L.; supervision, G.I.L.; project administration, A.Z. and G.I.L.; funding acquisition, A.Z. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Kohl, N.E.; Kanda, N.; Schreck, R.R.; Bruns, G.; Latt, S.A.; Gilbert, F.; Alt, F.W. Transposition and amplification of oncogene-related sequences in human neuroblastomas. Cell 1983, 35, 359–367. [Google Scholar] [CrossRef]
- Schwab, M.; Alitalo, K.; Klempnauer, K.H.; Varmus, H.E.; Bishop, J.M.; Gilbert, F.; Brodeur, G.; Goldstein, M.; Trent, J. Amplified DNA with limited homology to myc cellular oncogene is shared by human neuroblastoma cell lines and a neuroblastoma tumour. Nature 1983, 305, 245–248. [Google Scholar] [CrossRef] [PubMed]
- Huang, M.; Weiss, W.A. Neuroblastoma and mycn. Cold Spring Harb. Perspect. Med. 2013, 3, a014415. [Google Scholar] [CrossRef]
- Schwab, M.; Varmus, H.E.; Bishop, J.M. Human n-myc gene contributes to neoplastic transformation of mammalian cells in culture. Nature 1985, 316, 160–162. [Google Scholar] [CrossRef]
- Schwab, M.; Bishop, J.M. Sustained expression of the human protooncogene mycn rescues rat embryo cells from senescence. Proc. Natl. Acad. Sci. USA 1988, 85, 9585–9589. [Google Scholar] [CrossRef]
- Schweigerer, L.; Scheurich, P.; Fotsis, T. Enhanced mycn oncogene expression in human neuroblastoma cells results in increased susceptibility to growth inhibition by tnf alpha. Biochem. Biophys. Res. Commun. 1990, 170, 1301–1307. [Google Scholar] [CrossRef]
- VanDevanter, D.R.; Piaskowski, V.D.; Casper, J.T.; Douglass, E.C.; Von Hoff, D.D. Ability of circular extrachromosomal DNA molecules to carry amplified mycn proto-oncogenes in human neuroblastomas in vivo. J. Natl. Cancer Inst. 1990, 82, 1815–1821. [Google Scholar] [CrossRef]
- Brady, S.W.; Liu, Y.; Ma, X.; Gout, A.M.; Hagiwara, K.; Zhou, X.; Wang, J.; Macias, M.; Chen, X.; Easton, J.; et al. Pan-neuroblastoma analysis reveals age- and signature-associated driver alterations. Nat. Commun. 2020, 11, 5183. [Google Scholar] [CrossRef] [PubMed]
- Borgenvik, A.; Čančer, M.; Hutter, S.; Swartling, F.J. Targeting mycn in molecularly defined malignant brain tumors. Front. Oncol. 2020, 10, 626751. [Google Scholar] [CrossRef] [PubMed]
- Gruszka, R.; Zakrzewski, K.; Liberski, P.P.; Zakrzewska, M. Mrna and mirna expression analyses of the myc/e2f/mir-17-92 network in the most common pediatric brain tumors. Int. J. Mol. Sci. 2021, 22, 543. [Google Scholar] [CrossRef] [PubMed]
- Raghuram, N.; Khan, S.; Mumal, I.; Bouffet, E.; Huang, A. Embryonal tumors with multi-layered rosettes: A disease of dysregulated mirnas. J. Neuro-Oncol. 2020, 150, 63–73. [Google Scholar] [CrossRef]
- Kumar, R.; Smith, K.S.; Deng, M.; Terhune, C.; Robinson, G.W.; Orr, B.A.; Liu, A.P.Y.; Lin, T.; Billups, C.A.; Chintagumpala, M.; et al. Clinical outcomes and patient-matched molecular composition of relapsed medulloblastoma. J. Clin. Oncol. 2021, 39, 807–821. [Google Scholar] [CrossRef] [PubMed]
- Skowron, P.; Farooq, H.; Cavalli, F.M.G.; Morrissy, A.S.; Ly, M.; Hendrikse, L.D.; Wang, E.Y.; Djambazian, H.; Zhu, H.; Mungall, K.L.; et al. The transcriptional landscape of shh medulloblastoma. Nat. Commun. 2021, 12, 1749. [Google Scholar] [CrossRef]
- Estiar, M.A.; Javan, F.; Zekri, A.; Mehrazin, M.; Mehdipour, P. Prognostic significance of mycn gene amplification and protein expression in primary brain tumors: Astrocytoma and meningioma. Cancer Biomark. 2017, 19, 341–351. [Google Scholar] [CrossRef] [PubMed]
- Čančer, M.; Drews, L.F.; Bengtsson, J.; Bolin, S.; Rosén, G.; Westermark, B.; Nelander, S.; Forsberg-Nilsson, K.; Uhrbom, L.; Weishaupt, H.; et al. Bet and aurora kinase a inhibitors synergize against mycn-positive human glioblastoma cells. Cell Death Dis. 2019, 10, 881. [Google Scholar] [CrossRef]
- Wang, B.; Wu, Z.H.; Lou, P.Y.; Chai, C.; Han, S.Y.; Ning, J.F.; Li, M. Human bone marrow-derived mesenchymal stem cell-secreted exosomes overexpressing microrna-34a ameliorate glioblastoma development via down-regulating mycn. Cell. Oncol. 2019, 42, 783–799. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Chen, S.S.; Clarke, S.; Veschi, V.; Thiele, C.J. Targeting mycn in pediatric and adult cancers. Front. Oncol. 2020, 10, 623679. [Google Scholar] [CrossRef]
- Nakazawa, A. Biological categories of neuroblastoma based on the international neuroblastoma pathology classification for treatment stratification. Pathol. Int. 2021, 71, 232–244. [Google Scholar] [CrossRef] [PubMed]
- Alaminos, M.; Gerald, W.L.; Cheung, N.K. Prognostic value of mycn and id2 overexpression in neuroblastoma. Pediatr. Blood Cancer 2005, 45, 909–915. [Google Scholar] [CrossRef]
- Michalowski, M.B.; de Fraipont, F.; Plantaz, D.; Michelland, S.; Combaret, V.; Favrot, M.C. Methylation of tumor-suppressor genes in neuroblastoma: The rassf1a gene is almost always methylated in primary tumors. Pediatr. Blood Cancer 2008, 50, 29–32. [Google Scholar] [CrossRef]
- Zafar, A.; Wang, W.; Liu, G.; Xian, W.; McKeon, F.; Zhou, J.; Zhang, R. Targeting the p53-mdm2 pathway for neuroblastoma therapy: Rays of hope. Cancer Lett. 2021, 496, 16–29. [Google Scholar] [CrossRef]
- Jiang, C.Y.; Xu, X.; Jian, B.L.; Zhang, X.; Yue, Z.X.; Guo, W.; Ma, X.L. Chromosome 10 abnormality predicts prognosis of neuroblastoma patients with bone marrow metastasis. Ital. J. Pediatr. 2021, 47, 134. [Google Scholar] [CrossRef]
- Koh, K.N.; Lee, J.Y.; Lim, J.; Shin, J.; Kang, S.H.; Suh, J.K.; Kim, H.; Im, H.J.; Namgoong, J.M.; Kim, D.Y.; et al. Genetic alterations detected by targeted next-generation sequencing and their clinical implications in neuroblastoma. Anticancer Res. 2020, 40, 7057–7065. [Google Scholar] [CrossRef]
- Higashi, M.; Sakai, K.; Fumino, S.; Aoi, S.; Furukawa, T.; Tajiri, T. The roles played by the mycn, trk, and alk genes in neuroblastoma and neural development. Surg. Today 2019, 49, 721–727. [Google Scholar] [CrossRef] [PubMed]
- Korshunov, A.; Remke, M.; Kool, M.; Hielscher, T.; Northcott, P.A.; Williamson, D.; Pfaff, E.; Witt, H.; Jones, D.T.; Ryzhova, M.; et al. Biological and clinical heterogeneity of mycn-amplified medulloblastoma. Acta Neuropathol. 2012, 123, 515–527. [Google Scholar] [CrossRef] [PubMed]
- Wright, J.H. Neurocytoma or neuroblastoma, a kind of tumor not generally recognized. J. Exp. Med. 1910, 12, 556–561. [Google Scholar] [CrossRef]
- Matthay, K.K. Neuroblastoma: Biology and therapy. Oncology 1997, 11, 1857–1866, discussion 1869–1872, 1875. [Google Scholar]
- Knudson, A.G., Jr.; Strong, L. Mutation and cancer: Neuroblastoma and pheochromocytoma. Am. J. Hum. Genet. 1972, 24, 514. [Google Scholar] [PubMed]
- Smith, E.I.; Haase, G.M.; Seeger, R.C.; Brodeur, G.M. A surgical perspective on the current staging in neuroblastoma—The international neuroblastoma staging system proposal. J. Pediatr. Surg. 1989, 24, 386–390. [Google Scholar] [CrossRef]
- Wood, L.; Lowis, S. An update on neuroblastoma. Paediatr. Child Health 2008, 18, 123–128. [Google Scholar] [CrossRef]
- Seeger, R.C.; Brodeur, G.M.; Sather, H.; Dalton, A.; Siegel, S.E.; Wong, K.Y.; Hammond, D. Association of multiple copies of the n-myc oncogene with rapid progression of neuroblastomas. N. Engl. J. Med. 1985, 313, 1111–1116. [Google Scholar] [CrossRef]
- Ludwig, J.A.; Weinstein, J.N. Biomarkers in cancer staging, prognosis and treatment selection. Nat. Rev. Cancer 2005, 5, 845–856. [Google Scholar] [CrossRef]
- Brodeur, G.M. Neuroblastoma: Biological insights into a clinical enigma. Nat. Rev. Cancer 2003, 3, 203–216. [Google Scholar] [CrossRef]
- Lapenna, S.; Giordano, A. Cell cycle kinases as therapeutic targets for cancer. Nat. Rev. Drug Discov. 2009, 8, 547–566. [Google Scholar] [CrossRef] [PubMed]
- Beltran, H. The n-myc oncogene: Maximizing its targets, regulation, and therapeutic potential. Mol. Cancer Res. 2014, 12, 815–822. [Google Scholar] [CrossRef] [PubMed]
- Futreal, P.A.; Coin, L.; Marshall, M.; Down, T.; Hubbard, T.; Wooster, R.; Rahman, N.; Stratton, M.R. A census of human cancer genes. Nat. Rev. Cancer 2004, 4, 177–183. [Google Scholar] [CrossRef] [PubMed]
- Knuutila, S.; Björkqvist, A.-M.; Autio, K.; Tarkkanen, M.; Wolf, M.; Monni, O.; Szymanska, J.; Larramendy, M.L.; Tapper, J.; Pere, H.; et al. DNA copy number amplifications in human neoplasms: Review of comparative genomic hybridization studies. Am. J. Pathol. 1998, 152, 1107. [Google Scholar]
- Wieland, L.; Engel, K.; Volkmer, I.; Krüger, A.; Posern, G.; Kornhuber, M.E.; Staege, M.S.; Emmer, A. Overexpression of endogenous retroviruses and malignancy markers in neuroblastoma cell lines by medium-induced microenvironmental changes. Front. Oncol. 2021, 11, 637522. [Google Scholar] [CrossRef]
- Sooksawasdi Na Ayudhya, S.; Meijer, A.; Bauer, L.; Oude Munnink, B.; Embregts, C.; Leijten, L.; Siegers, J.Y.; Laksono, B.M.; van Kuppeveld, F.; Kuiken, T.; et al. Enhanced enterovirus d68 replication in neuroblastoma cells is associated with a cell culture-adaptive amino acid substitution in vp1. mSphere 2020, 5, e00941-20. [Google Scholar] [CrossRef]
- Dang, C.V. C-myc target genes involved in cell growth, apoptosis, and metabolism. Mol. Cell. Biol. 1999, 19, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Bordow, S.B.; Norris, M.D.; Haber, P.S.; Marshall, G.M.; Haber, M. Prognostic significance of mycn oncogene expression in childhood neuroblastoma. J. Clin. Oncol. 1998, 16, 3286–3294. [Google Scholar] [CrossRef]
- Strieder, V.; Lutz, W. Regulation of n-myc expression in development and disease. Cancer Lett. 2002, 180, 107–119. [Google Scholar] [CrossRef]
- Maris, J.M.; Weiss, M.J.; Guo, C.; Gerbing, R.B.; Stram, D.O.; White, P.S.; Hogarty, M.D.; Sulman, E.P.; Thompson, P.M.; Lukens, J.N.; et al. Loss of heterozygosity at 1p36 independently predicts for disease progression but not decreased overall survival probability in neuroblastoma patients: A children’s cancer group study. J. Clin. Oncol. 2000, 18, 1888–1899. [Google Scholar] [CrossRef] [PubMed]
- Cohn, S.L.; London, W.B.; Huang, D.; Katzenstein, H.M.; Salwen, H.R.; Reinhart, T.; Madafiglio, J.; Marshall, G.M.; Norris, M.D.; Haber, M. Mycn expression is not prognostic of adverse outcome in advanced-stage neuroblastoma with nonamplified mycn. J. Clin. Oncol. 2000, 18, 3604–3613. [Google Scholar] [CrossRef]
- Sato, Y.; Kobayashi, Y.; Sasaki, H.; Toyama, T.; Kondo, S.; Kiriyama, M.; Fujii, Y. Expression of id2 mrna in neuroblastoma and normal ganglion. Eur. J. Surg. Oncol. 2003, 29, 284–287. [Google Scholar] [CrossRef] [PubMed]
- Pan, L.J.; Chen, J.L.; Wu, Z.X.; Wu, Y.M. Exportin-t: A novel prognostic predictor and potential therapeutic target for neuroblastoma. Technol. Cancer Res. Treat. 2021, 20, 15330338211039132. [Google Scholar] [CrossRef]
- Irwin, M.S.; Naranjo, A.; Zhang, F.F.; Cohn, S.L.; London, W.B.; Gastier-Foster, J.M.; Ramirez, N.C.; Pfau, R.; Reshmi, S.; Wagner, E.; et al. Revised neuroblastoma risk classification system: A report from the children’s oncology group. J. Clin. Oncol. 2021, 39, 3229–3241. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Tian, Z.; Li, T.; Hu, X.; Zhu, J. Prognostic value of c-myc expression in patients with peripheral neuroblastic tumors. Int. J. Gen. Med. 2021, 14, 2901–2907. [Google Scholar] [CrossRef]
- Wang, H.; Wang, X.; Xu, L.; Zhang, J.; Cao, H. Age related gene dst represents an independent prognostic factor for mycn non-amplified neuroblastoma. BMC Pediatr. 2021, 21, 272. [Google Scholar] [CrossRef]
- Liao, Y.M.; Hung, T.H.; Tung, J.K.; Yu, J.; Hsu, Y.L.; Hung, J.T.; Yu, A.L. Low expression of il-15 and nkt in tumor microenvironment predicts poor outcome of mycn-non-amplified neuroblastoma. J. Pers. Med. 2021, 11, 122. [Google Scholar] [CrossRef] [PubMed]
- Wolf, E.; Eilers, M. Targeting myc proteins for tumor therapy. Annu. Rev. Cancer Biol. 2020, 4, 61–75. [Google Scholar] [CrossRef]
- Liao, Y.; Wang, J.; Jaehnig, E.J.; Shi, Z.; Zhang, B. Webgestalt 2019: Gene set analysis toolkit with revamped uis and apis. Nucleic Acids Res. 2019, 47, W199–W205. [Google Scholar] [CrossRef]
- Wang, J.; Vasaikar, S.; Shi, Z.; Greer, M.; Zhang, B. Webgestalt 2017: A more comprehensive, powerful, flexible and interactive gene set enrichment analysis toolkit. Nucleic Acids Res. 2017, 45, W130–W137. [Google Scholar] [CrossRef]
- Wang, J.; Duncan, D.; Shi, Z.; Zhang, B. Web-based gene set analysis toolkit (webgestalt): Update 2013. Nucleic Acids Res. 2013, 41, W77–W83. [Google Scholar] [CrossRef]
- Zhang, B.; Kirov, S.; Snoddy, J. Webgestalt: An integrated system for exploring gene sets in various biological contexts. Nucleic Acids Res. 2005, 33, W741–W748. [Google Scholar] [CrossRef]
- Richards, M.W.; Burgess, S.G.; Poon, E.; Carstensen, A.; Eilers, M.; Chesler, L.; Bayliss, R. Structural basis of n-myc binding by aurora-a and its destabilization by kinase inhibitors. Proc. Natl. Acad. Sci. USA 2016, 113, 13726–13731. [Google Scholar] [CrossRef]
- Bannasch, D.; Weis, I.; Schwab, M. Nmi protein interacts with regions that differ between mycn and myc and is localized in the cytoplasm of neuroblastoma cells in contrast to nuclear mycn. Oncogene 1999, 18, 6810–6817. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Beierle, E.A.; Trujillo, A.; Nagaram, A.; Kurenova, E.V.; Finch, R.; Ma, X.; Vella, J.; Cance, W.G.; Golubovskaya, V.M. N-myc regulates focal adhesion kinase expression in human neuroblastoma. J. Biol. Chem. 2007, 282, 12503–12516. [Google Scholar] [CrossRef] [PubMed]
- Ramsay, G.; Stanton, L.; Schwab, M.; Bishop, J.M. Human proto-oncogene n-myc encodes nuclear proteins that bind DNA. Mol. Cell. Biol. 1986, 6, 4450–4457. [Google Scholar] [CrossRef]
- Gaudet, P.; Livstone, M.S.; Lewis, S.E.; Thomas, P.D. Phylogenetic-based propagation of functional annotations within the gene ontology consortium. Brief. Bioinform. 2011, 12, 449–462. [Google Scholar] [CrossRef]
- Amente, S.; Gargano, B.; Diolaiti, D.; Della Valle, G.; Lania, L.; Majello, B. P14arf interacts with n-myc and inhibits its transcriptional activity. FEBS Lett. 2007, 581, 821–825. [Google Scholar] [CrossRef] [PubMed]
- Bannasch, D.; Mädge, B.; Schwab, M. Functional interaction of yaf2 with the central region of mycn. Oncogene 2001, 20, 5913–5919. [Google Scholar] [CrossRef][Green Version]
- Brockmann, M.; Poon, E.; Berry, T.; Carstensen, A.; Deubzer, H.E.; Rycak, L.; Jamin, Y.; Thway, K.; Robinson, S.P.; Roels, F.; et al. Small molecule inhibitors of aurora-a induce proteasomal degradation of n-myc in childhood neuroblastoma. Cancer Cell 2013, 24, 75–89. [Google Scholar] [CrossRef]
- Dardenne, E.; Beltran, H.; Benelli, M.; Gayvert, K.; Berger, A.; Puca, L.; Cyrta, J.; Sboner, A.; Noorzad, Z.; MacDonald, T.; et al. N-myc induces an ezh2-mediated transcriptional program driving neuroendocrine prostate cancer. Cancer Cell 2016, 30, 563–577. [Google Scholar] [CrossRef]
- Gustafson, W.C.; Meyerowitz, J.G.; Nekritz, E.A.; Chen, J.; Benes, C.; Charron, E.; Simonds, E.F.; Seeger, R.; Matthay, K.K.; Hertz, N.T.; et al. Drugging mycn through an allosteric transition in aurora kinase a. Cancer Cell 2014, 26, 414–427. [Google Scholar] [CrossRef]
- Liu, T.; Tee, A.E.; Porro, A.; Smith, S.A.; Dwarte, T.; Liu, P.Y.; Iraci, N.; Sekyere, E.; Haber, M.; Norris, M.D.; et al. Activation of tissue transglutaminase transcription by histone deacetylase inhibition as a therapeutic approach for myc oncogenesis. Proc. Natl. Acad. Sci. USA 2007, 104, 18682–18687. [Google Scholar] [CrossRef] [PubMed]
- Marshall, G.M.; Liu, P.Y.; Gherardi, S.; Scarlett, C.J.; Bedalov, A.; Xu, N.; Iraci, N.; Valli, E.; Ling, D.; Thomas, W.; et al. Sirt1 promotes n-myc oncogenesis through a positive feedback loop involving the effects of mkp3 and erk on n-myc protein stability. PLoS Genet. 2011, 7, e1002135. [Google Scholar] [CrossRef]
- Otto, T.; Horn, S.; Brockmann, M.; Eilers, U.; Schüttrumpf, L.; Popov, N.; Kenney, A.M.; Schulte, J.H.; Beijersbergen, R.; Christiansen, H.; et al. Stabilization of n-myc is a critical function of aurora a in human neuroblastoma. Cancer Cell 2009, 15, 67–78. [Google Scholar] [CrossRef] [PubMed]
- Vo, B.T.; Wolf, E.; Kawauchi, D.; Gebhardt, A.; Rehg, J.E.; Finkelstein, D.; Walz, S.; Murphy, B.L.; Youn, Y.H.; Han, Y.G.; et al. The interaction of myc with miz1 defines medulloblastoma subgroup identity. Cancer Cell 2016, 29, 5–16. [Google Scholar] [CrossRef] [PubMed]
- Yin, Y.; Morgunova, E.; Jolma, A.; Kaasinen, E.; Sahu, B.; Khund-Sayeed, S.; Das, P.K.; Kivioja, T.; Dave, K.; Zhong, F.; et al. Impact of cytosine methylation on DNA binding specificities of human transcription factors. Science 2017, 356, eaaj2239. [Google Scholar] [CrossRef]
- De Brouwer, S.; Mestdagh, P.; Lambertz, I.; Pattyn, F.; De Paepe, A.; Westermann, F.; Schroeder, C.; Schulte, J.H.; Schramm, A.; De Preter, K.; et al. Dickkopf-3 is regulated by the mycn-induced mir-17-92 cluster in neuroblastoma. Int. J. Cancer 2012, 130, 2591–2598. [Google Scholar] [CrossRef]
- Kumps, C.; Fieuw, A.; Mestdagh, P.; Menten, B.; Lefever, S.; Pattyn, F.; De Brouwer, S.; Sante, T.; Schulte, J.H.; Schramm, A.; et al. Focal DNA copy number changes in neuroblastoma target mycn regulated genes. PLoS ONE 2013, 8, e52321. [Google Scholar] [CrossRef]
- Suenaga, Y.; Islam, S.M.; Alagu, J.; Kaneko, Y.; Kato, M.; Tanaka, Y.; Kawana, H.; Hossain, S.; Matsumoto, D.; Yamamoto, M.; et al. Ncym, a cis-antisense gene of mycn, encodes a de novo evolved protein that inhibits gsk3β resulting in the stabilization of mycn in human neuroblastomas. PLoS Genet. 2014, 10, e1003996. [Google Scholar] [CrossRef]
- Gellert, M.; Lipsett, M.N.; Davies, D.R. Helix formation by guanylic acid. Proc. Natl. Acad. Sci. USA 1962, 48, 2013–2018. [Google Scholar] [CrossRef] [PubMed]
- Routh, E.D.; Creacy, S.D.; Beerbower, P.E.; Akman, S.A.; Vaughn, J.P.; Smaldino, P.J. A g-quadruplex DNA-affinity approach for purification of enzymatically active g4 resolvase1. J. Vis. Exp 2017, 121, e55496. [Google Scholar] [CrossRef]
- Largy, E.; Mergny, J.L.; Gabelica, V. Role of alkali metal ions in g-quadruplex nucleic acid structure and stability. Met. Ions Life Sci. 2016, 16, 203–258. [Google Scholar]
- Sundquist, W.I.; Klug, A. Telomeric DNA dimerizes by formation of guanine tetrads between hairpin loops. Nature 1989, 342, 825–829. [Google Scholar] [CrossRef] [PubMed]
- Sen, D.; Gilbert, W. Formation of parallel four-stranded complexes by guanine-rich motifs in DNA and its implications for meiosis. Nature 1988, 334, 364–366. [Google Scholar] [CrossRef]
- Han, H.; Hurley, L.H. G-quadruplex DNA: A potential target for anti-cancer drug design. Trends Pharmacol. Sci. 2000, 21, 136–142. [Google Scholar] [CrossRef]
- Wang, W.; Hu, S.; Gu, Y.; Yan, Y.; Stovall, D.B.; Li, D.; Sui, G. Human myc g-quadruplex: From discovery to a cancer therapeutic target. Biochim. Biophys. Acta. Rev. Cancer 2020, 1874, 188410. [Google Scholar] [CrossRef]
- Kikin, O.; D’Antonio, L.; Bagga, P.S. Qgrs mapper: A web-based server for predicting g-quadruplexes in nucleotide sequences. Nucleic Acids Res. 2006, 34, W676–W682. [Google Scholar] [CrossRef]
- Trajkovski, M.; da Silva, M.W.; Plavec, J. Unique structural features of interconverting monomeric and dimeric g-quadruplexes adopted by a sequence from the intron of the n-myc gene. J. Am. Chem. Soc. 2012, 134, 4132–4141. [Google Scholar] [CrossRef]
- Benabou, S.; Ferreira, R.; Aviñó, A.; González, C.; Lyonnais, S.; Solà, M.; Eritja, R.; Jaumot, J.; Gargallo, R. Solution equilibria of cytosine- and guanine-rich sequences near the promoter region of the n-myc gene that contain stable hairpins within lateral loops. Biochim. Biophys. Acta 2014, 1840, 41–52. [Google Scholar] [CrossRef]
- Li, F.; Guo, D.; Kang, L. Study on the recognition of g-quadruplexes by two stereoisomers of alkaloids. Anal. Bioanal. Chem. 2019, 411, 5555–5561. [Google Scholar] [CrossRef]
- Prosperini, A.; Berrada, H.; Ruiz, M.J.; Caloni, F.; Coccini, T.; Spicer, L.J.; Perego, M.C.; Lafranconi, A. A review of the mycotoxin enniatin b. Front. Public Health 2017, 5, 304. [Google Scholar] [CrossRef]
- Li, F.; Chen, H.; Zhou, J.; Yuan, G. Exploration of the selective recognition of the g-quadruplex in the n-myc oncogene by electrospray ionization mass spectrometry. Rapid Commun. Mass Spectrom. 2015, 29, 247–252. [Google Scholar] [CrossRef] [PubMed]
- Antczak, M.; Popenda, M.; Zok, T.; Sarzynska, J.; Ratajczak, T.; Tomczyk, K.; Adamiak, R.W.; Szachniuk, M. New functionality of rnacomposer: An application to shape the axis of mir160 precursor structure. Acta Biochim. Pol. 2016, 63, 737–744. [Google Scholar] [CrossRef] [PubMed]
- Popenda, M.; Szachniuk, M.; Antczak, M.; Purzycka, K.J.; Lukasiak, P.; Bartol, N.; Blazewicz, J.; Adamiak, R.W. Automated 3d structure composition for large rnas. Nucleic Acids Res. 2012, 40, e112. [Google Scholar] [CrossRef]
- Besançon, R.; Valsesia-Wittmann, S.; Locher, C.; Delloye-Bourgeois, C.; Furhman, L.; Tutrone, G.; Bertrand, C.; Jallas, A.C.; Garin, E.; Puisieux, A. Upstream orf affects mycn translation depending on exon 1b alternative splicing. BMC Cancer 2009, 9, 445. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Carter, S.; Parmar, S.; Bume, D.D.; Calabrese, D.R.; Liang, X.; Yazdani, K.; Xu, M.; Liu, Z.; Thiele, C.J.; et al. Targeting a noncanonical, hairpin-containing g-quadruplex structure from the mycn gene. Nucleic Acids Res. 2021, 49, 7856–7869. [Google Scholar] [CrossRef]
- Jacobs, J.F.; van Bokhoven, H.; van Leeuwen, F.N.; Hulsbergen-van de Kaa, C.A.; de Vries, I.J.; Adema, G.J.; Hoogerbrugge, P.M.; de Brouwer, A.P. Regulation of mycn expression in human neuroblastoma cells. BMC Cancer 2009, 9, 239. [Google Scholar] [CrossRef] [PubMed]
- Stanton, L.W.; Bishop, J.M. Alternative processing of rna transcribed from nmyc. Mol. Cell. Biol. 1987, 7, 4266–4272. [Google Scholar] [CrossRef] [PubMed]
- Peirce, S.K.; Findley, H.W. High level mycn expression in non-mycn amplified neuroblastoma is induced by the combination treatment nutlin-3 and doxorubicin and enhances chemosensitivity. Oncol. Rep. 2009, 22, 1443–1449. [Google Scholar] [CrossRef]
- Cage, T.A.; Chanthery, Y.; Chesler, L.; Grimmer, M.; Knight, Z.; Shokat, K.; Weiss, W.A.; Gustafson, W.C. Downregulation of mycn through pi3k inhibition in mouse models of pediatric neural cancer. Front. Oncol. 2015, 5, 111. [Google Scholar] [CrossRef]
- Ruiz-Pérez, M.V.; Henley, A.B.; Arsenian-Henriksson, M. The mycn protein in health and disease. Genes 2017, 8, 113. [Google Scholar] [CrossRef]
- Bell, E.; Chen, L.; Liu, T.; Marshall, G.M.; Lunec, J.; Tweddle, D.A. Mycn oncoprotein targets and their therapeutic potential. Cancer Lett. 2010, 293, 144–157. [Google Scholar] [CrossRef]
- Ham, J.; Costa, C.; Sano, R.; Lochmann, T.L.; Sennott, E.M.; Patel, N.U.; Dastur, A.; Gomez-Caraballo, M.; Krytska, K.; Hata, A.N.; et al. Exploitation of the apoptosis-primed state of mycn-amplified neuroblastoma to develop a potent and specific targeted therapy combination. Cancer Cell 2016, 29, 159–172. [Google Scholar] [CrossRef]
- Cortés, C.; Kozma, S.C.; Tauler, A.; Ambrosio, S. Mycn concurrence with saha-induced cell death in human neuroblastoma cells. Cell. Oncol. 2015, 38, 341–352. [Google Scholar] [CrossRef]
- DeBerardinis, R.J.; Mancuso, A.; Daikhin, E.; Nissim, I.; Yudkoff, M.; Wehrli, S.; Thompson, C.B. Beyond aerobic glycolysis: Transformed cells can engage in glutamine metabolism that exceeds the requirement for protein and nucleotide synthesis. Proc. Natl. Acad. Sci. USA 2007, 104, 19345–19350. [Google Scholar] [CrossRef] [PubMed]
- Qing, G.; Li, B.; Vu, A.; Skuli, N.; Walton, Z.E.; Liu, X.; Mayes, P.A.; Wise, D.R.; Thompson, C.B.; Maris, J.M.; et al. Atf4 regulates myc-mediated neuroblastoma cell death upon glutamine deprivation. Cancer Cell 2012, 22, 631–644. [Google Scholar] [CrossRef]
- Pinkel, D.; Segraves, R.; Sudar, D.; Clark, S.; Poole, I.; Kowbel, D.; Collins, C.; Kuo, W.L.; Chen, C.; Zhai, Y.; et al. High resolution analysis of DNA copy number variation using comparative genomic hybridization to microarrays. Nat. Genet. 1998, 20, 207–211. [Google Scholar] [CrossRef]
- Walker, C.; Joyce, K.A.; Thompson-Hehir, J.; Davies, M.P.; Gibbs, F.E.; Halliwell, N.; Lloyd, B.H.; Machell, Y.; Roebuck, M.M.; Salisbury, J.; et al. Characterisation of molecular alterations in microdissected archival gliomas. Acta Neuropathol. 2001, 101, 321–333. [Google Scholar] [CrossRef] [PubMed]
- Kaneko, Y.; Knudson, A.G. Mechanism and relevance of ploidy in neuroblastoma. Genes Chromosomes Cancer 2000, 29, 89–95. [Google Scholar] [CrossRef]
- Ladenstein, R.; Ambros, I.M.; Potschger, U.; Amann, G.; Urban, C.; Fink, F.M.; Schmitt, K.; Jones, R.; Slociak, M.; Schilling, F.; et al. Prognostic significance of DNA di-tetraploidy in neuroblastoma. Med. Pediatr. Oncol. 2001, 36, 83–92. [Google Scholar] [CrossRef]
- Fujita, T.; Igarashi, J.; Okawa, E.R.; Gotoh, T.; Manne, J.; Kolla, V.; Kim, J.; Zhao, H.; Pawel, B.R.; London, W.B.; et al. Chd5, a tumor suppressor gene deleted from 1p36.31 in neuroblastomas. J. Natl. Cancer Inst. 2008, 100, 940–949. [Google Scholar] [CrossRef]
- Schwab, M.; Westermann, F.; Hero, B.; Berthold, F. Neuroblastoma: Biology and molecular and chromosomal pathology. Lancet Oncol 2003, 4, 472–480. [Google Scholar] [CrossRef]
- Van Roy, N.; De Preter, K.; Hoebeeck, J.; Van Maerken, T.; Pattyn, F.; Mestdagh, P.; Vermeulen, J.; Vandesompele, J.; Speleman, F. The emerging molecular pathogenesis of neuroblastoma: Implications for improved risk assessment and targeted therapy. Genome Med 2009, 1, 74. [Google Scholar] [CrossRef]
- White, P.S.; Thompson, P.M.; Gotoh, T.; Okawa, E.R.; Igarashi, J.; Kok, M.; Winter, C.; Gregory, S.G.; Hogarty, M.D.; Maris, J.M.; et al. Definition and characterization of a region of 1p36.3 consistently deleted in neuroblastoma. Oncogene 2005, 24, 2684–2694. [Google Scholar] [CrossRef][Green Version]
- Michels, E.; Hoebeeck, J.; De Preter, K.; Schramm, A.; Brichard, B.; De Paepe, A.; Eggert, A.; Laureys, G.; Vandesompele, J.; Speleman, F. Cadm1 is a strong neuroblastoma candidate gene that maps within a 3.72 mb critical region of loss on 11q23. BMC Cancer 2008, 8, 173. [Google Scholar] [CrossRef] [PubMed]
- Mosse, Y.P.; Diskin, S.J.; Wasserman, N.; Rinaldi, K.; Attiyeh, E.F.; Cole, K.; Jagannathan, J.; Bhambhani, K.; Winter, C.; Maris, J.M. Neuroblastomas have distinct genomic DNA profiles that predict clinical phenotype and regional gene expression. Genes Chromosomes Cancer 2007, 46, 936–949. [Google Scholar] [CrossRef]
- Berry, T.; Luther, W.; Bhatnagar, N.; Jamin, Y.; Poon, E.; Sanda, T.; Pei, D.; Sharma, B.; Vetharoy, W.R.; Hallsworth, A.; et al. The alk(f1174l) mutation potentiates the oncogenic activity of mycn in neuroblastoma. Cancer Cell 2012, 22, 117–130. [Google Scholar] [CrossRef]
- Cazes, A.; Lopez-Delisle, L.; Tsarovina, K.; Pierre-Eugene, C.; De Preter, K.; Peuchmaur, M.; Nicolas, A.; Provost, C.; Louis-Brennetot, C.; Daveau, R.; et al. Activated alk triggers prolonged neurogenesis and ret upregulation providing a therapeutic target in alk-mutated neuroblastoma. Oncotarget 2014, 5, 2688–2702. [Google Scholar] [CrossRef]
- George, R.E.; Sanda, T.; Hanna, M.; Frohling, S.; Luther, W., 2nd; Zhang, J.; Ahn, Y.; Zhou, W.; London, W.B.; McGrady, P.; et al. Activating mutations in alk provide a therapeutic target in neuroblastoma. Nature 2008, 455, 975–978. [Google Scholar] [CrossRef] [PubMed]
- Janoueix-Lerosey, I.; Lequin, D.; Brugieres, L.; Ribeiro, A.; de Pontual, L.; Combaret, V.; Raynal, V.; Puisieux, A.; Schleiermacher, G.; Pierron, G.; et al. Somatic and germline activating mutations of the alk kinase receptor in neuroblastoma. Nature 2008, 455, 967–970. [Google Scholar] [CrossRef]
- Mosse, Y.P.; Laudenslager, M.; Longo, L.; Cole, K.A.; Wood, A.; Attiyeh, E.F.; Laquaglia, M.J.; Sennett, R.; Lynch, J.E.; Perri, P.; et al. Identification of alk as a major familial neuroblastoma predisposition gene. Nature 2008, 455, 930–935. [Google Scholar] [CrossRef] [PubMed]
- Ogawa, S.; Takita, J.; Sanada, M.; Hayashi, Y. Oncogenic mutations of alk in neuroblastoma. Cancer Sci 2011, 102, 302–308. [Google Scholar] [CrossRef]
- Pugh, T.J.; Morozova, O.; Attiyeh, E.F.; Asgharzadeh, S.; Wei, J.S.; Auclair, D.; Carter, S.L.; Cibulskis, K.; Hanna, M.; Kiezun, A.; et al. The genetic landscape of high-risk neuroblastoma. Nat. Genet. 2013, 45, 279–284. [Google Scholar] [CrossRef] [PubMed]
- Schulte, J.H.; Lindner, S.; Bohrer, A.; Maurer, J.; De Preter, K.; Lefever, S.; Heukamp, L.; Schulte, S.; Molenaar, J.; Versteeg, R.; et al. Mycn and alkf1174l are sufficient to drive neuroblastoma development from neural crest progenitor cells. Oncogene 2013, 32, 1059–1065. [Google Scholar] [CrossRef] [PubMed]
- Abel, F.; Dalevi, D.; Nethander, M.; Jornsten, R.; De Preter, K.; Vermeulen, J.; Stallings, R.; Kogner, P.; Maris, J.; Nilsson, S. A 6-gene signature identifies four molecular subgroups of neuroblastoma. Cancer Cell Int. 2011, 11, 9. [Google Scholar] [CrossRef]
- Brodeur, G.M.; Iyer, R.; Croucher, J.L.; Zhuang, T.; Higashi, M.; Kolla, V. Therapeutic targets for neuroblastomas. Expert Opin. Targets 2014, 18, 277–292. [Google Scholar] [CrossRef]
- Bartel, D.P. Metazoan micrornas. Cell 2018, 173, 20–51. [Google Scholar] [CrossRef] [PubMed]
- Bartel, D.P. Micrornas: Target recognition and regulatory functions. Cell 2009, 136, 215–233. [Google Scholar] [CrossRef] [PubMed]
- Fabian, M.R.; Sonenberg, N.; Filipowicz, W. Regulation of mrna translation and stability by micrornas. Annu. Rev. Biochem. 2010, 79, 351–379. [Google Scholar] [CrossRef]
- Misiak, D.; Hagemann, S.; Bell, J.L.; Busch, B.; Lederer, M.; Bley, N.; Schulte, J.H.; Hüttelmaier, S. The microrna landscape of mycn-amplified neuroblastoma. Front. Oncol. 2021, 11, 647737. [Google Scholar] [CrossRef] [PubMed]
- Wang, X. Improving microrna target prediction by modeling with unambiguously identified microrna-target pairs from clip-ligation studies. Bioinformatics 2016, 32, 1316–1322. [Google Scholar] [CrossRef]
- Wong, N.; Wang, X. Mirdb: An online resource for microrna target prediction and functional annotations. Nucleic Acids Res. 2015, 43, D146–D152. [Google Scholar] [CrossRef]
- Wang, X.; El Naqa, I.M. Prediction of both conserved and nonconserved microrna targets in animals. Bioinformatics 2008, 24, 325–332. [Google Scholar] [CrossRef]
- Wang, X. Mirdb: A microrna target prediction and functional annotation database with a wiki interface. RNA 2008, 14, 1012–1017. [Google Scholar] [CrossRef]
- Kozomara, A.; Birgaoanu, M.; Griffiths-Jones, S. Mirbase: From microrna sequences to function. Nucleic Acids Res. 2019, 47, D155–D162. [Google Scholar] [CrossRef] [PubMed]
- Kozomara, A.; Griffiths-Jones, S. Mirbase: Annotating high confidence micrornas using deep sequencing data. Nucleic Acids Res. 2014, 42, D68–D73. [Google Scholar] [CrossRef] [PubMed]
- Kozomara, A.; Griffiths-Jones, S. Mirbase: Integrating microrna annotation and deep-sequencing data. Nucleic Acids Res. 2011, 39, D152–D157. [Google Scholar] [CrossRef]
- Griffiths-Jones, S.; Saini, H.K.; van Dongen, S.; Enright, A.J. Mirbase: Tools for microrna genomics. Nucleic Acids Res. 2008, 36, D154–D158. [Google Scholar] [CrossRef]
- Griffiths-Jones, S.; Grocock, R.J.; van Dongen, S.; Bateman, A.; Enright, A.J. Mirbase: Microrna sequences, targets and gene nomenclature. Nucleic Acids Res. 2006, 34, D140–D144. [Google Scholar] [CrossRef] [PubMed]
- Griffiths-Jones, S. The microrna registry. Nucleic Acids Res. 2004, 32, D109–D111. [Google Scholar] [CrossRef]
- Mogilyansky, E.; Rigoutsos, I. The mir-17/92 cluster: A comprehensive update on its genomics, genetics, functions and increasingly important and numerous roles in health and disease. Cell Death Differ. 2013, 20, 1603–1614. [Google Scholar] [CrossRef] [PubMed]
- Buechner, J.; Tømte, E.; Haug, B.H.; Henriksen, J.R.; Løkke, C.; Flægstad, T.; Einvik, C. Tumour-suppressor micrornas let-7 and mir-101 target the proto-oncogene mycn and inhibit cell proliferation in mycn-amplified neuroblastoma. Br. J. Cancer 2011, 105, 296–303. [Google Scholar] [CrossRef]
- Zhao, Z.; Ma, X.; Shelton, S.D.; Sung, D.C.; Li, M.; Hernandez, D.; Zhang, M.; Losiewicz, M.D.; Chen, Y.; Pertsemlidis, A.; et al. A combined gene expression and functional study reveals the crosstalk between n-myc and differentiation-inducing micrornas in neuroblastoma cells. Oncotarget 2016, 7, 79372–79387. [Google Scholar] [CrossRef]
- Cole, K.A.; Attiyeh, E.F.; Mosse, Y.P.; Laquaglia, M.J.; Diskin, S.J.; Brodeur, G.M.; Maris, J.M. A functional screen identifies mir-34a as a candidate neuroblastoma tumor suppressor gene. Mol. Cancer Res. 2008, 6, 735–742. [Google Scholar] [CrossRef]
- Di Paolo, D.; Pastorino, F.; Brignole, C.; Corrias, M.V.; Emionite, L.; Cilli, M.; Tamma, R.; Priddy, L.; Amaro, A.; Ferrari, D.; et al. Combined replenishment of mir-34a and let-7b by targeted nanoparticles inhibits tumor growth in neuroblastoma preclinical models. Small 2020, 16, e1906426. [Google Scholar] [CrossRef] [PubMed]
- Soni, K.; Gupta, S.; Gokhale, S.S.; Dey, R.; Gunjal, A.D.; Kumar, V.A.; Pillai, B. Detection and knockdown of microrna-34a using thioacetamido nucleic acid. Nucleic Acid Ther. 2013, 23, 195–202. [Google Scholar] [CrossRef]
- Stallings, R.L. Microrna involvement in the pathogenesis of neuroblastoma: Potential for microrna mediated therapeutics. Curr. Pharm. Des. 2009, 15, 456–462. [Google Scholar] [CrossRef] [PubMed]
- Wei, J.S.; Song, Y.K.; Durinck, S.; Chen, Q.R.; Cheuk, A.T.; Tsang, P.; Zhang, Q.; Thiele, C.J.; Slack, A.; Shohet, J.; et al. The mycn oncogene is a direct target of mir-34a. Oncogene 2008, 27, 5204–5213. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.G.; He, J.H.; Yu, L.; Hang, Z.P.; Li, W.; Shun, W.H.; Huang, G.X. Microrna-202 suppresses mycn expression under the control of e2f1 in the neuroblastoma cell line lan-5. Mol. Med. Rep. 2014, 9, 541–546. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Lynch, J.; Fay, J.; Meehan, M.; Bryan, K.; Watters, K.M.; Murphy, D.M.; Stallings, R.L. Mirna-335 suppresses neuroblastoma cell invasiveness by direct targeting of multiple genes from the non-canonical tgf-β signalling pathway. Carcinogenesis 2012, 33, 976–985. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.J.; Li, G.L.; Lei, P.C. [effects of mir-144 on proliferation, apoptosis and cisplatin resistance by targeting mycn in pediatric neuroblastoma]. Zhonghua Zhong Liu Za Zhi 2019, 41, 516–521. [Google Scholar]
- Ramaiah, M.J.; Pushpavalli, S.N.; Lavanya, A.; Bhadra, K.; Haritha, V.; Patel, N.; Tamboli, J.R.; Kamal, A.; Bhadra, U.; Pal-Bhadra, M. Novel anthranilamide-pyrazolo [1,5-a]pyrimidine conjugates modulate the expression of p53-mycn associated micro rnas in neuroblastoma cells and cause cell cycle arrest and apoptosis. Bioorgan. Med. Chem. Lett. 2013, 23, 5699–5706. [Google Scholar] [CrossRef]
- Gan, L.; Xiu, R.; Ren, P.; Yue, M.; Su, H.; Guo, G.; Xiao, D.; Yu, J.; Jiang, H.; Liu, H.; et al. Metabolic targeting of oncogene myc by selective activation of the proton-coupled monocarboxylate family of transporters. Oncogene 2016, 35, 3037–3048. [Google Scholar] [CrossRef]
- Cheung, I.Y.; Farazi, T.A.; Ostrovnaya, I.; Xu, H.; Tran, H.; Mihailovic, A.; Tuschl, T.; Cheung, N.K. Deep microrna sequencing reveals downregulation of mir-29a in neuroblastoma central nervous system metastasis. Genes Chromosomes Cancer 2014, 53, 803–814. [Google Scholar] [CrossRef]
- Teitz, T.; Inoue, M.; Valentine, M.B.; Zhu, K.; Rehg, J.E.; Zhao, W.; Finkelstein, D.; Wang, Y.D.; Johnson, M.D.; Calabrese, C.; et al. Th-mycn mice with caspase-8 deficiency develop advanced neuroblastoma with bone marrow metastasis. Cancer Res. 2013, 73, 4086–4097. [Google Scholar] [CrossRef]
- Cheng, J.; Xu, L.; Deng, L.; Xue, L.; Meng, Q.; Wei, F.; Wang, J. Rna n(6)-methyladenosine modification is required for mir-98/mycn axis-mediated inhibition of neuroblastoma progression. Sci. Rep. 2020, 10, 13624. [Google Scholar] [CrossRef]
- Zhao, J.; Zhou, K.; Ma, L.; Zhang, H. Microrna-145 overexpression inhibits neuroblastoma tumorigenesis in vitro and in vivo. Bioengineered 2020, 11, 219–228. [Google Scholar] [CrossRef]
- Lovén, J.; Zinin, N.; Wahlström, T.; Müller, I.; Brodin, P.; Fredlund, E.; Ribacke, U.; Pivarcsi, A.; Påhlman, S.; Henriksson, M. Mycn-regulated micrornas repress estrogen receptor-alpha (esr1) expression and neuronal differentiation in human neuroblastoma. Proc. Natl. Acad. Sci. USA 2010, 107, 1553–1558. [Google Scholar] [CrossRef]
- Roth, S.A.; Hald, Ø.H.; Fuchs, S.; Løkke, C.; Mikkola, I.; Flægstad, T.; Schulte, J.; Einvik, C. Microrna-193b-3p represses neuroblastoma cell growth via downregulation of cyclin d1, mcl-1 and mycn. Oncotarget 2018, 9, 18160–18179. [Google Scholar] [CrossRef]
- Schulte, J.H.; Horn, S.; Otto, T.; Samans, B.; Heukamp, L.C.; Eilers, U.C.; Krause, M.; Astrahantseff, K.; Klein-Hitpass, L.; Buettner, R.; et al. Mycn regulates oncogenic micrornas in neuroblastoma. Int. J. Cancer 2008, 122, 699–704. [Google Scholar] [CrossRef] [PubMed]
- Samaraweera, L.; Spengler, B.A.; Ross, R.A. Reciprocal antagonistic regulation of n-myc mrna by mir-17 and the neuronal-specific rna-binding protein hud. Oncol. Rep. 2017, 38, 545–550. [Google Scholar] [CrossRef][Green Version]
- Claeys, S.; Denecker, G.; Durinck, K.; Decaesteker, B.; Mus, L.M.; Loontiens, S.; Vanhauwaert, S.; Althoff, K.; Wigerup, C.; Bexell, D.; et al. Alk positively regulates mycn activity through repression of hbp1 expression. Oncogene 2019, 38, 2690–2705. [Google Scholar] [CrossRef]
- Naraparaju, K.; Kolla, V.; Zhuang, T.; Higashi, M.; Iyer, R.; Kolla, S.; Okawa, E.R.; Blobel, G.A.; Brodeur, G.M. Role of micrornas in epigenetic silencing of the chd5 tumor suppressor gene in neuroblastomas. Oncotarget 2016, 7, 15977–15985. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Buechner, J.; Henriksen, J.R.; Haug, B.H.; Tømte, E.; Flaegstad, T.; Einvik, C. Inhibition of mir-21, which is up-regulated during mycn knockdown-mediated differentiation, does not prevent differentiation of neuroblastoma cells. Differ. Res. Biol. Divers. 2011, 81, 25–34. [Google Scholar] [CrossRef][Green Version]
- Westerlund, I.; Shi, Y.; Toskas, K.; Fell, S.M.; Li, S.; Surova, O.; Södersten, E.; Kogner, P.; Nyman, U.; Schlisio, S.; et al. Combined epigenetic and differentiation-based treatment inhibits neuroblastoma tumor growth and links hif2α to tumor suppression. Proc. Natl. Acad. Sci. USA 2017, 114, E6137–E6146. [Google Scholar] [CrossRef]
- Parodi, F.; Carosio, R.; Ragusa, M.; Di Pietro, C.; Maugeri, M.; Barbagallo, D.; Sallustio, F.; Allemanni, G.; Pistillo, M.P.; Casciano, I.; et al. Epigenetic dysregulation in neuroblastoma: A tale of mirnas and DNA methylation. Biochim. Biophys. Acta 2016, 1859, 1502–1514. [Google Scholar] [CrossRef] [PubMed]
- Uekusa, S.; Kawashima, H.; Sugito, K.; Yoshizawa, S.; Shinojima, Y.; Igarashi, J.; Ghosh, S.; Wang, X.; Fujiwara, K.; Ikeda, T.; et al. Nr4a3, a possibile oncogenic factor for neuroblastoma associated with cpgi methylation within the third exon. Int. J. Oncol. 2014, 44, 1669–1677. [Google Scholar] [CrossRef] [PubMed]
- Asada, K.; Watanabe, N.; Nakamura, Y.; Ohira, M.; Westermann, F.; Schwab, M.; Nakagawara, A.; Ushijima, T. Stronger prognostic power of the cpg island methylator phenotype than methylation of individual genes in neuroblastomas. JPN J. Clin. Oncol. 2013, 43, 641–645. [Google Scholar] [CrossRef][Green Version]
- Hoebeeck, J.; Michels, E.; Pattyn, F.; Combaret, V.; Vermeulen, J.; Yigit, N.; Hoyoux, C.; Laureys, G.; De Paepe, A.; Speleman, F.; et al. Aberrant methylation of candidate tumor suppressor genes in neuroblastoma. Cancer Lett. 2009, 273, 336–346. [Google Scholar] [CrossRef]
- Yan, P.; Mühlethaler, A.; Bourloud, K.B.; Beck, M.N.; Gross, N. Hypermethylation-mediated regulation of cd44 gene expression in human neuroblastoma. Genes Chromosomes Cancer 2003, 36, 129–138. [Google Scholar] [CrossRef]
- Korshunov, A.; Okonechnikov, K.; Schmitt-Hoffner, F.; Ryzhova, M.; Sahm, F.; Stichel, D.; Schrimpf, D.; Reuss, D.E.; Sievers, P.; Suwala, A.K.; et al. Molecular analysis of pediatric cns-pnet revealed nosologic heterogeneity and potent diagnostic markers for cns neuroblastoma with foxr2-activation. Acta Neuropathol. Commun. 2021, 9, 20. [Google Scholar] [CrossRef]
- Schmitt-Hoffner, F.; van Rijn, S.; Toprak, U.H.; Mauermann, M.; Rosemann, F.; Heit-Mondrzyk, A.; Hübner, J.M.; Camgöz, A.; Hartlieb, S.; Pfister, S.M.; et al. Foxr2 stabilizes mycn protein and identifies non-mycn-amplified neuroblastoma patients with unfavorable outcome. J. Clin. Oncol. 2021, 39, 3217–3228. [Google Scholar] [CrossRef]
- Colmenero-Repiso, A.; Gómez-Muñoz, M.A.; Rodríguez-Prieto, I.; Amador-Álvarez, A.; Henrich, K.O.; Pascual-Vaca, D.; Okonechnikov, K.; Rivas, E.; Westermann, F.; Pardal, R.; et al. Identification of vrk1 as a new neuroblastoma tumor progression marker regulating cell proliferation. Cancers 2020, 12, 3465. [Google Scholar] [CrossRef]
- Lin, H.Y.; Chuang, J.H.; Wang, P.W.; Lin, T.K.; Wu, M.T.; Hsu, W.M.; Chuang, H.C. 5-aza-2’-deoxycytidine induces a rig-i-related innate immune response by modulating mitochondria stress in neuroblastoma. Cells 2020, 9, 1920. [Google Scholar] [CrossRef] [PubMed]
- Gupta, S.; Doyle, K.; Mosbruger, T.L.; Butterfield, A.; Weston, A.; Ast, A.; Kaadige, M.; Verma, A.; Sharma, S. Reversible lsd1 inhibition with hci-2509 induces the p53 gene expression signature and disrupts the mycn signature in high-risk neuroblastoma cells. Oncotarget 2018, 9, 9907–9924. [Google Scholar] [CrossRef]
- Wong, M.; Tee, A.E.L.; Milazzo, G.; Bell, J.L.; Poulos, R.C.; Atmadibrata, B.; Sun, Y.; Jing, D.; Ho, N.; Ling, D.; et al. The histone methyltransferase dot1l promotes neuroblastoma by regulating gene transcription. Cancer Res. 2017, 77, 2522–2533. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.Y.; Atmadibrata, B.; Mondal, S.; Tee, A.E.; Liu, T. Ncym is upregulated by lncusmycn and modulates n-myc expression. Int. J. Oncol. 2016, 49, 2464–2470. [Google Scholar] [CrossRef] [PubMed]
- Henrich, K.O.; Bender, S.; Saadati, M.; Dreidax, D.; Gartlgruber, M.; Shao, C.; Herrmann, C.; Wiesenfarth, M.; Parzonka, M.; Wehrmann, L.; et al. Integrative genome-scale analysis identifies epigenetic mechanisms of transcriptional deregulation in unfavorable neuroblastomas. Cancer Res. 2016, 76, 5523–5537. [Google Scholar] [CrossRef] [PubMed]
- Fabian, J.; Opitz, D.; Althoff, K.; Lodrini, M.; Hero, B.; Volland, R.; Beckers, A.; de Preter, K.; Decock, A.; Patil, N.; et al. Mycn and hdac5 transcriptionally repress cd9 to trigger invasion and metastasis in neuroblastoma. Oncotarget 2016, 7, 66344–66359. [Google Scholar] [CrossRef] [PubMed]
- Ikram, F.; Ackermann, S.; Kahlert, Y.; Volland, R.; Roels, F.; Engesser, A.; Hertwig, F.; Kocak, H.; Hero, B.; Dreidax, D.; et al. Transcription factor activating protein 2 beta (tfap2b) mediates noradrenergic neuronal differentiation in neuroblastoma. Mol. Oncol. 2016, 10, 344–359. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Bell, J.L.; Carter, D.; Gherardi, S.; Poulos, R.C.; Milazzo, G.; Wong, J.W.; Al-Awar, R.; Tee, A.E.; Liu, P.Y.; et al. Wdr5 supports an n-myc transcriptional complex that drives a protumorigenic gene expression signature in neuroblastoma. Cancer Res. 2015, 75, 5143–5154. [Google Scholar] [CrossRef]
- Yáñez, Y.; Grau, E.; Rodríguez-Cortez, V.C.; Hervás, D.; Vidal, E.; Noguera, R.; Hernández, M.; Segura, V.; Cañete, A.; Conesa, A.; et al. Two independent epigenetic biomarkers predict survival in neuroblastoma. Clin. Epigenetics 2015, 7, 16. [Google Scholar] [CrossRef]
- Gonzalez-Gomez, P.; Bello, M.J.; Lomas, J.; Arjona, D.; Alonso, M.E.; Amiñoso, C.; Lopez-Marin, I.; Anselmo, N.P.; Sarasa, J.L.; Gutierrez, M.; et al. Aberrant methylation of multiple genes in neuroblastic tumours. Relationship with mycn amplification and allelic status at 1p. Eur. J. Cancer 2003, 39, 1478–1485. [Google Scholar] [CrossRef]
- Dreidax, D.; Bannert, S.; Henrich, K.O.; Schröder, C.; Bender, S.; Oakes, C.C.; Lindner, S.; Schulte, J.H.; Duffy, D.; Schwarzl, T.; et al. P19-ink4d inhibits neuroblastoma cell growth, induces differentiation and is hypermethylated and downregulated in mycn-amplified neuroblastomas. Hum. Mol. Genet. 2014, 23, 6826–6837. [Google Scholar] [CrossRef]
- Haruta, M.; Kamijo, T.; Nakagawara, A.; Kaneko, Y. Rassf1a methylation may have two biological roles in neuroblastoma tumorigenesis depending on the ploidy status and age of patients. Cancer Lett. 2014, 348, 167–176. [Google Scholar] [CrossRef]
- Lázcoz, P.; Muñoz, J.; Nistal, M.; Pestaña, A.; Encío, I.; Castresana, J.S. Frequent promoter hypermethylation of rassf1a and casp8 in neuroblastoma. BMC Cancer 2006, 6, 254. [Google Scholar] [CrossRef]
- Yang, Q.; Zage, P.; Kagan, D.; Tian, Y.; Seshadri, R.; Salwen, H.R.; Liu, S.; Chlenski, A.; Cohn, S.L. Association of epigenetic inactivation of rassf1a with poor outcome in human neuroblastoma. Clin. Cancer Res. 2004, 10, 8493–8500. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Fulda, S.; Poremba, C.; Berwanger, B.; Häcker, S.; Eilers, M.; Christiansen, H.; Hero, B.; Debatin, K.M. Loss of caspase-8 expression does not correlate with mycn amplification, aggressive disease, or prognosis in neuroblastoma. Cancer Res. 2006, 66, 10016–10023. [Google Scholar] [CrossRef] [PubMed]
- Casciano, I.; Banelli, B.; Croce, M.; De Ambrosis, A.; di Vinci, A.; Gelvi, I.; Pagnan, G.; Brignole, C.; Allemanni, G.; Ferrini, S.; et al. Caspase-8 gene expression in neuroblastoma. Ann. N. Y. Acad. Sci. 2004, 1028, 157–167. [Google Scholar] [CrossRef]
- Sugito, K.; Kawashima, H.; Yoshizawa, S.; Uekusa, S.; Hoshi, R.; Furuya, T.; Kaneda, H.; Hosoda, T.; Konuma, N.; Masuko, T.; et al. Non-promoter DNA hypermethylation of zygote arrest 1 (zar1) in neuroblastomas. J. Pediatr. Surg. 2013, 48, 782–788. [Google Scholar] [CrossRef]
- Casalà, C.; Gil-Guiñón, E.; Ordóñez, J.L.; Miguel-Queralt, S.; Rodríguez, E.; Galván, P.; Lavarino, C.; Munell, F.; de Alava, E.; Mora, J.; et al. The calcium-sensing receptor is silenced by genetic and epigenetic mechanisms in unfavorable neuroblastomas and its reactivation induces erk1/2-dependent apoptosis. Carcinogenesis 2013, 34, 268–276. [Google Scholar] [CrossRef]
- Decock, A.; Ongenaert, M.; Hoebeeck, J.; De Preter, K.; Van Peer, G.; Van Criekinge, W.; Ladenstein, R.; Schulte, J.H.; Noguera, R.; Stallings, R.L.; et al. Genome-wide promoter methylation analysis in neuroblastoma identifies prognostic methylation biomarkers. Genome Biol. 2012, 13, R95. [Google Scholar] [CrossRef]
- Lau, D.T.; Hesson, L.B.; Norris, M.D.; Marshall, G.M.; Haber, M.; Ashton, L.J. Prognostic significance of promoter DNA methylation in patients with childhood neuroblastoma. Clin. Cancer Res. 2012, 18, 5690–5700. [Google Scholar] [CrossRef]
- Sugito, K.; Kawashima, H.; Uekusa, S.; Yoshizawa, S.; Hoshi, R.; Furuya, T.; Kaneda, H.; Hosoda, T.; Masuko, T.; Ohashi, K.; et al. Identification of aberrant methylation regions in neuroblastoma by screening of tissue-specific differentially methylated regions. Pediatr. Blood Cancer 2013, 60, 383–389. [Google Scholar] [CrossRef] [PubMed]
- Djos, A.; Martinsson, T.; Kogner, P.; Carén, H. The rassf gene family members rassf5, rassf6 and rassf7 show frequent DNA methylation in neuroblastoma. Mol. Cancer 2012, 11, 40. [Google Scholar] [CrossRef]
- Misawa, A.; Tanaka, S.; Yagyu, S.; Tsuchiya, K.; Iehara, T.; Sugimoto, T.; Hosoi, H. Rassf1a hypermethylation in pretreatment serum DNA of neuroblastoma patients: A prognostic marker. Br. J. Cancer 2009, 100, 399–404. [Google Scholar] [CrossRef]
- Gumy-Pause, F.; Pardo, B.; Khoshbeen-Boudal, M.; Ansari, M.; Gayet-Ageron, A.; Sappino, A.P.; Attiyeh, E.F.; Ozsahin, H. Gstp1 hypermethylation is associated with reduced protein expression, aggressive disease and prognosis in neuroblastoma. Genes Chromosomes Cancer 2012, 51, 174–185. [Google Scholar] [CrossRef]
- Nair, P.N.; McArdle, L.; Cornell, J.; Cohn, S.L.; Stallings, R.L. High-resolution analysis of 3p deletion in neuroblastoma and differential methylation of the sema3b tumor suppressor gene. Cancer Genet. Cytogenet. 2007, 174, 100–110. [Google Scholar] [CrossRef] [PubMed]
- Lázcoz, P.; Muñoz, J.; Nistal, M.; Pestaña, A.; Encío, I.J.; Castresana, J.S. Loss of heterozygosity and microsatellite instability on chromosome arm 10q in neuroblastoma. Cancer Genet. Cytogenet. 2007, 174, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Erdreich-Epstein, A.; Singh, A.R.; Joshi, S.; Vega, F.M.; Guo, P.; Xu, J.; Groshen, S.; Ye, W.; Millard, M.; Campan, M.; et al. Association of high microvessel alphavbeta3 and low pten with poor outcome in stage 3 neuroblastoma: Rationale for using first in class dual pi3k/brd4 inhibitor, sf1126. Oncotarget 2017, 8, 52193–52210. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Choi, J.H.; Jeong, Y.J.; Yu, A.R.; Yoon, K.S.; Choe, W.; Ha, J.; Kim, S.S.; Yeo, E.J.; Kang, I. Fluoxetine induces apoptosis through endoplasmic reticulum stress via mitogen-activated protein kinase activation and histone hyperacetylation in sk-n-be(2)-m17 human neuroblastoma cells. Apoptosis Int. J. Program. Cell Death 2017, 22, 1079–1097. [Google Scholar] [CrossRef]
- Wang, S.S.; Hsiao, R.; Limpar, M.M.; Lomahan, S.; Tran, T.A.; Maloney, N.J.; Ikegaki, N.; Tang, X.X. Destabilization of myc/mycn by the mitochondrial inhibitors, metaiodobenzylguanidine, metformin and phenformin. Int. J. Mol. Med. 2014, 33, 35–42. [Google Scholar] [CrossRef] [PubMed]
- Hegarty, S.V.; Togher, K.L.; O’Leary, E.; Solger, F.; Sullivan, A.M.; O’Keeffe, G.W. Romidepsin induces caspase-dependent cell death in human neuroblastoma cells. Neurosci. Lett. 2017, 653, 12–18. [Google Scholar] [CrossRef] [PubMed]
- Fabian, J.; Lodrini, M.; Oehme, I.; Schier, M.C.; Thole, T.M.; Hielscher, T.; Kopp-Schneider, A.; Opitz, L.; Capper, D.; von Deimling, A.; et al. Grhl1 acts as tumor suppressor in neuroblastoma and is negatively regulated by mycn and hdac3. Cancer Res. 2014, 74, 2604–2616. [Google Scholar] [CrossRef] [PubMed]
- Cheung, N.K.; Dyer, M.A. Neuroblastoma: Developmental biology, cancer genomics and immunotherapy. Nat. Rev. Cancer 2013, 13, 397–411. [Google Scholar] [CrossRef]
- Brodeur, G.M.; Seeger, R.C.; Schwab, M.; Varmus, H.E.; Bishop, J.M. Amplification of n-myc in untreated human neuroblastomas correlates with advanced disease stage. Science 1984, 224, 1121–1124. [Google Scholar] [CrossRef]
- Chen, L.; Iraci, N.; Gherardi, S.; Gamble, L.D.; Wood, K.M.; Perini, G.; Lunec, J.; Tweddle, D.A. P53 is a direct transcriptional target of mycn in neuroblastoma. Cancer Res. 2010, 70, 1377–1388. [Google Scholar] [CrossRef]
- Helmsauer, K.; Valieva, M.E.; Ali, S.; Chamorro González, R.; Schöpflin, R.; Röefzaad, C.; Bei, Y.; Dorado Garcia, H.; Rodriguez-Fos, E.; Puiggròs, M.; et al. Enhancer hijacking determines extrachromosomal circular mycn amplicon architecture in neuroblastoma. Nat. Commun. 2020, 11, 5823. [Google Scholar] [CrossRef]
- George, R.E.; London, W.B.; Cohn, S.L.; Maris, J.M.; Kretschmar, C.; Diller, L.; Brodeur, G.M.; Castleberry, R.P.; Look, A.T. Hyperdiploidy plus nonamplified mycn confers a favorable prognosis in children 12 to 18 months old with disseminated neuroblastoma: A pediatric oncology group study. J. Clin. 2005, 23, 6466–6473. [Google Scholar] [CrossRef] [PubMed]
- Maris, J.M. The biologic basis for neuroblastoma heterogeneity and risk stratification. Curr. Opin. Pediatr. 2005, 17, 7–13. [Google Scholar] [CrossRef]
- Schmidt, M.L.; Lal, A.; Seeger, R.C.; Maris, J.M.; Shimada, H.; O’Leary, M.; Gerbing, R.B.; Matthay, K.K. Favorable prognosis for patients 12 to 18 months of age with stage 4 nonamplified mycn neuroblastoma: A children’s cancer group study. J. Clin. Oncol. 2005, 23, 6474–6480. [Google Scholar] [CrossRef] [PubMed]
- Cohn, S.L.; Pearson, A.D.; London, W.B.; Monclair, T.; Ambros, P.F.; Brodeur, G.M.; Faldum, A.; Hero, B.; Iehara, T.; Machin, D.; et al. The international neuroblastoma risk group (inrg) classification system: An inrg task force report. J. Clin. Oncol. 2009, 27, 289–297. [Google Scholar] [CrossRef] [PubMed]
- Godfried, M.B.; Veenstra, M.; v Sluis, P.; Boon, K.; v Asperen, R.; Hermus, M.C.; v Schaik, B.D.; Voute, T.P.; Schwab, M.; Versteeg, R.; et al. The n-myc and c-myc downstream pathways include the chromosome 17q genes nm23-h1 and nm23-h2. Oncogene 2002, 21, 2097–2101. [Google Scholar] [CrossRef] [PubMed]
- Akhavanfard, S.; Nohr, E.; AlNajjar, M.; Haughn, M.; Hashimoto, S.; Deeg, C.; Pfau, R.; Brundler, M.A.; Reshmi, S.C. 5′ alk amplification in neuroblastoma: A case report. Case Rep. Oncol. 2021, 14, 585–591. [Google Scholar] [CrossRef]
- Bellini, A.; Pötschger, U.; Bernard, V.; Lapouble, E.; Baulande, S.; Ambros, P.F.; Auger, N.; Beiske, K.; Bernkopf, M.; Betts, D.R.; et al. Frequency and prognostic impact of alk amplifications and mutations in the european neuroblastoma study group (siopen) high-risk neuroblastoma trial (hr-nbl1). J. Clin. Oncol. 2021, 39, 3377–3390. [Google Scholar] [CrossRef]
- Javanmardi, N.; Fransson, S.; Djos, A.; Umapathy, G.; Östensson, M.; Milosevic, J.; Borenäs, M.; Hallberg, B.; Kogner, P.; Martinsson, T.; et al. Analysis of alk, mycn, and the alk ligand alkal2 (fam150b/augalpha) in neuroblastoma patient samples with chromosome arm 2p rearrangements. Genes Chromosomes Cancer 2019, 59, 50–57. [Google Scholar] [CrossRef]
- Janoueix-Lerosey, I.; Schleiermacher, G.; Michels, E.; Mosseri, V.; Ribeiro, A.; Lequin, D.; Vermeulen, J.; Couturier, J.; Peuchmaur, M.; Valent, A.; et al. Overall genomic pattern is a predictor of outcome in neuroblastoma. J. Clin. Oncol. 2009, 27, 1026–1033. [Google Scholar] [CrossRef] [PubMed]
- Lastowska, M.; Viprey, V.; Santibanez-Koref, M.; Wappler, I.; Peters, H.; Cullinane, C.; Roberts, P.; Hall, A.G.; Tweddle, D.A.; Pearson, A.D.; et al. Identification of candidate genes involved in neuroblastoma progression by combining genomic and expression microarrays with survival data. Oncogene 2007, 26, 7432–7444. [Google Scholar] [CrossRef] [PubMed]
- Vandesompele, J.; Baudis, M.; De Preter, K.; Van Roy, N.; Ambros, P.; Bown, N.; Brinkschmidt, C.; Christiansen, H.; Combaret, V.; Lastowska, M.; et al. Unequivocal delineation of clinicogenetic subgroups and development of a new model for improved outcome prediction in neuroblastoma. J. Clin. Oncol. 2005, 23, 2280–2299. [Google Scholar] [CrossRef] [PubMed]
- Spitz, R.; Hero, B.; Ernestus, K.; Berthold, F. Fish analyses for alterations in chromosomes 1, 2, 3, and 11 define high-risk groups in neuroblastoma. Med. Pediatr. Oncol. 2003, 41, 30–35. [Google Scholar] [CrossRef]
- Chen, Q.R.; Bilke, S.; Wei, J.S.; Whiteford, C.C.; Cenacchi, N.; Krasnoselsky, A.L.; Greer, B.T.; Son, C.G.; Westermann, F.; Berthold, F.; et al. Cdna array-cgh profiling identifies genomic alterations specific to stage and mycn-amplification in neuroblastoma. BMC Genom. 2004, 5, 70. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Fang, Y.F.; Wu, D.M.; Lin, Y.; Zhang, B.; Liu, M.K.; Bai, J.X.; Chen, F. Comparison of the efficacy of minimally invasive and open surgery on children with neuroblastoma: A meta-analysis. J. Laparoendosc. Adv. Surg. Tech. Part A 2021, 31, 829–838. [Google Scholar] [CrossRef] [PubMed]
- Brisse, H.J.; McCarville, M.B.; Granata, C.; Krug, K.B.; Wootton-Gorges, S.L.; Kanegawa, K.; Giammarile, F.; Schmidt, M.; Shulkin, B.L.; Matthay, K.K.; et al. Guidelines for imaging and staging of neuroblastic tumors: Consensus report from the international neuroblastoma risk group project. Radiology 2011, 261, 243–257. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.; Pearson, A.; Lunec, J. The mycn oncoprotein as a drug development target. Cancer Lett. 2003, 197, 125–130. [Google Scholar] [CrossRef]
- Kling, M.J.; Griggs, C.N.; McIntyre, E.M.; Alexander, G.; Ray, S.; Challagundla, K.B.; Joshi, S.S.; Coulter, D.W.; Chaturvedi, N.K. Synergistic efficacy of inhibiting mycn and mtor signaling against neuroblastoma. BMC Cancer 2021, 21, 1061. [Google Scholar] [CrossRef] [PubMed]
- Di Giulio, S.; Colicchia, V.; Pastorino, F.; Pedretti, F.; Fabretti, F.; Nicolis di Robilant, V.; Ramponi, V.; Scafetta, G.; Moretti, M.; Licursi, V.; et al. A combination of parp and chk1 inhibitors efficiently antagonizes mycn-driven tumors. Oncogene 2021, 40, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Ding, J.; Zhu, S.; Alptekin, A.; Dong, Z.; Yan, C.; Zha, Y.; Ding, H.F. Therapeutic targeting of both dihydroorotate dehydrogenase and nucleoside transport in mycn-amplified neuroblastoma. Cell Death Dis. 2021, 12, 821. [Google Scholar] [CrossRef] [PubMed]
- Song, X.; Wang, L.; Wang, T.; Hu, J.; Wang, J.; Tu, R.; Su, H.; Jiang, J.; Qing, G.; Liu, H. Synergistic targeting of chk1 and mtor in myc-driven tumors. Carcinogenesis 2021, 42, 448–460. [Google Scholar] [CrossRef]
- Waetzig, R.; Matthes, M.; Leister, J.; Penkivech, G.; Heise, T.; Corbacioglu, S.; Sommer, G. Comparing mtor inhibitor rapamycin with torin-2 within the rist molecular-targeted regimen in neuroblastoma cells. Int. J. Med Sci. 2021, 18, 137–149. [Google Scholar] [CrossRef]
- Dedoni, S.; Marras, L.; Olianas, M.C.; Ingianni, A.; Onali, P. The neurotrophin receptor trkc as a novel molecular target of the antineuroblastoma action of valproic acid. Int. J. Mol. Sci. 2021, 22, 7790. [Google Scholar] [CrossRef]
- Takatori, A.; Hossain, M.S.; Ogura, A.; Akter, J.; Nakamura, Y.; Nakagawara, A. Nlrr1 is a potential therapeutic target in neuroblastoma and mycn-driven malignant cancers. Front. Oncol. 2021, 11, 669667. [Google Scholar] [CrossRef]
- Liu, C.; Gen, Y.; Tanimoto, K.; Muramatsu, T.; Inoue, J.; Inazawa, J. Concurrent targeting of map3k3 and brd4 by mir-3140-3p overcomes acquired resistance to bet inhibitors in neuroblastoma cells. Mol. Ther. Nucleic Acids 2021, 25, 83–92. [Google Scholar] [CrossRef] [PubMed]
- Schultz, C.R.; Swanson, M.A.; Dowling, T.C.; Bachmann, A.S. Probenecid increases renal retention and antitumor activity of dfmo in neuroblastoma. Cancer Chemother. Pharmacol. 2021, 88, 607–617. [Google Scholar] [CrossRef] [PubMed]
- Dalton, K.M.; Krytska, K.; Lochmann, T.L.; Sano, R.; Casey, C.; D’Aulerio, A.; Khan, Q.A.; Crowther, G.S.; Coon, C.; Cai, J.; et al. Venetoclax-based rational combinations are effective in models of mycn-amplified neuroblastoma. Mol. Cancer Ther. 2021, 20, 1400–1411. [Google Scholar] [CrossRef] [PubMed]
- Ando, K.; Nakagawara, A. Acceleration or brakes: Which is rational for cell cycle-targeting neuroblastoma therapy? Biomolecules 2021, 11, 750. [Google Scholar] [CrossRef]
- Umapathy, G.; Mendoza-Garcia, P.; Hallberg, B.; Palmer, R.H. Targeting anaplastic lymphoma kinase in neuroblastoma. Acta Pathol. Microbiol. Immunol. Scand. 2019, 127, 288–302. [Google Scholar] [CrossRef] [PubMed]
- Hu, Z.; Cheng, C.; Wang, Y.; Chen, T.; Tu, J.; Niu, C.; Xing, R.; Wang, Y.; Xu, Y. Synergistic effect of statins and abiraterone acetate on the growth inhibition of neuroblastoma via targeting androgen receptor. Front. Oncol. 2021, 11, 595285. [Google Scholar] [CrossRef] [PubMed]
- Dubiella, C.; Pinch, B.J.; Koikawa, K.; Zaidman, D.; Poon, E.; Manz, T.D.; Nabet, B.; He, S.; Resnick, E.; Rogel, A.; et al. Sulfopin is a covalent inhibitor of pin1 that blocks myc-driven tumors in vivo. Nat. Chem. Biol. 2021, 17, 954–963. [Google Scholar] [CrossRef] [PubMed]
- Wood, L.; Huang, M.; Zeki, J.; Gong, M.; Taylor, J.; Shimada, H.; Chiu, B. Combining inhibitors of brd4 and cyclin-dependent kinase can decrease tumor growth in neuroblastoma with mycn amplification. J. Pediatr. Surg. 2021, 56, 1199–1202. [Google Scholar] [CrossRef] [PubMed]
- Dalton, K.M.; Lochmann, T.L.; Floros, K.V.; Calbert, M.L.; Kurupi, R.; Stein, G.T.; McClanaghan, J.; Murchie, E.; Egan, R.K.; Greninger, P.; et al. Catastrophic atp loss underlies a metabolic combination therapy tailored for mycn-amplified neuroblastoma. Proc. Natl. Acad. Sci. USA 2021, 118, e2009620118. [Google Scholar] [CrossRef] [PubMed]
- Cheung, B.B.; Kleynhans, A.; Mittra, R.; Kim, P.Y.; Holien, J.K.; Nagy, Z.; Ciampa, O.C.; Seneviratne, J.A.; Mayoh, C.; Raipuria, M.; et al. A novel combination therapy targeting ubiquitin-specific protease 5 in mycn-driven neuroblastoma. Oncogene 2021, 40, 2367–2381. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).