Targeting NSP16 Methyltransferase for the Broad-Spectrum Clinical Management of Coronaviruses: Managing the Next Pandemic
Abstract
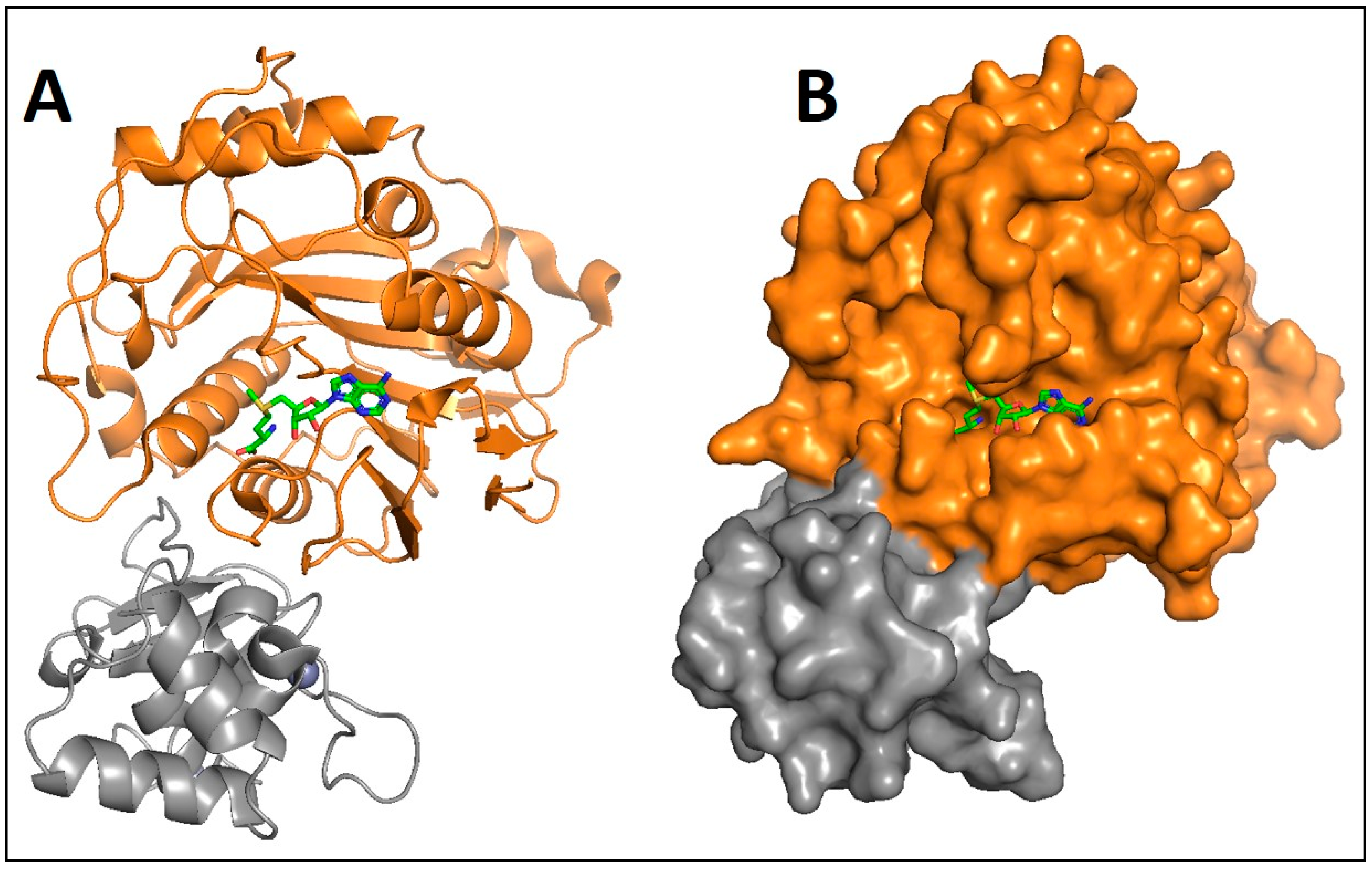
1. Introduction
2. Discussion and Conclusions
Conflicts of Interest
References
- Dong, S.; Sun, J.; Mao, Z.; Wang, L.; Lu, Y.-L.; Li, J. A guideline for homology modeling of the proteins from newly discovered betacoronavirus, 2019 novel coronavirus (2019-nCoV). J. Med. Virol. 2020, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Ke, M.; Chen, Y.; Wu, A.; Sun, Y.; Su, C.; Wu, H.; Jin, X.; Tao, J.; Wang, Y.; Ma, X.; et al. Short peptides derived from the interaction domain of SARS coronavirus nonstructural protein nsp10 can suppress the 2′-O-methyltransferase activity of nsp10/nsp16 complex. Virus Res. 2012, 167, 322–328. [Google Scholar] [CrossRef] [PubMed]
- Decroly, E.; Debarnot, C.; Ferron, F.; Bouvet, M.; Coutard, B.; Imbert, I.; Gluais, L.; Papageorgiou, N.; Sharff, A.; Bricogne, G.; et al. Crystal structure and functional analysis of the SARS-Coronavirus RNA cap 2′-O-Methyltransferase nsp10/nsp16 complex. PLoS Pathog. 2011, 7, e1002059. [Google Scholar] [CrossRef] [PubMed]
- Aouadi, W.; Blanjoie, A.; Vasseur, J.-J.; Debart, F.; Canard, B.; Decroly, E. Binding of the methyl donor S-Adenosyl-L-Methionine to Middle East respiratory syndrome Coronavirus 2′-O-Methyltransferase nsp16 promotes recruitment of the allosteric activator nsp10. J. Virol. 2017, 91, e02217-16. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Su, C.; Ke, M.; Jin, X.; Xu, L.; Zhang, Z.; Wu, A.; Sun, Y.; Yang, Z.; Tien, P.; et al. Biochemical and structural insights into the mechanisms of SARS coronavirus RNA ribose 2′-O-methylation by nsp16/nsp10 protein complex. PLoS Pathog. 2011, 7, e1002294. [Google Scholar] [CrossRef] [PubMed]
- Sohag, A.A.M.; Hannan, M.A.; Rahman, S.; Hossain, M.; Hasan, M.; Khan, M.K.; Khatun, A.; Dash, R.; Uddin, M.J. Revisiting potential druggable targets against SARS-CoV-2 and repurposing therapeutics under preclinical study and clinical trials: A comprehensive review. Drug Dev Res. 2020, 81, 919–941. [Google Scholar] [CrossRef] [PubMed]
- Chéron, N.; Yu, C.; Kolawole, A.; Shakhnovich, E.; Wobus, C. Repur-Posing of rutin for the inhibition of norovirus replication. In Archives of Virology; Springer: Berlin, Germany, 2015; Volume 160, pp. 2353–2358. [Google Scholar]
- Irwin, J.J.; Shoichet, B.K. ZINC—A free database of commercially available compounds for virtual screening. J. Chem. Inf. Model. 2015, 45, 177–182. [Google Scholar] [CrossRef] [PubMed]
- Tazikeh-Lemeski, E.; Moradi, S.; Raoufi, R.; Shahlaei, M.; Janlou, M.A.M.; Zolghadri, S. Targeting SARS-COV-2 non-structural protein 16: A virtual drug repurposing study. J. Biomol. Struct. Dyn. 2020, 23, 1–14. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alshiraihi, I.M.; Klein, G.L.; Brown, M.A. Targeting NSP16 Methyltransferase for the Broad-Spectrum Clinical Management of Coronaviruses: Managing the Next Pandemic. Diseases 2021, 9, 12. https://doi.org/10.3390/diseases9010012
Alshiraihi IM, Klein GL, Brown MA. Targeting NSP16 Methyltransferase for the Broad-Spectrum Clinical Management of Coronaviruses: Managing the Next Pandemic. Diseases. 2021; 9(1):12. https://doi.org/10.3390/diseases9010012
Chicago/Turabian StyleAlshiraihi, Ilham M., Gerald L. Klein, and Mark A. Brown. 2021. "Targeting NSP16 Methyltransferase for the Broad-Spectrum Clinical Management of Coronaviruses: Managing the Next Pandemic" Diseases 9, no. 1: 12. https://doi.org/10.3390/diseases9010012
APA StyleAlshiraihi, I. M., Klein, G. L., & Brown, M. A. (2021). Targeting NSP16 Methyltransferase for the Broad-Spectrum Clinical Management of Coronaviruses: Managing the Next Pandemic. Diseases, 9(1), 12. https://doi.org/10.3390/diseases9010012





