Abstract
The bioactivity of glucosinolates (GSs), and more specifically their hydrolysis products (GSHPs), has been well documented. These secondary metabolites evolved in the order Brassicales as plant defense compounds with proven ability to deter or impede the growth of several biotic challenges including insect infestation, fungal and bacterial infection, and competition from other plants. However, the bioactivity of GSHPs is not limited to activity that inhibits these kingdoms of life. Many of these compounds have been shown to have bioactivity in mammalian systems as well, with epidemiological links to cancer chemoprevention in humans supported by in vitro, in vivo, and small clinical studies. Although other chemopreventive mechanisms have been identified, the primary mechanism believed to be responsible for the observed chemoprevention from GSHPs is the induction of antioxidant enzymes, such as NAD(P)H quinone reductase (NQO1), heme oxygenase 1 (HO-1), glutamate-cysteine ligase catalytic subunit (GCLC), and glutathione S transferases (GSTs), through the Keap1-Nrf2-ARE signaling pathway. Induction of this pathway is generally associated with aliphatic isothiocyanate GSHPs, although some indole-derived GSHPs have also been associated with induction of one or more of these enzymes.
1. Introduction
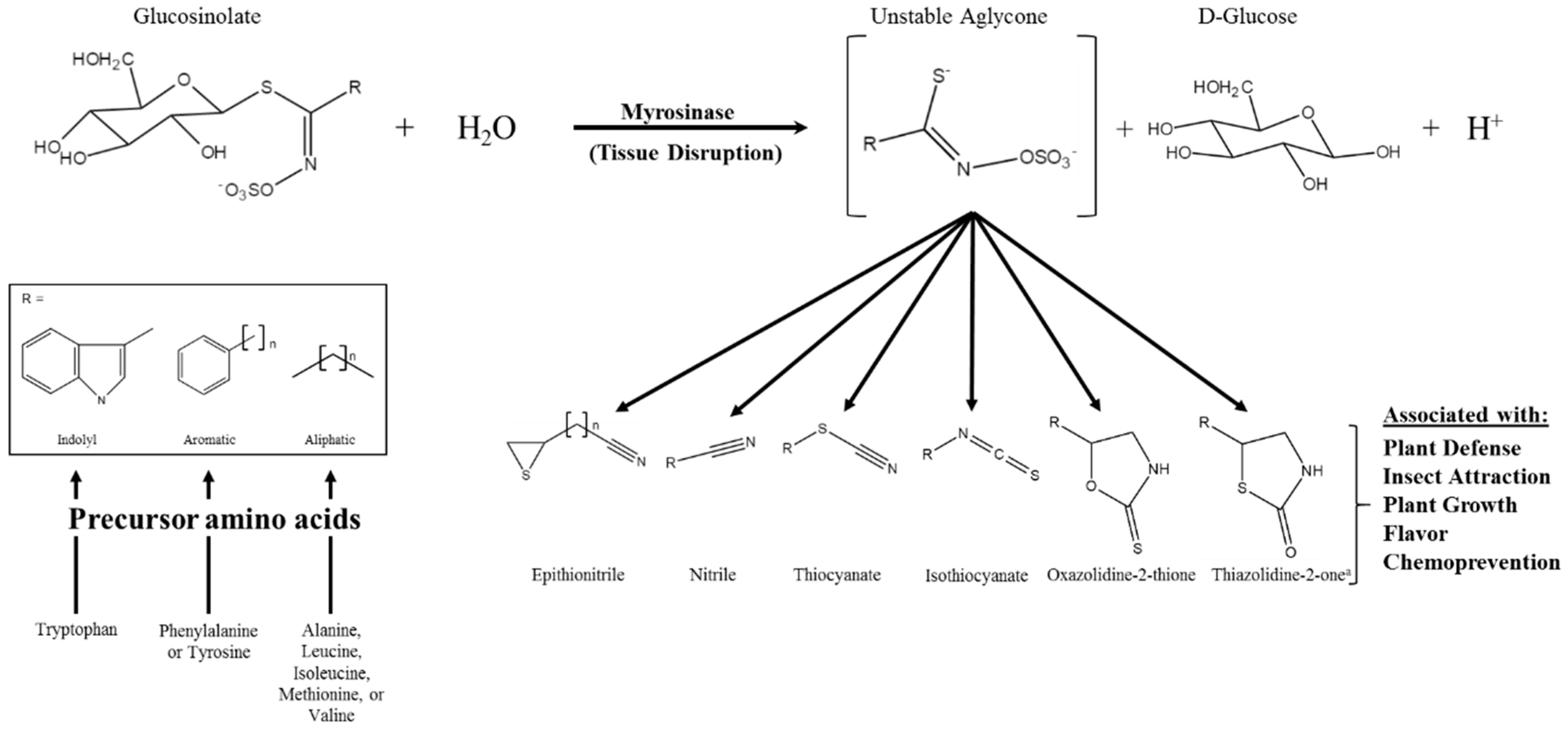
Glucosinolates (GSs) are a class of amino acid-derived, sulfur-rich secondary metabolites found in the order Brassicales, which includes the scientifically and economically important genera of Arabidopsis and Brassica [1,2]. Glucosinolates are a well-defined diverse class of secondary metabolites with approximately 132 documented GSs structures by 2011 and at least a dozen additional natural structures awaiting elucidation [3,4]. Glucosinolates are generally classified based on the structure of their precursor amino acid, with aliphatic, indole, and aromatic GSs derived from methionine, tryptophan, and phenylalanine/tyrosine, respectively, being the major classes found in the Brassica genus [5,6]. Glucosinolates are “activated” by a class of hydrolytic enzymes called myrosinases, which, for the most part [7,8], are physically separated from GSs in intact cells [9,10,11]. Upon tissue disruption, the hydrolysis reaction mediated by myrosinase results in the formation of GS hydrolysis products (GSHPs), which are considered the bioactive component of this system (Figure 1; [12]). These compounds are well known for their role in plant defense against herbivory and pathogens [13,14,15,16]. However, perhaps more importantly to humans, they have also been associated with cancer chemoprevention [17,18].
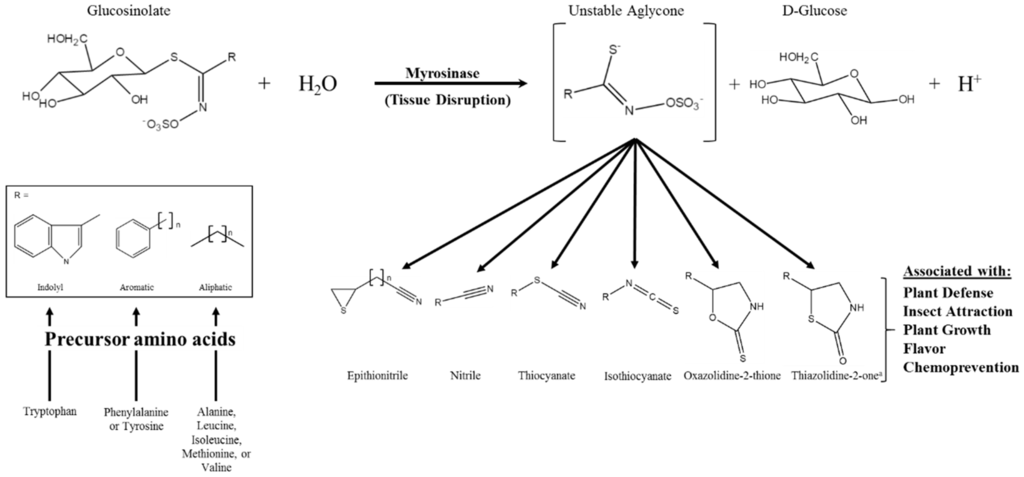
Figure 1.
Glucosinolate hydrolysis by myrosinase, possible hydrolysis products, and the various bioactivities associated with those products; modifed from [12,19]. The generic glucosinolate side chain structures (R) can undergo further modifications at virtually every position. a Novel hydrolysis product structure recently described in [20].
Chemoprevention of cancer has been an active field of research since the seminal review by Wattenberg [21] in which he described early in vivo experiments with compounds that displayed an ability to inhibit cancer formation in animal models. To describe this observed result, he coined the term “chemoprophylaxis of carcinogenesis”, which has since evolved into “cancer chemoprevention” [22]. In the almost five decades since, the research of chemical compounds that can inhibit the formation of neoplasms and/or aid in ridding the body of neoplastic cells, through apoptosis or other means, has been steadily increasing in popularity and promise.
Among the compounds that have shown chemopreventive qualities are the GSHPs, particularly isothiocyanates (ITCs) derived from aliphatic GS precursors [23]. From the indole GS precursor, glucobrassicin, the ITC-derived hydrolysis products indole-3-carbinol (I3C) and 3,3′-diindolylmethane (DIM) [24] have shown promising chemopreventive effects against hormone-responsive cancers, such as those of the breast, prostate, and ovaries [25]. Also, several ITCs have been linked to a number of chemopreventive mechanisms, such as: induction of cytoprotective proteins through the Keap1/Nrf2/ARE pathway, inhibition of proinflammatory responses through the regulation of the NFκB pathway, induction of cell cycle arrest and apoptosis, effects on heat shock proteins, and inhibition of angiogenesis and metastasis [26]. Inhibition of histone deacetylase (HDAC) enzymes by a number of ITCs has been implicated as at least one of the mechanisms responsible for the observed apoptotic properties of these GSHPs [26,27]. Though a significant amount of research has been done on these and other compounds from cruciferous vegetables, there is still much to be learned about the specific chemopreventive mechanisms and synergistic relationships of these compounds [28,29]. Of the several chemopreventive mechanisms of GSHPs, the best studied is probably their action on the Keap1/Nrf2/ARE (antioxidant response element) pathway, which is discussed in more detail below as it is a central mechanism by which GSHPs reduce oxidative stress in human cells.
Other compounds known to affect cellular oxidative stress are the vitamins A, C, and E, which are also relatively abundant in Brassica vegetables. An excellent review by Bodupalli et al. [30] discusses how these known antioxidant compounds may contribute to reduction of oxidative stress in humans synergistically with ITCs. This is proposed to be achieved through the induction of ARE-dependent enzymes by GSHPs that maintain the free radical scavenging capabilities of the vitamins, increasing their biologically effective life span.
2. The Influence of Diet on Cancer Development
The twentieth century saw many great medical revolutions, from penicillin to the polio vaccine, which led to a severe decline in the number and proportion of total deaths due to infectious disease in the U.S. However, as infectious diseases were better managed, death due to chronic health problems, such as heart disease and cancer, became more common. In 1900, cancer and heart disease reportedly caused 64 and 137.4 of every 100,000 deaths, respectively. By 2010, those numbers had increased to 185.9 for cancer and 192.9 for heart disease, making these the most common causes of death among Americans as a whole [31]. The increased prevalence of these diseases has led to increased research into their causes and modes of prevention. Prevention and treatment of heart disease has experienced great strides, as mortality from heart disease has been on a steady decline since the 1970s [32]. In fact, cancer has surpassed heart disease as the leading cause of death in individuals between the ages of 40 and 79 [33]. For this reason, cancer is now a major focus of medical research. Specifically, understanding the mechanisms of carcinogenesis, metastasis, and the overall progression of cancer is of the utmost importance in accomplishing the goal of decreasing new cancer incidence. It is known that there are certain genetic factors that can lead to increased risk for several types of cancer. However, the effect of these genetic defects can only be attributed to a relatively minor proportion of total cancer incidence for most types of neoplasms [34]. Whereas, diet has been estimated to be associated with 20%–42% of cancer cases [35]. Many studies have supported this conclusion, showing positive associations between cancer rates and certain foods such as meat (red meat in particular), saturated fats, trans-fats, and eggs [36,37]. However, not all foods have a positive association with cancer risk. Certain components of an individual’s diet, such as consumption of vegetables, fruits, cereals, and olive oil as well as a high ratio of monounsaturated to saturated lipids, can have a negative association with cancer risk [37]. In other words, there is evidence that certain foods and dietary choices can affect an individual’s risk for cancer (reviewed by Erdman Jr. et al. [38]).
The link between certain dietary components and lower cancer risk helps to corroborate the results of early research indicating that people who live in the Mediterranean, and more specifically, adhere to the classic Mediterranean diet, have a lower cancer incidence [37,39,40]. Although there are many characteristics of the Mediterranean diet that have been shown to influence cancer rates, consumption of vegetables from the Brassica genus may be one of the most important contributing factors [41]. The cancer-preventative qualities associated with Brassica vegetables could be partially linked to the high levels of vitamins A, C, E, and other antioxidants found in these vegetables [30]. However, the general scientific consensus due to a number of epidemiological, in vitro, and in vivo studies is that the chemopreventive effects of Brassica vegetable consumption are in large part due to the presence of GSs, and more specifically, their GSHPs [42,43].
3. Glucosinolate Hydrolysis Products and Their Chemopreventive Bioactivity
Chemopreventive bioactivity often refers to the ability of a chemical compound, or mixture of compounds, to induce phase I (PI)/phase II (PII) detoxification enzymes and/or antioxidant enzymes in the human body. Antioxidant (AO) enzymes generally act in the regulation of glutathione metabolism and quenching of free radicals via one- and two-electron reductions, thereby contributing to the reduction of oxidative stress. AO enzymes include catalases (CAT), superoxide dismutases (SOD), glutathione reductases (GSR), glutathione peroxidases (GPX), glutaredoxins (GLRX), thioredoxins (TXN), thioredoxin reductases (TXNRD), heme-oxygenase 1 (HO-1), and NAD(P)H:quinone oxidoreductase 1 (NQO1) [30,44]. Phase I/phase II detoxification enzymes are often called biotransformation enzymes because, as a team, they transform toxic xenobiotics into non-toxic forms that can be excreted from the body. In this process, PI enzymes perform various reactions that alter the lipophilic xenobiotic target compound in a way that allows it to react with PII enzymes [45]. Phase II enzymes can directly act on some xenobiotics as well as perform conjugation reactions on the PI products. Most PII enzymes are transferases: UDP-glucuronosyltransferases (UGTs), sulfotransferases (SULTs), glutathione S-transferases (GSTs), N-acetyltransferases (NATs), and S- and O-methyltransferases (MTs) [30]. NQO1 is often included in the list of PII enzymes [44], although it is also often referred to it as an AO enzyme. The product of the PII reaction is a more polar compound that can be readily excreted by the body either through passive or active transport. In general, PII/AO enzymes maintain a balanced redox state in mammalian cells by controlling levels of reactive oxygen species (ROS), which is done by maintaining glutathione (GSH) and thioredoxin levels as well as helping to maintain the equilibrium between NAD+/NADH and NADP+/NADPH [46]. Despite their role in detoxification, PI enzymes can also activate compounds known as procarcinogens [30]. When a procarcinogen reacts with a PI enzyme, such as one of many cytochrome P450s (CYPs), the non-toxic procarcinogen is transformed into a toxic, carcinogenic substance. For this reason, the induction of PI enzymes by a given compound may decrease the overall chemopreventive effect of that compound. On the other hand, PII enzymes typically perform conjugation reactions that result in a compound that is more polar, and subsequently less toxic, than the non-conjugated form [30,47,48,49]. For this reason, compounds that induce only PII enzymes are considered more beneficial, as there is little chance of negative effects. Compounds with this quality are known as monofunctional inducers, as opposed to bifunctional inducers that induce both PI and II enzyme activity. Sulforaphane (SF), the hydrolysis product of the GS called glucoraphanin, has been shown to be a PII monofunctional inducer [50]. In fact, ITCs like SF are generally considered to be PII monofunctional inducers [51]. Therefore, future prospects for Brassica breeders could be to increase SF or other ITC levels in their crops with little fear of affecting the safety of the crop’s consumption.
However, this may not be true for all GSs and GSHPs. While there is no strong evidence of negative effects from GS consumption in humans, this has been observed quite frequently in livestock that eat Brassica forages as a major part of their diet. The adverse symptoms seen in livestock from the consumption of high levels of GSs/GSHPs are often attributed to the consumption of goitrin, an oxazolidine-2-thione GSHP formed from the spontaneous cyclization of the ITC product of progoitrin hydrolysis [52]. Although, goitrin may not be the only GSHP that causes negative health effects upon ingestion of high doses. Research has shown teratogenic effects of other GSHPs in murine models, most notably allyl ITC (AITC) from sinigrin, 1-cyano-2-hydroxy-3,4-epithiobutane (CETB) from progoitrin, and iberin from glucoiberin [53]. The common symptoms seen in livestock often attributed to the overconsumption of GS/GSHPs are: slowed growth [54], impaired movement and general disorientation [55], impaired fertility [56], and damage to the thyroid, gastro-intestinal tract, and/or liver [57,58]. Because of what has been seen in animals, caution must be taken when manipulating GS and GSHP profiles. Although it seems that some of the most bioactive GSHPs show beneficial health effects at the normally consumed dosages, breeders and food scientists should take care to screen for any negative health effects from GS and GSHP profile manipulation of agricultural products.
Although there have been negative effects observed in livestock linked to high levels of GS and/or GSHP consumption, several GSHPs have displayed significant chemopreventive activity in a number of in vitro studies. Common Brassica ITCs, such as SF and phenethyl ITC (PEITC), have proven to either inhibit carcinogenesis or induce cancer cell growth arrest and apoptosis in several cell types including: breast [59], bladder [60], colon [61], ovary [62], blood [63], skin [64], and prostate cells [65]. The mechanism by which these compounds accomplish this task is not fully understood and probably not universal, but some of the known effects of ITC treatment on cell metabolism include modulation of gene expression and alternative gene splicing [66]. Perhaps most importantly, several ITCs have been shown to increase the activity of nuclear factor (erythroid-derived 2)-like 2, also known as NFE2L2 or Nrf2 [67]. When activated, Nrf2 increases transcription rates of a number of AO and PII genes, ultimately leading to cells that are less likely to develop neoplasia [68,69]. While it appears that most ITCs from Brassica or related vegetables induce PII enzymes, there may be differences between aromatic, indole, and aliphatic ITCs in their modulation of PI enzyme activity [70,71,72]. A summary of experiments utilizing ITCs, ITC-derived compounds, and GSHP mixtures as treatments testing their effect on PII/AO gene transcript/protein abundance and/or enzyme activity can be found in Table 1.

Table 1.
Summary of human, animal, and in vitro studies of the effects of glucosinolate hydrolysis products on phase II and antioxidant enzymes; updated from [73].
In addition to the direct benefits of Nrf2 induction on cellular xenobiotic metabolism, there is also evidence of possible crosstalk between the Nrf2 and NFκB pathways (reviewed by Li et al. [106]) as well as evidence that these compounds block the phosphorylation and subsequent degradation of the protein that acts to sequester NFκB in the cytosol. Degradation of this protein would lead to increased nuclear translocation of NFκB, which is associated with inducing transcription of pro-inflammatory genes commonly found to be upregulated in cancer cells (reviewed by Cheung and Kong [107]). This is just one example of multiple chemopreventive bioactivities of GSHPs, which demonstrates the complexity underlying the crosstalk and co-regulation that exists between cellular stress mechanisms and their effect on the initiation, promotion, and/or progression of cancer.
4. Nrf2/Keap1/ARE Signaling Cascade
Regarding chemopreventive bioactivity induced by GSHPs, one mechanism that has been well described is the Keap-1/Nrf2/ARE signaling cascade present in mammalian cells (reviewed by Jaramillo and Zhang [108] and Kansanen et al. [109]). In this signaling pathway, expression of many PII and AO enzymes is promoted by the binding of Nrf2 transcription factors to ARE sequences in the promoter regions of these genes [110]. It is believed that under basal conditions, the transcription factor Nrf2 is sequestered by Keap1 in the cytoplasm. The generally accepted mechanism by which this occurs is that two Keap1 proteins, which are part of a larger Keap1-Cul3-E3 ubiquitin ligase complex, bind to the ETGE and DLG motifs in the Neh2 domain of Nrf2 and promote polyubiquitination and subsequent proteasomal degradation of Nrf2 [69,111,112,113,114]. However, when Keap1 reacts with any of a number of bioactive molecules, polyubiquitination may be impeded.
It is hypothesized that the cysteine residues of Keap1 can react with a number of electrophilic compounds. The modification of the thiol groups of these cysteine residues is thought to alter the conformation of Keap1 [115,116,117]. This, in turn, causes the Keap1 dimer to release the DLG motif of Nrf2, which is suggested to prevent Nrf2 polyubiquitination and degradation [111,112,113]. This proposed mechanism results in Keap1 becoming saturated with Nrf2, allowing newly synthesized Nrf2 to be freely translocated to the nucleus. Once in the nucleus, Nrf2 forms a heterodimer with one of a number of small masculoaponeurotic fibrosarcoma (sMaf) proteins [118] allowing for the binding of the heterodimer to antioxidant response elements (AREs) in the promoter regions of a number of genes involved in cell metabolism and detoxification [118,119,120], while other Maf proteins can act as repressors [121]. In the absence of Nrf2, sMaf proteins form homodimers and bind to AREs, but effectively act as repressors due to their inability to stimulate transcriptional activation [122]. This mechanism for Nrf2/Keap1 regulation of ARE-dependent genes, including those encoding PII and AO enzymes, has been well supported by a number of other studies not already mentioned [123,124,125]. The Nrf2/Keap1/ARE signaling cascade is discussed in more detail by a number of reviews [30,126,127,128].
5. In Vitro Evidence of ARE-Dependent Gene Induction by ITCs
The utility of ITCs for inducing ARE-dependent gene transcription and activity has been thoroughly studied for several years. Popular approaches for such research have been reviewed by Fuentes et al. [129]. There have been a number of ITCs found in Brassica crops that have shown the ability to induce PII and/or AO gene expression/activity, although perhaps through slightly different mechanisms. These include but are not limited to SF, PEITC, AITC, benzyl ITC (BITC), iberin, erucin, and the ITC-derived compounds, I3C and DIM. These GSHPs can be found in a number of Brassica crops, although with variable abundance due to environmental and genetic factors. A few of the Brassica crops considered to be good sources of one or several of these GSHPs are broccoli, various mustards, cabbage (most types), and gai-lan (Chinese broccoli) (Becker et al., in preparation for submission [130]).
SF induces ARE-dependent gene expression through reaction with Keap1. The reaction of SF with Keap1 cysteine residues (primarily C38, C151, C368, and C489; [131]) results in the formation of thionoacyl adducts. However, unlike some other inducers of Nrf2-dependent genes, SF was shown not to result in polyubiquinaton of Keap1 [132,133]. It is believed that the formation of thionoacyl adducts on Keap1 in the presence of SF reduces the binding affinity of Keap1 for Cul3, resulting in an inability to eliminate Nrf2 through proteasomal degradation and migration of free Nrf2 to the nucleus [131,134,135]. Additionally, there is some evidence that a transcriptional coregulator called SPBP may be involved in SF perception and Nrf2 induction [136]. The effects of SF treatment on AO and PII enzymes in vitro have been studied in a wide variety of human and murine cell lines, including LNCaP, PC-3, TSU-Pr1, MDA PCa 2a, MDA PCa 2b, MDA-MB-231, transgenic adenocarcinoma of mouse prostate (TRAMP) C1, HeLa, HT-29, CaCo-2, HepG2, Hepa1c1c7 and MCF-7 [85,102,137,138,139]. The protective effect of SF against oxidative stress has been well documented as this compound was identified several years ago as a strong inducer of PII and AO enzymes, and subsequently, an effective chemopreventive agent.
Much of the early work with SF was reviewed by Fahey and Talalay [140], with a more recent review conducted by Guerrero-Beltran et al. [141]. The studies reviewed by these authors generally attributed the protective effect of SF, no matter the tissue, to increases in transcription and/or activity of one or several PII and/or AO enzymes. Some studies also went as far as to show these effects to be Nrf2-dependent using Nrf2 inhibitors or Nrf2-deficient cell lines. Other studies not included in the review by Guerrero-Beltran et al. have shown similar results. For example, Mizuno et al. [76] showed SF treatment (1 µM) to increase nuclear translocation of Nrf2 as well as expression of γ-glutamylcysteine synthetase (γ-GCS) and HO-1 in rat neuronal cells. In this experiment, the researchers hypothesized that the protective effect of SF observed is primarily due to the increase in γ-GCS expression and subsequent increase in intercellular reduced GSH, based on results from experiments with inhibitors of γ-GCS and HO-1. Also, as a side note, the level of SF used in this and many SF bioactivity studies is well within the range of 0.943–2.27 µM reported to be present in human plasma and erythrocytes 1 h following ingestion of approximately 200 µmol of ITCs in the form of broccoli sprouts [142].
While results from several studies using different tissues show similar results from SF treatment, there is evidence that there are mechanisms other than the Keap1 pathway by which SF induces Nrf2 expression/activity. This is shown in a study using mouse TRAMP cells, a prostate cancer model that was previously reported as containing an epigenetic mechanism that leads to decreased Nrf2 and downstream gene expression [143]. Using these cells, Zhang et al. [102] reports that SF treatment results in the demethylation of the first five CpG sites in the Nrf2 promoter, leading to an increase in Nrf2 mRNA and protein expression. The beneficial effect of SF on oxidative stress may not be limited to induction of Nrf2-dependent genes. In addition to the well-demonstrated effectiveness of SF in that capacity, there is evidence that SF may act directly as a radical scavenger from superoxide through the action of SOD and from hydrogen peroxide without enzymatic interaction [144].
The mechanism for PEITC induction of ARE-dependent genes is believed to occur via increased extracellular signal-regulated kinase (ERK)- and/or c-Jun-NH2-kinase (JNK)-dependent phosphorylation of Nrf2. Upon treatment with PEITC, increased phosphorylation by ERK and/or JNK results in improved migration of Nrf2 to the nucleus and ARE-dependent gene expression. Support for this mechanism comes from results that show an attenuation of the effects of PEITC treatment by ERK/JNK inhibition and or genetic knockout [145,146]. This mechanism also appears to be partially responsible for the Nrf2 induction by SF and AITC [146]. In a recent study, PEITC-induced Nrf2-dependent PII and AO enzyme induction was tested in HepG2 cells. At concentrations of 1 µM and 5 µM, PEITC was shown to significantly increase Nrf2 mRNA, nuclear Nrf2 protein, and phosphorylated nuclear Nrf2 by approximately 2-fold, 1.3-fold, and 1.5-fold, respectively. Both PII and AO enzymes showed increases in expression and protein abundance following PEITC treatment. Both tested concentrations of PEITC increased expression and protein abundance of all four major classes of GSTs while NQO1 expression and abundance was only significantly increased by 5 µM PEITC treatment. Several AO enzymes (SOD, CAT, GPX, and GSR) also responded to both tested concentrations of PEITC with significant increases in both transcript and protein abundance [44]. In another study, PEITC was also shown to increase expression of HO-1 and an ARE-driven luciferase reporter gene in PC-3 cells. Even though no statistical analysis was presented in this study, a clear positive trend was observed in both HO-1 and ARE/luciferase up to concentrations of 7.5–10 µM PEITC. However, ARE/luciferase activity decreased at 20 µM, and there were signs of cytotoxicity with long term exposure to higher concentrations [147].
In addition to SF and PEITC, other ITCs common in Brassica vegetables have shown evidence of the ability to induce the ARE-dependent genes. For example, SF, PEITC, BITC, and AITC were generally shown to increase transcript and protein abundance of γ-GCS, HO-1, and NQO1 when applied to NIH3T3 fibroblast cells at the lowest dose tested (5 µM) [74]. A similar study using the same four ITCs and mouse skin papilloma cells found analogous results, with significant increases in NQO1 and GST activity. The authors also showed that the measured area under time-concentration curve of intracellular concentrations of a given ITC were strongly correlated with the induction of the measured enzymes as well as glutathione (GSH) content. Comparable correlations were found for all four ITCs using human HepG2 cells and an ARE-luciferase reporter [148]. While Nrf2 signaling is no doubt one of the major contributors to the chemopreventive bioactivity of PEITC, several other mechanisms besides Nrf2 activation have been proposed to contribute, including induction of apoptosis and cell cycle arrest. Several of these potential mechanisms are discussed in depth in a review by Qin et al. [149].
A few studies also exist exploring the effect of I3C treatment on AO and PII enzymes in vitro. One such study was briefly discussed above for its evaluation of PEITC induction of AO and PII enzymes. I3C showed similar results to PEITC in this study, although the tested concentrations were double that of PEITC (2 and 10 µM). Even though I3C was shown to significantly increase transcript and protein abundance of many of the same PII and AO enzymes as PEITC, some differences existed. In general, PEITC treatments resulted in greater increases in transcript and protein for the genes tested, even at lower concentrations. These differences were most extreme for transcript abundance of a number of genes (SOD, GPX, and all tested GSTs), but were less evident when evaluating the corresponding protein abundance [44]. In yet another study, TRAMP C1 cells modified to contain an ARE-luciferase reporter were treated with I3C at concentrations of 25, 50, 75, and 100 µM. Treatments were shown to increase luciferase reporter activity in a dose-dependent manner, with an effective dose (statistically different from control) of 50 µM. Likewise, Nrf2 and several downstream genes (glutamate-cysteine ligase catalytic subunit [GCLC], NQO1, and HO-1) were upregulated, in terms of mRNA levels. Induction was, again, dose-dependent with effective doses being reached at 75, 25, 25, and 75 µM for Nrf2, GCLC, NQO1, and HO-1, respectively [77]. The mechanism(s) for I3C induction of Nrf2 have yet to be fully elucidated, but one study suggests that at least one mechanism is indirect, acting through suppression of the production of reactive oxygen species (ROS) upon treatment of the cytotoxic compounds, such as dexamethasone (Dex). In this study, I3C treatments as low as 5 µM resulted in a negation (statistically speaking) of cytotoxic effects of Dex as determined by cell viability comparisons to a control in MC3T3-E1 osteoblastic cells. Also, 10 µM and 20 µM I3C pretreatment prior to treatment with Dex reduced the production of superoxide by 30% and 40%, respectively, when measured as percent of the control. Superoxide levels were also found, roughly, to negatively associate with Nrf2 and downstream gene expression in the Dex and Dex+I3C treatments. This led the authors to hypothesize that in their study, the induction of Nrf2 and downstream genes by I3C was largely due to the attenuation of Dex-induced ROS production and any Nrf2-repression mechanisms that those ROS levels may have imposed [150].
While the experiments discussed above use I3C as a treatment, there is evidence that I3C undergoes oligomerization in cell culture [151], just as it does in acidic conditions like the stomach [152]. This means that other compounds, such as 3,3′-diindolylmethane (DIM), may be responsible for some of the bioactivity associated with I3C. Yet, a reminder of just how complex the mechanisms of bioactivity underlying the chemopreventive properties of these compounds can be seen in an experiment done by Szaefer et al. [94]. In this study, three different breast cell lines, two of which were tumorigenic, were treated with both I3C (10, 30, and 50 µM) and DIM (5 and 10 µM). Levels of transcript and protein were measured for a number of genes, including NQO1 and GST-π (GSTP). I3C and DIM showed similar patterns of induction for these genes, but some slight differences are seen depending on the gene being measured and the cell line, an indication of possible interactions between treatment and genotype. However, it is difficult to determine if the observed differences are real, as no ANOVA was reported for this study to test for statistical significance of any of these effects. One thing that is suggested from the results of the Szaefer et al. study is that the effective dose of DIM for induction of Nrf2 and downstream genes is much lower than I3C, as treatments of 5 µM and 10 µM DIM showed similar levels of induction of both mRNA and protein of NQO1 and GSTP as 30 µM and 50 µM I3C, respectively. Along with Nrf2 induction, I3C and its derivatives exhibit chemopreventive bioactivity through a number of other mechanisms, including suppression of proliferation of cancer cells from various tissues through regulatory repression of cyclin-dependent kinases, induction of apoptosis, and modulation of expression of several transcription factors (reviewed by Aggarwal and Ichikawa [153]). However, these indole-derived GSHPs are generally considered to be inferior to the aliphatic-derived ITCs in terms of chemopreventive potential, mainly due to their bifunctional nature [70,71].
6. Response of AO and PII Enzymes to ITC Treatment in Animal Studies
As with in vitro studies, SF has been the most studied ITC in animal models. SF has generally been shown to inhibit PI enzymes while inducing PII enzymes, such as NQO1, TXNRD, GST, and HO-1, in a variety of tissues/cell types [23,73]. However, the inhibitory effect of SF on PI enzymes may not be universal for all genetic backgrounds. A report by Hu et al. [81] detected increases in PI gene expression following treatment with SF in Nrf2 wild-type mice compared to Nrf2 knockouts. This inconsistency in results from different studies concerning the action of SF on PI enzymes is indicative that the mechanisms of bioactivity of SF, probably the most-studied ITC, are still not fully understood.
One factor that has been considered as research on SF and other GSHPs moved from in vitro to in vivo studies is that the bioactivity of a given GSHP depends on the level of bioavailability, or ease of absorption by the body or a given tissue, of that compound [154]. Studies show high bioavailability of SF in CD-1 mice following oral administration of 2.5 mg/kg broccoli sprout preparations. Modeling of SF uptake in this experiment indicated that SF absorption is principally perfusion-limited [155]. Similarly high levels of bioavailability have been measured in humans [156,157,158], independent of GSTP1 polymorphism [156]. It has also been observed that the bioavailability of SF is greatly dependent upon whether or not functional myrosinase is included with SF’s precursor GS, glucoraphanin. Use of fresh Brassica tissue and/or supplementation with myrosinase results in a higher bioavailability of SF in both mice [155] and humans [156,157,158].
The AO and PII inductive effects of PEITC have also been tested in animal models. One such study treated Nrf2 wild-type and knockout mice with a dosage of 40 mg/kg PEITC. Microarray analysis of liver mRNA revealed upregulation for a large number of genes both 3 h and 12 h after treatment. These included a number of AO and PII genes along with several genes associated with heat shock proteins, ubiquitin/26 S proteasome subunits, and lipid metabolism [88]. Like SF, PEITC appears to be highly bioavailable. Oral bioavailability in rats was calculated to be at or near 100% for doses of 10 and 100 μmol/kg [159]. In addition, half-life of PEITC in rat is reported to be between 4 and 22 h, depending on the tissue [160]. The high bioavailability of PEITC, much like SF, contributes to the promise of this phytochemical being absorbed in effective doses through dietary means. A disadvantage of PEITC compared to SF is that this compound is much more volatile [161], less water soluble [162], and therefore more likely to be lost from the Brassica vegetable during preparation (i.e., after cutting of plant tissue leads to hydrolysis of gluconasturtiin, the precursor GS of PEITC). However, consumption of salad crops like watercress, from a genus closely related to Brassica, can deliver large doses of gluconasturtiin, active myrosinase, and subsequently, PEITC at estimated levels of 37 µmol per ounce [163]. Another aliphatic ITC common to Brassica crops that is gaining attention is erucin, from the precursor GS glucoerucin. This ITC was recently found to induce HO-1 expression in vitro and in vivo, possibly acting through a similar mechanism of Nrf2 activation as PEITC [96]. Interestingly, erucin can also undergo interconversion to SF in vivo, although the interconversion ratio of SF and erucin is not consistent across individuals [156].
As mentioned above, indole-derived GSHPs are considered less effective chemopreventive compounds due to their bifunctional induction capabilities. Both I3C and DIM have been tested for their high dose toxicity and PI enzyme induction in Sprague-Dawley rats using doses equivalent to 5–7x the maximal human dose for I3C and 1 or 10x for DIM. After 12 months of feeding, no major indicators of toxicity were observed, but the I3C diet was shown to significantly increase PI protein levels. The high dosage DIM diet also caused PI protein induction, but generally at a lower level than I3C and with some differences between tissues for both compounds [70]. These results along with those of previously discussed studies indicate the superiority of DIM as a chemopreventive agent compared to I3C due to its lower bifunctional induction [70] and lower effective dose for the induction of PII enzymes in vitro [94]. In some trials, DIM treatments showed results similar to those of in vitro studies. In one such study, DIM administered intravenously to male Sprague-Dawley rats at a dose of 10 mg/kg resulted in significant increases in mRNA levels of NQO1, GSTP1, and UGT1A1 in blood lymphocytes, peaking approximately 1–2 h after DIM treatment. Concentrations of DIM in plasma were also measured, but due to the poor bioavailability of DIM, calculated in this study as 1%–3%, plasma concentrations were relatively low, peaking around 9 µg/mL five minutes after administration. DIM was also shown to be quickly metabolized, with a terminal half-life of 0.737 h [164]. The low bioavailability of DIM has been corroborated by other research [165,166]. This fact has raised concerns that the levels of DIM needed to elicit a response in mice may not be achievable through dietary intake of Brassica crops. Wu et al. [164] calculated that the dosage used in their experiment would translate to a human dosage of 1.6 mg/kg. Concentrations of DIM in both green and red kale leaf tissue, a crop containing relatively high amounts of DIM’s precursor GS (glucobrassicin), have been shown to be less than 1 µg/mg dried tissue [167]. One caveat is that liver concentrations of DIM have been shown to be six to eight times that of plasma in mice [166], indicating that a smaller dose may be necessary to reach effective dose levels in liver cells [164].
7. Clinical and Epidemiological Evidence for the Importance of ITCs in Chemoprevention
Although there has been significant evidence that a diet rich in GSHPs can have chemopreventive effects, this relationship often is not found to be significant in epidemiological studies. A review by Higdon et al. [42] discusses this common observation. The authors looked at several epidemiological studies for the four types of cancer with the highest mortality rates in the U.S.: lung, colorectal, breast, and prostate. Though there are obvious differences between these cancers, there seemed to be some commonalities in the results of the different epidemiological studies. For each type of cancer, the authors concluded that even though many small case-control studies found a significant inverse relationship between cruciferous vegetable intake and cancer rates, the larger prospective cohort studies often did not find the same significant relationship. There were a few exceptions to this generalization. Some of the prospective cohort studies found significant inverse relationships, but usually in specific populations within the full data set of the study [168,169,170,171,172]. The different results between the two types of studies may be due to participation bias in case-control studies, wherein control groups who choose to participate are more health conscious and have better eating habits compared to non-participating controls [173]. However, as the authors of the review indicate, the inconsistency of results may be due to a more complex relationship between cruciferous vegetable intake and cancer risk than what was previously thought.
Although results of different epidemiological studies can often be contradictory, a review of 87 case-control studies that were performed prior to 1996 indicated that a majority (67%) show an inverse relationship between cancer risk and cruciferous vegetable consumption [174]. Building on this, Jeffery and Keck [175] make the case that there is enough evidence to proceed with larger clinical trials testing the efficacy of purified SF, semi-purified SF, and/or whole broccoli for inducing detoxification enzyme activity. The authors reviewed a number of epidemiological, in vitro, and in vivo studies examining the link between cruciferous vegetable intake, or bioactive GSHPs, and cancer. In this article, all epidemiological studies reviewed showed a significant (p < 0.05) decrease in cancer risk associated with cruciferous vegetable consumption for various types of cancer including: bladder [176], lung [177], lymphoma [178], prostate [179,180], breast [181], kidney [182], and ovarian [183]. These results were further supported with a number of in vitro studies showing several anti-tumorigenic activities in various mammalian cancer cell types after treatment with SF. Also reported were several in vivo animal model studies showing decreased tumor growth, incidence, and/or multiplicity using broccoli, SF, and ITCs as treatments. Finally, several small clinical studies were reported that further corroborated the observed results in epidemiological, in vitro, and animal model studies described above. These studies reported positive associations between broccoli or broccoli sprout consumption and several different biomarkers linked to efficacy of the treatment for inducing cellular detoxification mechanisms. More recent reviews and/or meta-analyses have also corroborated the conclusion that consumption of cruciferous (Brassica) vegetables, in general, results in a decreased risk of several cancer types [184,185,186,187,188,189,190,191].
Singh and Singh [23] expanded on the case made by Jeffery and Keck, widening the scope to include PEITC and BITC. Again, the authors cited evidence supporting the chemopreventive potential of these compounds through a number of mechanisms, including cellular ROS modulation, and called for a transition into large-scale clinical research of dietary ITCs. However, the authors of both reviews discussed above admitted to challenges that exist when considering the transition into human clinical trials with ITCs, including formulation of ITC treatments that are repeatable and suitable for oral administration as well as identifying proper dosage and treatment scheduling due to the rapid metabolism and excretion of ITC derivatives/conjugates.
Another major factor to be considered when planning clinical trials testing the chemopreventive properties of ITCs, possibly through the use of dietary intervention with Brassica vegetables, is the inter- and intra-individual variation in bioavailability of SF, and likely other GSHPs. Although the mechanism of ITC uptake is likely complex, bioavailability of these compounds is generally attributed to differences in the GST genotype and/or microbiota composition of the subject [158,192,193]. Yet, the magnitude of the impact of those factors on SF and other GSHP bioavailability/bioactivity is still not fully clear. It is hypothesized that due to the major role of GSH in the conjugation and elimination of SF, GSTM1-null individuals would benefit more from SF consumption due to the increased time that SF would be in the body [42,194]; however, this has not been fully confirmed by epidemiological evidence [73]. A number of studies have shown no significant effect of GST genotype on ITC uptake and/or Nrf2 and downstream gene activation [156,195], while other research shows a significant genotype effect [196,197,198]. The effect of GST genotype may depend on the specific GST being considered as well as the level of dietary ITC consumption [197]. The true influence of microbiota on GSHP bioavailability has also been questioned, as germ-free mice show only slightly altered profiles of excreted GS metabolites [199]. Also, even though high and low ITC excretors have been described [200], specific gut microorganisms could not be associated with this capability [201].
There is still work to be done towards implementing large-scale fully randomized placebo control trials (reviewed by Fahey et al. [202]). However, several smaller clinical trials have occurred over the past couple decades, generally using Brassica vegetables as the ITC delivery matrix. Several of these studies were reviewed by Boddupalli et al. [30] and James et al. [73]. In general, the research discussed in these reviews showed induction of ARE-mediated enzymes and/or reduction of biomarkers for oxidative stress accompanying treatment with GSHPs. One study in particular showed that ingestion of a broccoli soup made with broccoli containing three times the normal glucoraphanin resulted in increased SF uptake in a dose-dependent manner [80]. This result indicates that improvement of Brassica crops for increased content of certain favorable GSs would be worthwhile and would result in higher intake of dietary ITCs for those who consumed said crops. A series of studies by Riso et al. not included in the aforementioned reviews reported similar beneficial results following dietary intervention with broccoli. Specifically, two of these studies reported a reduction of oxidized purines in smokers and a reduction in H2O2-induced strand breaks following broccoli consumption [203,204]. Also, this group reported an increase in GST activity only for GSTM1-positive individuals 6 h following broccoli consumption compared to 3 h and 24 h [205].
8. Conclusions
The results of these reports discussed above suggest that the level of the chemopreventive effect of Brassica vegetables may depend on the interaction of several variables, including the level of consumption of other dietary factors (vitamins, lipids, other phytochemicals, etc.) [30,206] and the individual’s genotype/metabolism [177,196,207,208]. In addition, the GS and subsequent GSHP profiles (presence/absence of certain GS/GSHPs) [42], as well as the presence/absence of active myrosinase [156,157,158], are also important variables in determining the chemopreventive effect of consuming a given Brassica vegetable. The activation of Nrf2 and downstream genes by Brassica ITCs and ITC-derived carbinols is a major factor in the chemopreventive properties displayed by these compounds. However, it is not the only chemopreventive mechanism stimulated by these compounds (e.g., apoptosis, cell cycle arrest, epigenetic regulation).
While there is a bulk of in vitro and in vivo evidence that many aliphatic-derived ITCs and perhaps some indole-derived GSHPs can be delivered at effective levels through modest increases in dietary intake of Brassica vegetables containing these compounds, caution has still been advised by the academic community when considering how large the ITC doses used in human trials can safely be. In high concentrations, these compounds have been shown to have genotoxic potential [209] and lead to cellular stress through alkylation and depletion of cellular thiols, damage to mitochondria, and elevated levels of reactive oxygen species (reviewed by Zhang et al. [210]). Also, there have been a few reports of increased cancer incidence in some animal models following ITC treatment, possibly depending on the specific ITC and timing of ITC treatment (pre- or post-initiation) [23]. Furthermore, constitutive activation of Nrf2 in cancer cells may be associated with drug resistance (reviewed by Huang et al. [211]). Despite these possible complications, the case has been made, partly in the two reviews by Jeffery and Keck [175] and Singh and Singh [23], for progression into larger-scale human clinical trials testing the chemopreventive and post-initiation cancer suppression potential of dietary ITCs. While many of the early studies show promising results in the modulation of Nrf2 and other chemopreventive molecular mechanisms with GSHPs, there is still much to learn about how these phytochemicals alter cellular metabolism in humans. Specifically, understanding the reasons behind inter- and intra-individual variation for ITC uptake, the effect of ITC mixture and/or delivery matrix, and the correct treatment dosage/timing for desired metabolic response are major goals of ongoing and future research, including human clinical trials.
Acknowledgements
The authors would like to acknowledge and thank Dr. E.H. Jeffery for her critical review and comments.
Author Contributions
T.B. authored the majority of the manuscript with critical review and additions by J.J. Both authors approved the final manuscript
Conflicts of Interest
The authors declare no conflict of interest.
References
- Halkier, B.A.; Gershenzon, J. Biology and biochemistry of glucosinolates. Annu. Rev. Plant Biol. 2006, 57, 303–333. [Google Scholar] [CrossRef] [PubMed]
- Sonderby, I.E.; Geu-Flores, F.; Halkier, B.A. Biosynthesis of glucosinolates—Gene discovery and beyond. Trends Plant Sci. 2010, 15, 283–290. [Google Scholar] [CrossRef] [PubMed]
- Fahey, J.W.; Zalcmann, A.T.; Talalay, P. The chemical diversity and distribution of glucosinolates and isothiocyanates among plants. Phytochemistry 2001, 56, 5–51. [Google Scholar] [CrossRef]
- Agerbirk, N.; Olsen, C.E. Glucosinolate structures in evolution. Phytochemistry 2012, 77, 16–45. [Google Scholar] [CrossRef] [PubMed]
- Padilla, G.; Cartea, M.E.; Velasco, P.; de Haro, A.; Ordás, A. Variation of glucosinolates in vegetable crops of Brassica rapa. Phytochemistry 2007, 68, 536–545. [Google Scholar] [CrossRef] [PubMed]
- Gómez-Campo, C. Biology of Brassica coenospecies, 1st ed.; Elsevier: Amsterdam, The Netherlands, 1999; Volume 4. [Google Scholar]
- Bednarek, P.; Piślewska-Bednarek, M.; Ver Loren van Themaat, E.; Maddula, R.K.; Svatoš, A.; Schulze-Lefert, P. Conservation and clade-specific diversification of pathogen-inducible tryptophan and indole glucosinolate metabolism in Arabidopsis thaliana relatives. New Phytol. 2011, 192, 713–726. [Google Scholar] [CrossRef] [PubMed]
- Wittstock, U.; Burow, M. Glucosinolate breakdown in Arabidopsis: Mechanism, regulation and biological significance. Arab. Book Am. Soc. Plant Biol. 2010, 8, e0134. [Google Scholar] [CrossRef] [PubMed]
- Kelly, P.; Bones, A.; Rossiter, J. Sub-cellular immunolocalization of the glucosinolate sinigrin in seedlings of Brassica juncea. Planta 1998, 206, 370–377. [Google Scholar] [CrossRef] [PubMed]
- Koroleva, O.A.; Davies, A.; Deeken, R.; Thorpe, M.R.; Tomos, A.D.; Hedrich, R. Identification of a new glucosinolate-rich cell type in Arabidopsis flower stalk. Plant Physiol. 2000, 124, 599–608. [Google Scholar] [CrossRef] [PubMed]
- Andreasson, E.; Jorgensen, L.B.; Hoglund, A.S.; Rask, L.; Meijer, J. Different myrosinase and idioblast distribution in Arabidopsis and Brassica napus. Plant Physiol. 2001, 127, 1750–1763. [Google Scholar] [CrossRef] [PubMed]
- Vig, A.P.; Rampal, G.; Thind, T.S.; Arora, S. Bio-protective effects of glucosinolates–A review. LWT-Food Sci. Technol. 2009, 42, 1561–1572. [Google Scholar] [CrossRef]
- Wittstock, U.; Gershenzon, J. Constitutive plant toxins and their role in defense against herbivores and pathogens. Curr. Opin. Plant Biol. 2002, 5, 300–307. [Google Scholar] [CrossRef]
- Burow, M.; Wittstock, U.; Gershenzon, J. Sulfur-containing secondary metabolites and their role in plant defense. In Sulfur Metabolism in Phototrophic Organisms; Springer: Dordrecht, The Netherlands, 2008; Volume 27, pp. 201–222. [Google Scholar]
- Hopkins, R.J.; van Dam, N.M.; van Loon, J.J.A. Role of Glucosinolates in Insect-Plant Relationships and Multitrophic Interactions. Annu. Rev. Entomol. 2009, 54, 57–83. [Google Scholar] [CrossRef] [PubMed]
- Ahuja, I.; Rohloff, J.; Bones, A.M. Defence mechanisms of Brassicaceae: Implications for plant-insect interactions and potential for integrated pest management. A review. Agron. Sustain. Dev. 2010, 30, 311–348. [Google Scholar] [CrossRef]
- Talalay, P.; Fahey, J.W. Phytochemicals from cruciferous plants protect against cancer by modulating carcinogen metabolism. J. Nutr. 2001, 131, 3027S–3033S. [Google Scholar] [PubMed]
- Razis, A.F.A.; Noor, N.M. Cruciferous vegetables: Dietary phytochemicals for cancer prevention. Asian PacJCancer Prev. 2013, 14, 1565–1570. [Google Scholar] [CrossRef]
- Yan, X.; Chen, S. Regulation of plant glucosinolate metabolism. Planta 2007, 226, 1343–1352. [Google Scholar] [CrossRef] [PubMed]
- Agerbirk, N.; Olsen, C.E. Glucosinolate hydrolysis products in the crucifer Barbarea vulgaris include a thiazolidine-2-one from a specific phenolic isomer as well as oxazolidine-2-thiones. Phytochemistry 2015, 115, 143–151. [Google Scholar] [CrossRef] [PubMed]
- Wattenberg, L.W. Chemoprophylaxis of carcinogenesis: A review. Cancer Res. 1966, 26, 1520–1526. [Google Scholar] [PubMed]
- Sporn, M.B. Chemoprevention of cancer. Nature 1978, 272, 402–403. [Google Scholar] [CrossRef]
- Singh, S.V.; Singh, K. Cancer chemoprevention with dietary isothiocyanates mature for clinical translational research. Carcinogenesis 2012, 33, 1833–1842. [Google Scholar] [CrossRef] [PubMed]
- Agerbirk, N.; Vos, M.D.; Kim, J.H.; Jander, G. Indole glucosinolate breakdown and its biological effects. Phytochem. Rev. 2009, 8, 101–120. [Google Scholar] [CrossRef]
- Acharya, A.; Das, I.; Singh, S.; Saha, T. Chemopreventive properties of indole-3-carbinol, diindolylmethane and other constituents of cardamom against carcinogenesis. Recent Pat. Food Nutr. Agric. 2010, 2, 166–177. [Google Scholar] [CrossRef] [PubMed]
- Dinkova-Kostova, A.T. Chemoprotection against cancer by isothiocyanates: A focus on the animal models and the protective mechanisms. In Natural Products in Cancer Prevention and Therapy; Springer: Heidelberg, Germany, 2013; pp. 179–201. [Google Scholar]
- Gerhauser, C. Epigenetic impact of dietary isothiocyanates in cancer chemoprevention: Curr. Curr. Opin. Clin. Nutr. Metab. Care 2013, 16, 405–410. [Google Scholar] [CrossRef] [PubMed]
- Nho, C.W.; Jeffery, E. The synergistic upregulation of phase II detoxification enzymes by glucosinolate breakdown products in cruciferous vegetables. Toxicol. Appl. Pharmacol. 2001, 174, 146–152. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.H. Potential synergy of phytochemicals in cancer prevention: Mechanism of action. J. Nutr. 2004, 134, 3479S–3485S. [Google Scholar] [PubMed]
- Boddupalli, S.; Mein, J.R.; Lakkanna, S.; James, D.R. Induction of Phase 2 Antioxidant Enzymes by Broccoli Sulforaphane: Perspectives in Maintaining the Antioxidant Activity of Vitamins A, C, and E. Front. Genet. 2012, 3, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Jones, D.S.; Podolsky, S.H.; Greene, J.A. The Burden of Disease and the Changing Task of Medicine. N. Engl. J. Med. 2012, 366, 2333–2338. [Google Scholar] [CrossRef] [PubMed]
- Jemal, A.; Siegel, R.; Ward, E.; Hao, Y.; Xu, J.; Murray, T.; Thun, M.J. Cancer statistics, 2008. CA-Cancer J. Clin. 2008, 58, 71–96. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.; Naishadham, D.; Jemal, A. Cancer statistics, 2013. CA-Cancer J. Clin. 2013, 63, 11–30. [Google Scholar] [CrossRef] [PubMed]
- Lichtenstein, P.; Holm, N.; Verkasalo, P.; Iliadou, A.; Kaprio, J.; Koskenvuo, M.; Pukkala, E.; Skytthe, A.; Hemminki, K. Environmental and heritable factors in the causation of cancer—Analyses of cohorts of twins from Sweden, Denmark, and Finland. N. Engl. J. Med. 2000, 343, 78–85. [Google Scholar] [CrossRef] [PubMed]
- Willett, W.C. Diet, nutrition, and avoidable cancer. Environ. Health Perspect. 1995, 103, 165–170. [Google Scholar] [CrossRef] [PubMed]
- Anand, P.; Kunnumakara, A.B.; Sundaram, C.; Harikumar, K.B.; Tharakan, S.T.; Lai, O.S.; Sung, B.; Aggarwal, B.B. Cancer is a Preventable Disease that Requires Major Lifestyle Changes. Pharm. Res. 2008, 25, 2097–2116. [Google Scholar] [CrossRef] [PubMed]
- Benetou, V.; Trichopoulou, A.; Orfanos, P.; Naska, A.; Lagiou, P.; Boffetta, P.; Trichopoulos, D. Conformity to traditional Mediterranean diet and cancer incidence: The Greek EPIC cohort. Br. J. Cancer 2008, 99, 191–195. [Google Scholar] [CrossRef] [PubMed]
- Erdman, J.W., Jr.; Jeffery, E.; Hendrickx, M.; Cross, A.J.; Lampe, J.W. Can Food Processing Enhance Cancer Protection? Nutr. Today 2014, 49, 230–234. [Google Scholar] [CrossRef] [PubMed]
- Kontou, N.; Psatopouou, T.; Panagiotakos, D.; Dimopouos, M.A.; Linos, A. The Mediterranean Diet in Cancer Prevention: A Review. J. Med. Food 2011, 14, 1065–1078. [Google Scholar] [CrossRef] [PubMed]
- Pauwels, E.K.J. The Protective Effect of the Mediterranean Diet: Focus on Cancer and Cardiovascular Risk. Med. Princ. Pract. 2010, 20, 103–111. [Google Scholar] [CrossRef] [PubMed]
- Van Poppel, G.; Verhoeven, D.T.; Verhagen, H.; Goldbohm, R.A. Brassica vegetables and cancer prevention. In Advances in Nutrition and Cancer 2; Springer: New York, NY, USA, 1999; pp. 159–168. [Google Scholar]
- Higdon, J.V.; Delage, B.; Williams, D.E.; Dashwood, R.H. Cruciferous vegetables and human cancer risk: Epidemiologic evidence and mechanistic basis. Pharmacol. Res. 2007, 55, 224–236. [Google Scholar] [CrossRef] [PubMed]
- Kapusta-Duch, J.; Kopec, A.; Piatkowska, E.; Borczak, B.; Leszczynska, T. The beneficial effects of Brassica vegetables on human health. Rocz. Panstwowego Zakadu Hig. 2012, 63, 389–395. [Google Scholar]
- Krajka-Kuźniak, V.; Paluszczak, J.; Szaefer, H.; Baer-Dubowska, W. The activation of the Nrf2/ARE pathway in HepG2 hepatoma cells by phytochemicals and subsequent modulation of phase II and antioxidant enzyme expression. J. Physiol. Biochem. 2015, 71, 227–238. [Google Scholar] [CrossRef] [PubMed]
- Mansuy, D. Brief historical overview and recent progress on cytochromes P450: Adaptation of aerobic organisms to their chemical environment and new mechanisms of prodrug bioactivation. Ann. Pharm. Fr. 2011, 69, 62–69. [Google Scholar] [CrossRef] [PubMed]
- Dinkova-Kostova, A.T.; Talalay, P. Direct and indirect antioxidant properties of inducers of cytoprotective proteins. Mol. Nutr. Food Res. 2008, 52, S128–S138. [Google Scholar] [CrossRef] [PubMed]
- Mithen, R.F.; Dekker, M.; Verkerk, R.; Rabot, S.; Johnson, I.T. The nutritional significance, biosynthesis and bioavailability of glucosinolates in human foods. J. Sci. Food Agric. 2000, 80, 967–984. [Google Scholar] [CrossRef]
- Croom, E. 3 Metabolism of Xenobiotics of Human Environments. Prog. Mol. Biol. Transl. Sci. 2012, 112, 31. [Google Scholar] [PubMed]
- Omiecinski, C.J.; Vanden Heuvel, J.P.; Perdew, G.H.; Peters, J.M. Xenobiotic metabolism, disposition, and regulation by receptors: From biochemical phenomenon to predictors of major toxicities. Toxicol. Sci. Off. J. Soc. Toxicol. 2011, 120 Suppl 1, S49–S75. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Talalay, P.; Cho, C.; Posner, G. A Major Inducer of Anticarcinogenic Protective Enzymes from Broccoli—Isolation and Elucidation of Structure. Proc. Natl. Acad. Sci. USA. 1992, 89, 2399–2403. [Google Scholar] [CrossRef] [PubMed]
- Prochaska, H.J.; Talalay, P. Regulatory Mechanisms of Monofunctional and Bifunctional Anticarcinogenic Enzyme Inducers in Murine Liver. Cancer Res. 1988, 48, 4776–4782. [Google Scholar] [PubMed]
- Greer, M.A. Isolation from Rutabaga Seed of Progoitrin, the Precursor of the Naturally Occurring Antithyroid Compound, Goitrin (L-5-vinyl-2-thiooxazolidone). J. Am. Chem. Soc. 1956, 78, 1260–1261. [Google Scholar] [CrossRef]
- Nishie, K.; Daxenbichler, M. Toxicology of glucosinolates, related compounds (nitriles, R-goitrin, isothiocyanates) and vitamin U found in Cruciferae. Food Cosmet. Toxicol. 1980, 18, 159–172. [Google Scholar] [CrossRef]
- Schone, F.; Rudolph, B.; Kirchheim, U.; Knapp, G. Counteracting the negative effects of rapeseed and rapeseed press cake in pig diets. Br. J. Nutr. 1997, 78, 947–962. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, R.A.; Maciel, M.G.; de Ochoteco, M.B.; Zimmermann, R.N.; Lavinia, F.M.; Formentini, E.A.; de Ochoteco, M.B. Blindness in Holando Argentine calves due to ingestion of turnip (Brassica campestris). Vet. Argent. 1997, 14, 601–605. [Google Scholar]
- Ahlin, K.A.; Emanuelson, M.; Wiktorsson, H. Rapeseed Products from Double-Low Cultivars as Feed for Dairy-Cows—Effects of Long-Term Feeding on Thyroid-Function, Fertility and Animal Health. Acta Vet. Scand. 1994, 35, 37–53. [Google Scholar] [PubMed]
- Campbell, L.D. Incidence of Liver Hemorrhage among White Leghorn Strains Fed on Diets Containing Different Types of Rapeseed Meals. Br. Poult. Sci. 1979, 20, 239–246. [Google Scholar] [CrossRef] [PubMed]
- Martland, M.F.; Butler, E.J.; Fenwick, G.R. Rapeseed Induced Liver Hemorrhage, Reticulolysis and Biochemical-Changes in Laying Hens—The Effects of Feeding High and Low Glucosinolate Meals. Res. Vet. Sci. 1984, 36, 298–309. [Google Scholar] [PubMed]
- Li, Y.; Zhang, T.; Korkaya, H.; Liu, S.; Lee, H.-F.; Newman, B.; Yu, Y.; Clouthier, S.G.; Schwartz, S.J.; Wicha, M.S.; et al. Sulforaphane, a Dietary Component of Broccoli/Broccoli Sprouts, Inhibits Breast Cancer Stem Cells. Clin. Cancer Res. 2010, 16, 2580–2590. [Google Scholar] [CrossRef] [PubMed]
- Munday, R.; Mhawech-Fauceglia, P.; Munday, C.M.; Paonessa, J.D.; Tang, L.; Munday, J.S.; Lister, C.; Wilson, P.; Fahey, J.W.; Davis, W.; et al. Inhibition of urinary bladder carcinogenesis by broccoli sprouts. Cancer Res. 2008, 68, 1593–1600. [Google Scholar] [CrossRef] [PubMed]
- Gamet-Payrastre, L.; Li, P.F.; Lumeau, S.; Cassar, G.; Dupont, M.A.; Chevolleau, S.; Gasc, N.; Tulliez, J.; Terce, F. Sulforaphane, a naturally occurring isothiocyanate, induces cell cycle arrest and apoptosis in HT29 human colon cancer cells. Cancer Res. 2000, 60, 1426–1433. [Google Scholar] [PubMed]
- Chuang, L.T.; Moqattash, S.T.; Gretz, H.F.; Nezhat, F.; Rahaman, J.; Chiao, J.-W. Sulforaphane induces growth arrest and apoptosis in human ovarian cancer cells. Acta Obstet. Gynecol. Scand. 2007, 86, 1263–1268. [Google Scholar] [CrossRef] [PubMed]
- Suppipat, K.; Park, C.S.; Shen, Y.; Zhu, X.; Lacorazza, H.D. Sulforaphane Induces Cell Cycle Arrest and Apoptosis in Acute Lymphoblastic Leukemia Cells. PloS ONE 2012, 7, e51251. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Huang, M.-T.; Shen, G.; Yuan, X.; Lin, W.; Khor, T.O.; Conney, A.H.; Kong, A.-N.T. Inhibition of 7,12-dimethylbenz(a)anthracene-induced skin tumorigenesis in C57BL/6 mice by sulforaphane is mediated by nuclear factor E2-related factor 2. Cancer Res. 2006, 66, 8293–8296. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.V.; Xiao, D.; Lew, K.L.; Dhir, R.; Singh, S.V. Sulforaphane induces caspase-mediated apoptosis in cultured PC-3 human prostate cancer cells and retards growth of PC-3 xenografts in vivo. Carcinogenesis 2004, 25, 83–90. [Google Scholar] [CrossRef] [PubMed]
- Traka, M.H.; Spinks, C.A.; Doleman, J.F.; Melchini, A.; Ball, R.Y.; Mills, R.D.; Mithen, R.F. The dietary isothiocyanate sulforaphane modulates gene expression and alternative gene splicing in a PTEN null preclinical murine model of prostate cancer. Mol. Cancer 2010, 9, 189. [Google Scholar] [CrossRef] [PubMed]
- Saw, C.L.; Huang, M.-T.; Liu, Y.; Khor, T.O.; Conney, A.H.; Kong, A.-N. Impact of Nrf2 on UVB-Induced Skin Inflammation/Photoprotection and Photoprotective Effect of Sulforaphane. Mol. Carcinog. 2011, 50, 479–486. [Google Scholar] [CrossRef] [PubMed]
- Surh, Y.-J. Cancer chemoprevention with dietary phytochemicals. Nat. Rev. Cancer 2003, 3, 768–780. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Gordon, G.B. A strategy for cancer prevention: Stimulation of the Nrf2-ARE signaling pathway. Mol. Cancer Ther. 2004, 3, 885–893. [Google Scholar] [PubMed]
- Leibelt, D.A.; Hedstrom, O.R.; Fischer, K.A.; Pereira, C.B.; Williams, D.E. Evaluation of Chronic Dietary Exposure to Indole-3-Carbinol and Absorption-Enhanced 3,3′-Diindolylmethane in Sprague-Dawley Rats. Toxicol. Sci. 2003, 74, 10–21. [Google Scholar] [CrossRef] [PubMed]
- Jeffery, E.H.; Araya, M. Physiological effects of broccoli consumption. Phytochem. Rev. 2008, 8, 283–298. [Google Scholar] [CrossRef]
- La Marca, M.; Beffy, P.; Della Croce, C.; Gervasi, P.G.; Iori, R.; Puccinelli, E.; Longo, V. Structural influence of isothiocyanates on expression of cytochrome P450, phase II enzymes, and activation of Nrf2 in primary rat hepatocytes. Food Chem. Toxicol. 2012, 50, 2822–2830. [Google Scholar] [CrossRef] [PubMed]
- James, D.; Devaraj, S.; Bellur, P.; Lakkanna, S.; Vicini, J.; Boddupalli, S. Novel concepts of broccoli sulforaphanes and disease: induction of phase II antioxidant and detoxification enzymes by enhanced-glucoraphanin broccoli. Nutr. Rev. 2012, 70, 654–665. [Google Scholar] [CrossRef] [PubMed]
- Ernst, I.M.A.; Wagner, A.E.; Schuemann, C.; Storm, N.; Höppner, W.; Döring, F.; Stocker, A.; Rimbach, G. Allyl-, butyl- and phenylethyl-isothiocyanate activate Nrf2 in cultured fibroblasts. Pharmacol. Res. 2011, 63, 233–240. [Google Scholar] [CrossRef] [PubMed]
- Ernst, I.M.A.; Schuemann, C.; Wagner, A.E.; Rimbach, G. 3,3′-Diindolylmethane but not indole-3-carbinol activates Nrf2 and induces Nrf2 target gene expression in cultured murine fibroblasts. Free Radic. Res. 2011, 45, 941–949. [Google Scholar] [CrossRef] [PubMed]
- Mizuno, K.; Kume, T.; Muto, C.; Takada-Takatori, Y.; Izumi, Y.; Sugimoto, H.; Akaike, A. Glutathione Biosynthesis via Activation of the Nuclear Factor E2–Related Factor 2 (Nrf2) – Antioxidant-Response Element (ARE) Pathway Is Essential for Neuroprotective Effects of Sulforaphane and 6-(Methylsulfinyl) Hexyl Isothiocyanate. J. Pharmacol. Sci. 2011, 115, 320–328. [Google Scholar] [CrossRef] [PubMed]
- Wu, T.-Y.; Saw, C.L.-L.; Khor, T.O.; Pung, D.; Boyanapalli, S.S.S.; Kong, A.-N.T. In vivo pharmacodynamics of indole-3-carbinol in the inhibition of prostate cancer in transgenic adenocarcinoma of mouse prostate (TRAMP) mice: Involvement of Nrf2 and cell cycle/apoptosis signaling pathways. Mol. Carcinog. 2012, 51, 761–770. [Google Scholar] [CrossRef] [PubMed]
- McWalter, G.K.; Higgins, L.G.; McLellan, L.I.; Henderson, C.J.; Song, L.; Thornalley, P.J.; Itoh, K.; Yamamoto, M.; Hayes, J.D. Transcription Factor Nrf2 Is Essential for Induction of NAD(P)H:Quinone Oxidoreductase 1, Glutathione S-Transferases, and Glutamate Cysteine Ligase by Broccoli Seeds and Isothiocyanates. J. Nutr. 2004, 134, 3499S–3506S. [Google Scholar] [PubMed]
- Thimmulappa, R.K.; Mai, K.H.; Srisuma, S.; Kensler, T.W.; Yamamoto, M.; Biswal, S. Identification of Nrf2-regulated Genes Induced by the Chemopreventive Agent Sulforaphane by Oligonucleotide Microarray. Cancer Res. 2002, 62, 5196–5203. [Google Scholar] [PubMed]
- Gasper, A.V.; Traka, M.; Bacon, J.R.; Smith, J.A.; Taylor, M.A.; Hawkey, C.J.; Barrett, D.A.; Mithen, R.F. Consuming Broccoli Does Not Induce Genes Associated with Xenobiotic Metabolism and Cell Cycle Control in Human Gastric Mucosa. J. Nutr. 2007, 137, 1718–1724. [Google Scholar] [PubMed]
- Hu, R.; Xu, C.; Shen, G.; Jain, M.R.; Khor, T.O.; Gopalkrishnan, A.; Lin, W.; Reddy, B.; Chan, J.Y.; Kong, A.-N.T. Gene expression profiles induced by cancer chemopreventive isothiocyanate sulforaphane in the liver of C57BL/6J mice and C57BL/6J/Nrf2 (−/−) mice. Cancer Lett. 2006, 243, 170–192. [Google Scholar] [CrossRef] [PubMed]
- Angeloni, C.; Leoncini, E.; Malaguti, M.; Angelini, S.; Hrelia, P.; Hrelia, S. Modulation of Phase II Enzymes by Sulforaphane: Implications for Its Cardioprotective Potential. J. Agric. Food Chem. 2009, 57, 5615–5622. [Google Scholar] [CrossRef] [PubMed]
- Vauzour, D.; Buonfiglio, M.; Corona, G.; Chirafisi, J.; Vafeiadou, K.; Angeloni, C.; Hrelia, S.; Hrelia, P.; Spencer, J.P.E. Sulforaphane protects cortical neurons against 5-S-cysteinyl-dopamine-induced toxicity through the activation of ERK1/2, Nrf-2 and the upregulation of detoxification enzymes. Mol. Nutr. Food Res. 2010, 54, 532–542. [Google Scholar] [CrossRef] [PubMed]
- Lii, C.-K.; Liu, K.-L.; Cheng, Y.-P.; Lin, A.-H.; Chen, H.-W.; Tsai, C.-W. Sulforaphane and α-Lipoic Acid Upregulate the Expression of the π Class of Glutathione S-Transferase through c-Jun and Nrf2 Activation. J. Nutr. 2010, 140, 885–892. [Google Scholar] [CrossRef] [PubMed]
- Brooks, J.D.; Paton, V.G.; Vidanes, G. Potent induction of phase 2 enzymes in human prostate cells by sulforaphane. Cancer Epidemiol. Biomark. Prev. Publ. Am. Assoc. Cancer Res. Cosponsored Am. Soc. Prev. Oncol. 2001, 10, 949–954. [Google Scholar]
- Petri, N.; Tannergren, C.; Holst, B.; Mellon, F.A.; Bao, Y.; Plumb, G.W.; Bacon, J.; O’Leary, K.A.; Kroon, P.A.; Knutson, L.; et al. Absorption/Metabolism of Sulforaphane and Quercetin, and Regulation of Phase Ii Enzymes, in Human Jejunum in Vivo. Drug Metab. Dispos. 2003, 31, 805–813. [Google Scholar] [CrossRef] [PubMed]
- McMahon, M.; Itoh, K.; Yamamoto, M.; Chanas, S.A.; Henderson, C.J.; McLellan, L.I.; Wolf, C.R.; Cavin, C.; Hayes, J.D. The Cap “n” Collar Basic Leucine Zipper Transcription Factor Nrf2 (NF-E2 p45-related Factor 2) Controls Both Constitutive and Inducible Expression of Intestinal Detoxification and Glutathione Biosynthetic Enzymes. Cancer Res. 2001, 61, 3299–3307. [Google Scholar] [PubMed]
- Hu, R.; Xu, C.; Shen, G.; Jain, M.R.; Khor, T.O.; Gopalkrishnan, A.; Lin, W.; Reddy, B.; Chan, J.Y.; Kong, A.-N.T. Identification of Nrf2-regulated genes induced by chemopreventive isothiocyanate PEITC by oligonucleotide microarray. Life Sci. 2006, 79, 1944–1955. [Google Scholar] [CrossRef] [PubMed]
- Munday, R.; Munday, C.M. Induction of phase II detoxification enzymes in rats by plant-derived isothlocyanates: Comparison of allyl isothiocyanate with sulforaphane and related compounds. J. Agric. Food Chem. 2004, 52, 1867–1871. [Google Scholar] [CrossRef] [PubMed]
- Sørensen, M.; Jensen, B.R.; Poulsen, H.E.; Deng, X.-S.; Tygstrup, N.; Dalhoff, K.; Loft, S. Effects of a Brussels sprouts extract on oxidative DNA damage and metabolising enzymes in rat liver. Food Chem. Toxicol. 2001, 39, 533–540. [Google Scholar] [CrossRef]
- Zhang, Y.; Munday, R.; Jobson, H.E.; Munday, C.M.; Lister, C.; Wilson, P.; Fahey, J.W.; Mhawech-Fauceglia, P. Induction of GST and NQO1 in cultured bladder cells and in the urinary bladders of rats by an extract of broccoli (Brassica oleracea italica) sprouts. J. Agric. Food Chem. 2006, 54, 9370–9376. [Google Scholar] [CrossRef] [PubMed]
- Ritz, S.A.; Wan, J.; Diaz-Sanchez, D. Sulforaphane-stimulated phase II enzyme induction inhibits cytokine production by airway epithelial cells stimulated with diesel extract. Am. J. Physiol. Lung Cell. Mol. Physiol. 2007, 292, L33–L39. [Google Scholar] [CrossRef] [PubMed]
- Riedl, M.A.; Saxon, A.; Diaz-Sanchez, D. Oral Sulforaphane increases Phase II antioxidant enzymes in the human upper airway. Clin. Immunol. Orlando Fla 2009, 130, 244–251. [Google Scholar] [CrossRef] [PubMed]
- Szaefer, H.; Krajka-Kuźniak, V.; Licznerska, B.; Bartoszek, A.; Baer-Dubowska, W. Cabbage Juices and Indoles Modulate the Expression Profile of AhR, ERα, and Nrf2 in Human Breast Cell Lines. Nutr. Cancer 2015, 67, 1342–1354. [Google Scholar] [CrossRef] [PubMed]
- Tan, X.-L.; Shi, M.; Tang, H.; Han, W.; Spivack, S.D. Candidate Dietary Phytochemicals Modulate Expression of Phase II Enzymes GSTP1 and NQO1 in Human Lung Cells. J. Nutr. 2010, 140, 1404–1410. [Google Scholar] [CrossRef] [PubMed]
- Wagner, A.E.; Sturm, C.; Piegholdt, S.; Wolf, I.M.A.; Esatbeyoglu, T.; De Nicola, G.R.; Iori, R.; Rimbach, G. Myrosinase-treated glucoerucin is a potent inducer of the Nrf2 target gene heme oxygenase 1—Studies in cultured HT-29 cells and mice. J. Nutr. Biochem. 2015, 26, 661–666. [Google Scholar] [CrossRef] [PubMed]
- Traka, M.; Gasper, A.V.; Smith, J.A.; Hawkey, C.J.; Bao, Y.; Mithen, R.F. Transcriptome Analysis of Human Colon Caco-2 Cells Exposed to Sulforaphane. J. Nutr. 2005, 135, 1865–1872. [Google Scholar] [PubMed]
- Bonnesen, C.; Eggleston, I.M.; Hayes, J.D. Dietary Indoles and Isothiocyanates That Are Generated from Cruciferous Vegetables Can Both Stimulate Apoptosis and Confer Protection against DNA Damage in Human Colon Cell Lines. Cancer Res. 2001, 61, 6120–6130. [Google Scholar] [PubMed]
- Dinkova-Kostova, A.T.; Fahey, J.W.; Wade, K.L.; Jenkins, S.N.; Shapiro, T.A.; Fuchs, E.J.; Kerns, M.L.; Talalay, P. Induction of the Phase 2 Response in Mouse and Human Skin by Sulforaphane-containing Broccoli Sprout Extracts. Cancer Epidemiol. Biomarkers Prev. 2007, 16, 847–851. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Dinkova-Kostova, A.T.; Talalay, P. Powerful and prolonged protection of human retinal pigment epithelial cells, keratinocytes, and mouse leukemia cells against oxidative damage: The indirect antioxidant effects of sulforaphane. Proc. Natl. Acad. Sci. USA 2001, 98, 15221–15226. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Talalay, P. Induction of phase 2 genes by sulforaphane protects retinal pigment epithelial cells against photooxidative damage. Proc. Natl. Acad. Sci. USA 2004, 101, 10446–10451. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Su, Z.-Y.; Khor, T.O.; Shu, L.; Kong, A.-N.T. Sulforaphane Enhances Nrf2 Expression in Prostate Cancer TRAMP C1 Cells through Epigenetic Regulation. Biochem. Pharmacol. 2013, 85, 1398–1404. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Wang, S.R.; Howie, A.F.; Beckett, G.J.; Mithen, R.; Bao, Y.P. Sulforaphane, erucin, and lberin up-regulate thioredoxin reductase 1 expression in human MCF-7 cells. J. Agric. Food Chem. 2005, 53, 1417–1421. [Google Scholar] [CrossRef] [PubMed]
- Bacon, J.R.; Plumb, G.W.; Howie, A.F.; Beckett, G.J.; Wang, W.; Bao, Y. Dual Action of Sulforaphane in the Regulation of Thioredoxin Reductase and Thioredoxin in Human HepG2 and Caco-2 Cells. J. Agric. Food Chem. 2007, 55, 1170–1176. [Google Scholar] [CrossRef] [PubMed]
- Kong, L.; Tanito, M.; Huang, Z.; Li, F.; Zhou, X.; Zaharia, A.; Yodoi, J.; McGinnis, J.F.; Cao, W. Delay of photoreceptor degeneration in tubby mouse by sulforaphane. J. Neurochem. 2007, 101, 1041–1052. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Khor, T.O.; Xu, C.; Shen, G.; Jeong, W.-S.; Yu, S.; Kong, A.-N. Activation of Nrf2-antioxidant signaling attenuates NFκB-inflammatory response and elicits apoptosis. Biochem. Pharmacol. 2008, 76, 1485–1489. [Google Scholar] [CrossRef] [PubMed]
- Cheung, K.L.; Kong, A.-N. Molecular Targets of Dietary Phenethyl Isothiocyanate and Sulforaphane for Cancer Chemoprevention. AAPS J. 2009, 12, 87–97. [Google Scholar] [CrossRef] [PubMed]
- Jaramillo, M.C.; Zhang, D.D. The emerging role of the Nrf2-Keap1 signaling pathway in cancer. Genes Dev. 2013, 27, 2179–2191. [Google Scholar] [CrossRef] [PubMed]
- Kansanen, E.; Kuosmanen, S.M.; Leinonen, H.; Levonen, A.-L. The Keap1-Nrf2 pathway: Mechanisms of activation and dysregulation in cancer. Redox Biol. 2013, 1, 45–49. [Google Scholar] [CrossRef] [PubMed]
- Chorley, B.N.; Campbell, M.R.; Wang, X.; Karaca, M.; Sambandan, D.; Bangura, F.; Xue, P.; Pi, J.; Kleeberger, S.R.; Bell, D.A. Identification of novel NRF2-regulated genes by ChIP-Seq: Influence on retinoid X receptor alpha. Nucleic Acids Res. 2012, 40, 7416–7429. [Google Scholar] [CrossRef] [PubMed]
- McMahon, M.; Thomas, N.; Itoh, K.; Yamamoto, M.; Hayes, J.D. Dimerization of substrate adaptors can facilitate cullin-mediated ubiquitylation of proteins by a “Tethering” mechanism—A two-site interaction model for the Nrf2-Keap1 complex. J. Biol. Chem. 2006, 281, 24756–24768. [Google Scholar] [CrossRef] [PubMed]
- Tong, K.I.; Kobayashi, A.; Katsuoka, F.; Yamamoto, M. Two-site substrate recognition model for the Keap1-Nrf2 system: A hinge and latch mechanism. Biol. Chem. 2006, 387, 1311–1320. [Google Scholar] [CrossRef] [PubMed]
- Tong, K.I.; Katoh, Y.; Kusunoki, H.; Itoh, K.; Tanaka, T.; Yamamoto, M. Keap1 recruits Neh2 through binding to ETGE and DLG motifs: Characterization of the two-site molecular recognition model. Mol. Cell. Biol. 2006, 26, 2887–2900. [Google Scholar] [CrossRef] [PubMed]
- Tong, K.I.; Padmanabhan, B.; Kobayashi, A.; Shang, C.; Hirotsu, Y.; Yokoyama, S.; Yamamoto, M. Different electrostatic potentials define ETGE and DLG motifs as hinge and latch in oxidative stress response. Mol. Cell. Biol. 2007, 27, 7511–7521. [Google Scholar] [CrossRef] [PubMed]
- Dinkova-Kostova, A.T.; Holtzclaw, W.D.; Cole, R.N.; Itoh, K.; Wakabayashi, N.; Katoh, Y.; Yamamoto, M.; Talalay, P. Direct evidence that sulfhydryl groups of Keap1 are the sensors regulating induction of phase 2 enzymes that protect against carcinogens and oxidants. Proc. Natl. Acad. Sci. USA 2002, 99, 11908–11913. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.D.; Hannink, M. Distinct cysteine residues in Keap1 are required for Keap1-dependent ubiquitination of Nrf2 and for stabilization of Nrf2 by chemopreventive agents and oxidative stress. Mol. Cell. Biol. 2003, 23, 8137–8151. [Google Scholar] [CrossRef] [PubMed]
- McMahon, M.; Lamont, D.J.; Beattie, K.A.; Hayes, J.D. Keap1 perceives stress via three sensors for the endogenous signaling molecules nitric oxide, zinc, and alkenals. Proc. Natl. Acad. Sci. USA 2010, 107, 18838–18843. [Google Scholar] [CrossRef] [PubMed]
- Hirotsu, Y.; Katsuoka, F.; Funayama, R.; Nagashima, T.; Nishida, Y.; Nakayama, K.; Engel, J.D.; Yamamoto, M. Nrf2-MafG heterodimers contribute globally to antioxidant and metabolic networks. Nucleic Acids Res. 2012, 40, 10228–10239. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.; Huang, H.C.; Pickett, C.B. Transcriptional regulation of the antioxidant response element—Activation by Nrf2 and repression by MafK. J. Biol. Chem. 2000, 275, 15466–15473. [Google Scholar] [CrossRef] [PubMed]
- Motohashi, H.; Katsuoka, F.; Engel, J.D.; Yamamoto, M. Small Maf proteins serve as transcriptional cofactors for keratinocyte differentiation in the Keap1-Nrf2 regulatory pathway. Proc. Natl. Acad. Sci. USA 2004, 101, 6379–6384. [Google Scholar] [CrossRef] [PubMed]
- Jaiswal, A.K. Nrf2 signaling in coordinated activation of antioxidant gene expression. Free Radic. Biol. Med. 2004, 36, 1199–1207. [Google Scholar] [CrossRef] [PubMed]
- Motohashi, H.; Katsuoka, F.; Miyoshi, C.; Uchimura, Y.; Saitoh, H.; Francastel, C.; Engel, J.D.; Yamamoto, M. MafG Sumoylation Is Required for Active Transcriptional Repression. Mol. Cell. Biol. 2006, 26, 4652–4663. [Google Scholar] [CrossRef] [PubMed]
- Jaiswal, A.K. Regulation of Antioxidant Response Element–Dependent Induction of Detoxifying Enzyme Synthesis. In Quinones and Quinone Enzymes, Part A; Enzymology, B.-M., Ed.; Academic Press: San Diego, CA, USA, 2004; Volume 378, pp. 221–238. [Google Scholar]
- Alnouti, Y.; Klaassen, C.D. Regulation of Sulfotransferase Enzymes by Prototypical Microsomal Enzyme Inducers in Mice. J. Pharmacol. Exp. Ther. 2008, 324, 612–621. [Google Scholar] [CrossRef] [PubMed]
- Kalthoff, S.; Ehmer, U.; Freiberg, N.; Manns, M.P.; Strassburg, C.P. Interaction between Oxidative Stress Sensor Nrf2 and Xenobiotic-activated Aryl Hydrocarbon Receptor in the Regulation of the Human Phase II Detoxifying UDP-glucuronosyltransferase 1A10. J. Biol. Chem. 2010, 285, 5993–6002. [Google Scholar] [CrossRef] [PubMed]
- Hayes, J.D.; McMahon, M.; Chowdhry, S.; Dinkova-Kostova, A.T. Cancer Chemoprevention Mechanisms Mediated Through the Keap1-Nrf2 Pathway. Antioxid. Redox Signal. 2010, 13, 1713–1748. [Google Scholar] [CrossRef] [PubMed]
- Taguchi, K.; Motohashi, H.; Yamamoto, M. Molecular mechanisms of the Keap1–Nrf2 pathway in stress response and cancer evolution. Genes Cells 2011, 16, 123–140. [Google Scholar] [CrossRef] [PubMed]
- Zenkov, N.K.; Menshchikova, E.B.; Tkachev, V.O. Keap1/Nrf2/ARE redox-sensitive signaling system as a pharmacological target. Biochem. Mosc. 2013, 78, 19–36. [Google Scholar] [CrossRef] [PubMed]
- Fuentes, F.; Shu, L.; Lee, J.; Su, Z.-Y.; Lee, K.-R.; Kong, A.-N. Nrf2-Target Approaches in Cancer Chemoprevention Mediated by Dietary Phytochemicals. In Cancer Prevention; Methods in Pharmacology and Toxicology; Bode, A.M., Dong, Z., Eds.; Springer: New York, NY, USA, 2014; pp. 53–83. [Google Scholar]
- Becker, T.M.; Jeffery, E.H.; Juvik, J.A. Survey of variation for chemopreventive bioactivity, glucosinolate, and glucosinolate hydrolysis product profiles in common Brassica crop species. J. Agric. Food Chem. 2016. in preparation. [Google Scholar]
- Hu, C.; Eggler, A.L.; Mesecar, A.D.; van Breemen, R.B. Modification of Keap1 Cysteine Residues by Sulforaphane. Chem. Res. Toxicol. 2011, 24, 515–521. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.D.; Lo, S.-C.; Sun, Z.; Habib, G.M.; Lieberman, M.W.; Hannink, M. Ubiquitination of Keap1, a BTB-Kelch Substrate Adaptor Protein for Cul3, Targets Keap1 for Degradation by a Proteasome-independent Pathway. J. Biol. Chem. 2005, 280, 30091–30099. [Google Scholar] [CrossRef] [PubMed]
- Hong, F.; Freeman, M.L.; Liebler, D.C. Identification of Sensor Cysteines in Human Keap1 Modified by the Cancer Chemopreventive Agent Sulforaphane. Chem. Res. Toxicol. 2005, 18, 1917–1926. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.D.; Lo, S.C.; Cross, J.V.; Templeton, D.J.; Hannink, M. Keap1 is a redox-regulated substrate adaptor protein for a Cul3-dependent ubiquitin ligase complex. Mol. Cell. Biol. 2004, 24, 10941–10953. [Google Scholar] [CrossRef] [PubMed]
- Eggler, A.L.; Small, E.; Hannink, M.; Mesecar, A.D. Cul3-mediated Nrf2 ubiquitination and antioxidant response element (ARE) activation are dependent on the partial molar volume at position 151 of Keap1. Biochem. J. 2009, 422, 171–180. [Google Scholar] [CrossRef] [PubMed]
- Darvekar, S.R.; Elvenes, J.; Brenne, H.B.; Johansen, T.; Sjøttem, E. SPBP Is a Sulforaphane Induced Transcriptional Coactivator of NRF2 Regulating Expression of the Autophagy Receptor p62/SQSTM1. PLoS ONE 2014, 9, e85262. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Z.-Q.; Chen, C.; Yang, B.; Hebbar, V.; Kong, A.-N.T. Differential responses from seven mammalian cell lines to the treatments of detoxifying enzyme inducers. Life Sci. 2003, 72, 2243–2253. [Google Scholar] [CrossRef]
- Nair, S.; Barve, A.; Khor, T.-O.; Shen, G.; Lin, W.; Chan, J.Y.; Cai, L.; Kong, A.-N. Regulation of Nrf2- and AP-1-mediated gene expression by epigallocatechin-3-gallate and sulforaphane in prostate of Nrf2-knockout or C57BL/6J mice and PC-3 AP-1 human prostate cancer cells. Acta Pharmacol. Sin. 2010, 31, 1223–1240. [Google Scholar] [CrossRef] [PubMed]
- Švehlíková, V.; Wang, S.; Jakubíková, J.; Williamson, G.; Mithen, R.; Bao, Y. Interactions between sulforaphane and apigenin in the induction of UGT1A1 and GSTA1 in CaCo-2 cells. Carcinogenesis 2004, 25, 1629–1637. [Google Scholar] [CrossRef] [PubMed]
- Fahey, J.W.; Talalay, P. Antioxidant Functions of Sulforaphane: A Potent Inducer of Phase II Detoxication Enzymes. Food Chem. Toxicol. 1999, 37, 973–979. [Google Scholar] [CrossRef]
- Guerrero-Beltrán, C.E.; Calderón-Oliver, M.; Pedraza-Chaverri, J.; Chirino, Y.I. Protective effect of sulforaphane against oxidative stress: Recent advances. Exp. Toxicol. Pathol. 2012, 64, 503–508. [Google Scholar] [CrossRef] [PubMed]
- Ye, L.; Dinkova-Kostova, A.T.; Wade, K.L.; Zhang, Y.; Shapiro, T.A.; Talalay, P. Quantitative determination of dithiocarbamates in human plasma, serum, erythrocytes and urine: Pharmacokinetics of broccoli sprout isothiocyanates in humans. Clin. Chim. Acta 2002, 316, 43–53. [Google Scholar] [CrossRef]
- Yu, S.; Khor, T.O.; Cheung, K.-L.; Li, W.; Wu, T.-Y.; Huang, Y.; Foster, B.A.; Kan, Y.W.; Kong, A.-N. Nrf2 Expression Is Regulated by Epigenetic Mechanisms in Prostate Cancer of TRAMP Mice. PLoS ONE 2010, 5, e8579. [Google Scholar] [CrossRef] [PubMed]
- Prasad, A.K.; Mishra, P.C. Mechanism of Action of Sulforaphane as a Superoxide Radical Anion and Hydrogen Peroxide Scavenger by Double Hydrogen Transfer: A Model for Iron Superoxide Dismutase. J. Phys. Chem. B 2015, 119, 7825–7836. [Google Scholar] [CrossRef] [PubMed]
- Keum, Y.-S.; Owuor, E.D.; Kim, B.-R.; Hu, R.; Kong, A.-N.T. Involvement of Nrf2 and JNK1 in the Activation of Antioxidant Responsive Element (ARE) by Chemopreventive Agent Phenethyl Isothiocyanate (PEITC). Pharm. Res. 2003, 20, 1351–1356. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.J.; Shen, G.X.; Yuan, X.L.; Kim, J.H.; Gopalkrishnan, A.; Keum, Y.S.; Nair, S.; Kong, A.N.T. ERK and JNK signaling pathways are involved in the regulation of activator protein 1 and cell death elicited by three isothiocyanates in human prostate cancer PC-3 cells. Carcinogenesis 2006, 27, 437–445. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Yuan, X.; Pan, Z.; Shen, G.; Kim, J.-H.; Yu, S.; Khor, T.O.; Li, W.; Ma, J.; Kong, A.-N.T. Mechanism of action of isothiocyanates: The induction of ARE-regulated genes is associated with activation of ERK and JNK and the phosphorylation and nuclear translocation of Nrf2. Mol. Cancer Ther. 2006, 5, 1918–1926. [Google Scholar] [CrossRef] [PubMed]
- Ye, L.; Zhang, Y. Total intracellular accumulation levels of dietary isothiocyanates determine their activity in elevation of cellular glutathione and induction of Phase 2 detoxification enzymes. Carcinogenesis 2001, 22, 1987–1992. [Google Scholar] [CrossRef] [PubMed]
- Qin, C.-Z.; Zhang, X.; Wu, L.-X.; Wen, C.-J.; Hu, L.; Lv, Q.-L.; Shen, D.-Y.; Zhou, H.-H. Advances in Molecular Signaling Mechanisms of β-Phenethyl Isothiocyanate Antitumor Effects. J. Agric. Food Chem. 2015, 63, 3311–3322. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.; Gao, X.; Chen, G.; Sun, J.; Chu, J.; Jing, K.; Li, P.; Zeng, R.; Wei, B. Indole-3-carbinol as inhibitors of glucocorticoid-induced apoptosis in osteoblastic cells through blocking ROS-mediated Nrf2 pathway. Biochem. Biophys. Res. Commun. 2015, 460, 422–427. [Google Scholar] [CrossRef] [PubMed]
- Bradlow, H.L.; Zeligs, M.A. Diindolylmethane (DIM) Spontaneously Forms from Indole-3-carbinol (I3C) During Cell Culture Experiments. In Vivo 2010, 24, 387–391. [Google Scholar] [PubMed]
- De Kruif, C.A.; Marsman, J.W.; Venekamp, J.C.; Falke, H.E.; Noordhoek, J.; Blaauboer, B.J.; Wortelboer, H.M. Structure elucidation of acid reaction products of indole-3-carbinol: Detection in vivo and enzyme induction in vitro. Chem. Biol. Interact. 1991, 80, 303–315. [Google Scholar] [CrossRef]
- Aggarwal, B.B.; Ichikawa, H. Molecular Targets and Anticancer Potential of Indole-3-Carbinol and Its Derivatives. Cell Cycle 2005, 4, 1201–1215. [Google Scholar] [CrossRef] [PubMed]
- Holst, B.; Williamson, G. A critical review of the bioavailability of glucosinolates and related compounds. Nat. Prod. Rep. 2004, 21, 425–447. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhang, T.; Li, X.; Zou, P.; Schwartz, S.J.; Sun, D. Kinetics of Sulforaphane in Mice after Consumption of Sulforaphane-Enriched Broccoli Sprout Preparation. Mol. Nutr. Food Res. 2013, 57. [Google Scholar] [CrossRef] [PubMed]
- Clarke, J.D.; Hsu, A.; Riedl, K.; Bella, D.; Schwartz, S.J.; Stevens, J.F.; Ho, E. Bioavailability and inter-conversion of sulforaphane and erucin in human subjects consuming broccoli sprouts or broccoli supplement in a cross-over study design. Pharmacol. Res. 2011, 64, 456–463. [Google Scholar] [CrossRef] [PubMed]
- Egner, P.A.; Chen, J.G.; Wang, J.B.; Wu, Y.; Sun, Y.; Lu, J.H.; Zhu, J.; Zhang, Y.H.; Chen, Y.S.; Friesen, M.D.; et al. Bioavailability of Sulforaphane from two broccoli sprout beverages: Results of a short-term, cross-over clinical trial in Qidong, China. Cancer Prev. Res. Phila. Pa 2011, 4, 384–395. [Google Scholar] [CrossRef] [PubMed]
- Fahey, J.W.; Holtzclaw, W.D.; Wehage, S.L.; Wade, K.L.; Stephenson, K.K.; Talalay, P. Sulforaphane Bioavailability from Glucoraphanin-Rich Broccoli: Control by Active Endogenous Myrosinase. PLoS ONE 2015, 10, e0140963. [Google Scholar] [CrossRef] [PubMed]
- Ji, Y.; Kuo, Y.; Morris, M.E. Pharmacokinetics of Dietary Phenethyl Isothiocyanate in Rats. Pharm. Res. 2005, 22, 1658–1666. [Google Scholar] [CrossRef] [PubMed]
- Liebes, L.; Conaway, C.C.; Hochster, H.; Mendoza, S.; Hecht, S.S.; Crowell, J.; Chung, F.-L. High-Performance Liquid Chromatography-Based Determination of Total Isothiocyanate Levels in Human Plasma: Application to Studies with 2-Phenethyl Isothiocyanate. Anal. Biochem. 2001, 291, 279–289. [Google Scholar] [CrossRef] [PubMed]
- Buttery, R.G.; Guadagni, D.G.; Ling, L.C.; Seifert, R.M.; Lipton, W. Additional volatile components of cabbage, broccoli, and cauliflower. J. Agric. Food Chem. 1976, 24, 829–832. [Google Scholar] [CrossRef]
- Wilson, E.A.; Ennahar, S.; Marchioni, E.; Bergaentzle, M.; Bindler, F. Improvement in determination of isothiocyanates using high-temperature reversed-phase HPLC. J. Sep. Sci. 2012, 35, 2026–2031. [Google Scholar] [CrossRef] [PubMed]
- Hecht, S.S. Inhibition of Carcinogenesis by Isothiocyanates. Drug Metab. Rev. 2000, 32, 395–411. [Google Scholar] [CrossRef] [PubMed]
- Wu, T.-Y.; Huang, Y.; Zhang, C.; Su, Z.-Y.; Boyanapalli, S.; Khor, T.O.; Wang, H.; Lin, H.; Gounder, M.; Kagan, L.; et al. Pharmacokinetics and pharmacodynamics of 3,3′-diindolylmethane (DIM) in regulating gene expression of phase II drug metabolizing enzymes. J. Pharmacokinet. Pharmacodyn. 2015, 42, 401–408. [Google Scholar] [CrossRef] [PubMed]
- Paltsev, M.; Kiselev, V.; Muyzhnek, E.; Drukh, V.; Kuznetsov, I.; Pchelintseva, O. Comparative preclinical pharmacokinetics study of 3,3′-diindolylmethane formulations: Is personalized treatment and targeted chemoprevention in the horizon? EPMA J. 2013, 4, 25. [Google Scholar] [CrossRef] [PubMed]
- Anderton, M.J.; Manson, M.M.; Verschoyle, R.; Gescher, A.; Steward, W.P.; Williams, M.L.; Mager, D.E. Physiological Modeling of Formulated and Crystalline 3,3′-Diindolylmethane Pharmacokinetics Following Oral Administration in Mice. Drug Metab. Dispos. 2004, 32, 632–638. [Google Scholar] [CrossRef] [PubMed]
- Ku, K.-M.; Jeffery, E.H.; Juvik, J.A. Exogenous methyl jasmonate treatment increases glucosinolate biosynthesis and quinone reductase activity in kale leaf tissue. PloS ONE 2014. [Google Scholar] [CrossRef] [PubMed]
- Feskanich, D.; Ziegler, R.G.; Michaud, D.S.; Giovannucci, E.L.; Speizer, F.E.; Willett, W.C.; Colditz, G.A. Prospective study of fruit and vegetable consumption and risk of lung cancer among men and women. J. Natl. Cancer Inst. 2000, 92, 1812–1823. [Google Scholar] [CrossRef] [PubMed]
- Voorrips, L.E.; Goldbohm, R.A.; van Poppel, G.; Sturmans, F.; Hermus, R.J.J.; van den Brandt, P.A. Vegetable and fruit consumption and risks of colon and rectal cancer in a prospective cohort study - The Netherlands Cohort Study on Diet and Cancer. Am. J. Epidemiol. 2000, 152, 1081–1092. [Google Scholar] [CrossRef] [PubMed]
- Voorrips, L.; Goldbohm, R.; Verhoeven, D.; van Poppel, G.; Sturmans, F.; Hermus, R.; van den Brandt, P. Vegetable and fruit consumption and lung cancer risk in the Netherlands Cohort Study on diet and cancer. Cancer Causes Control 2000, 11, 101–115. [Google Scholar] [CrossRef] [PubMed]
- Giovannucci, E.; Rimm, E.; Liu, Y.; Stampfer, M.; Willett, W. A prospective study of cruciferous vegetables and prostate cancer. Cancer Epidemiol. Biomarkers Prev. 2003, 12, 1403–1409. [Google Scholar] [PubMed]
- Neuhouser, M.; Patterson, R.; Thornquist, M.; Omenn, G.; King, I.; Goodman, G. Fruits and vegetables are associated with lower lung cancer risk only in the placebo arm of the beta-Carotene and Retinol Efficacy Trial (CARET). Cancer Epidemiol. Biomarkers Prev. 2003, 12, 350–358. [Google Scholar] [PubMed]
- Willett, W. Nutritional Epidemiology, 3rd ed.; Oxford University Press: New York, NY, USA, 2012; Volume 40. [Google Scholar]
- Verhoeven, D.; Goldbohm, R.; vanPoppel, G.; Verhagen, H.; vandenBrandt, P. Epidemiological studies on brassica vegetables and cancer risk. Cancer Epidemiol. Biomarkers Prev. 1996, 5, 733–748. [Google Scholar] [PubMed]
- Jeffery, E.H.; Keck, A. Translating knowledge generated by epidemiological and in vitro studies into dietary cancer prevention. Mol. Nutr. Food Res. 2008, 52, S7–S17. [Google Scholar] [PubMed]
- Michaud, D.S.; Spiegelman, D.; Clinton, S.K.; Rimm, E.B.; Willett, W.C.; Giovannucci, E.L. Fruit and vegetable intake and incidence of bladder cancer in a male prospective cohort. J. Natl. Cancer Inst. 1999, 91, 605–613. [Google Scholar] [CrossRef] [PubMed]
- Zhao, B.; Seow, A.; Lee, E.J.; Poh, W.T.; Teh, M.; Eng, P.; Wang, Y.T.; Tan, W.C.; Yu, M.C.; Lee, H.P. Dietary isothiocyanates, glutathione S-transferase -M1, -T1 polymorphisms and lung cancer risk among Chinese women in Singapore. Cancer Epidemiol. Biomark. Prev. Publ. Am. Assoc. Cancer Res. Cosponsored Am. Soc. Prev. Oncol. 2001, 10, 1063–1067. [Google Scholar]
- Zhang, S.M.; Hunter, D.J.; Rosner, B.A.; Giovannucci, E.L.; Colditz, G.A.; Speizer, F.E.; Willett, W.C. Intakes of fruits, vegetables, and related nutrients and the risk of non-Hodgkin’s lymphoma among women. Cancer Epidemiol. Biomark. Prev. Publ. Am. Assoc. Cancer Res. Cosponsored Am. Soc. Prev. Oncol. 2000, 9, 477–485. [Google Scholar]
- Cohen, J.H.; Kristal, A.R.; Stanford, J.L. Fruit and vegetable intakes and prostate cancer risk. J. Natl. Cancer Inst. 2000, 92, 61–68. [Google Scholar] [CrossRef] [PubMed]
- Kolonel, L.N.; Hankin, J.H.; Whittemore, A.S.; Wu, A.H.; Gallagher, R.P.; Wilkens, L.R.; John, E.M.; Howe, G.R.; Dreon, D.M.; West, D.W.; et al. Vegetables, fruits, legumes and prostate cancer: A multiethnic case-control study. Cancer Epidemiol. Biomark. Prev. Publ. Am. Assoc. Cancer Res. Cosponsored Am. Soc. Prev. Oncol. 2000, 9, 795–804. [Google Scholar]
- Fowke, J.H.; Chung, F.L.; Jin, F.; Qi, D.; Cai, Q.; Conaway, C.; Cheng, J.R.; Shu, X.O.; Gao, Y.T.; Zheng, W. Urinary isothiocyanate levels, brassica, and human breast cancer. Cancer Res. 2003, 63, 3980–3986. [Google Scholar] [PubMed]
- Yuan, J.; Gago-Dominguez, M.; Castelao, J.E.; Hankin, J.H.; Ross, R.K.; Yu, M.C. Cruciferous vegetables in relation to renal cell carcinoma. Int. J. Cancer 1998, 77, 211–216. [Google Scholar] [CrossRef]
- Nagle, C.M.; Purdie, D.M.; Webb, P.M.; Green, A.; Harvey, P.W.; Bain, C.J. Dietary influences on survival after ovarian cancer. Int. J. Cancer 2003, 106, 264–269. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Mao, Q.; Lin, Y.; Zhou, F.; Xie, L. The association of cruciferous vegetables intake and risk of bladder cancer: A meta-analysis. World J. Urol. 2013, 31, 127–133. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Luo, Y.; Lu, M.; Xu, X.; Lin, H.; Zheng, Z. Cruciferous vegetable consumption and the risk of pancreatic cancer: A meta-analysis. World J. Surg. Oncol. 2015, 13, 44. [Google Scholar] [CrossRef] [PubMed]
- Veeranki, O.L.; Bhattacharya, A.; Tang, L.; Marshall, J.R.; Zhang, Y. Cruciferous Vegetables, Isothiocyanates, and Prevention of Bladder Cancer. Curr. Pharmacol. Rep. 2015, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Tse, G.; Eslick, G.D. Cruciferous Vegetables and Risk of Colorectal Neoplasms: A Systematic Review and Meta-Analysis. Nutr. Cancer 2014, 66, 128–139. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Mao, Q.; Cao, M.; Xie, L. Cruciferous vegetables intake and risk of prostate cancer: A meta-analysis. Int. J. Urol. 2011, 19, 134–141. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Zhao, L. Cruciferous Vegetables Intake Is Associated with Lower Risk of Renal Cell Carcinoma: Evidence from a Meta-Analysis of Observational Studies. PLoS ONE 2013, 8, e75732. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Hu, Y.; Hu, Y.; Zheng, S. Intake of cruciferous vegetables is associated with reduced risk of ovarian cancer: A meta-analysis. Asia Pac. J. Clin. Nutr. 2015, 24, 101. [Google Scholar] [PubMed]
- Bosetti, C.; Filomeno, M.; Riso, P.; Polesel, J.; Levi, F.; Talamini, R.; Montella, M.; Negri, E.; Franceschi, S.; La Vecchia, C. Cruciferous vegetables and cancer risk in a network of case-control studies. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. ESMO 2012, 23, 2198–2203. [Google Scholar] [CrossRef] [PubMed]
- Shapiro, T.A.; Fahey, J.W.; Dinkova-Kostova, A.T.; Holtzclaw, W.D.; Stephenson, K.K.; Wade, K.L.; Ye, L.; Talalay, P. Safety, Tolerance, and Metabolism of Broccoli Sprout Glucosinolates and Isothiocyanates: A Clinical Phase I Study. Nutr. Cancer 2006, 55, 53–62. [Google Scholar] [CrossRef] [PubMed]
- Fahey, J.W.; Wehage, S.L.; Holtzclaw, W.D.; Kensler, T.W.; Egner, P.A.; Shapiro, T.A.; Talalay, P. Protection of humans by plant glucosinolates: Efficiency of conversion of glucosinolates to isothiocyanates by the gastrointestinal microflora. Cancer Prev. Res. (Phila. Pa) 2012, 5, 603–611. [Google Scholar] [CrossRef] [PubMed]
- Lampe, J.W. Diet, genetic polymorphisms, detoxification, and health risks. Altern. Ther. Health Med. 2007, 13, S108–S111. [Google Scholar] [PubMed]
- Riso, P.; Brusamolino, A.; Moro, M.; Porrini, M. Absorption of bioactive compounds from steamed broccoli and their effect on plasma glutathione S-transferase activity. Int. J. Food Sci. Nutr. 2009, 60, 56–71. [Google Scholar] [CrossRef] [PubMed]
- Seow, A.; Yuan, J.M.; Sun, C.L.; Van Den Berg, D.; Lee, H.P.; Yu, M.C. Dietary isothiocyanates, glutathione S-transferase polymorphisms and colorectal cancer risk in the Singapore Chinese Health Study. Carcinogenesis 2002, 23, 2055–2061. [Google Scholar] [CrossRef] [PubMed]
- Seow, A.; Shi, C.Y.; Chung, F.L.; Jiao, D.; Hankin, J.H.; Lee, H.P.; Coetzee, G.A.; Yu, M.C. Urinary total isothiocyanate (ITC) in a population-based sample of middle-aged and older Chinese in Singapore: Relationship with dietary total ITC and glutathione S-transferase M1/T1/P1 genotypes. Cancer Epidemiol. Biomark. Prev. 1998, 7, 775–781. [Google Scholar]
- Gasper, A.V.; Al-Janobi, A.; Smith, J.A.; Bacon, J.R.; Fortun, P.; Atherton, C.; Taylor, M.A.; Hawkey, C.J.; Barrett, D.A.; Mithen, R.F. Glutathione S-transferase M1 polymorphism and metabolism of sulforaphane from standard and high-glucosinolate broccoli. Am. J. Clin. Nutr. 2005, 82, 1283–1291. [Google Scholar] [PubMed]
- Budnowski, J.; Hanske, L.; Schumacher, F.; Glatt, H.; Platz, S.; Rohn, S.; Blaut, M. Glucosinolates Are Mainly Absorbed Intact in Germfree and Human Microbiota-Associated Mice. J. Agric. Food Chem. 2015, 63, 8418–8428. [Google Scholar] [CrossRef] [PubMed]
- Kensler, T.W.; Chen, J.-G.; Egner, P.A.; Fahey, J.W.; Jacobson, L.P.; Stephenson, K.K.; Ye, L.; Coady, J.L.; Wang, J.-B.; Wu, Y.; et al. Effects of Glucosinolate-Rich Broccoli Sprouts on Urinary Levels of Aflatoxin-DNA Adducts and Phenanthrene Tetraols in a Randomized Clinical Trial in He Zuo Township, Qidong, People’s Republic of China. Cancer Epidemiol. Biomark. Prev. 2005, 14, 2605–2613. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Hullar, M.A.; Beresford, S.A.; Lampe, J.W. Variation of glucoraphanin metabolism in vivo and ex vivo by human gut bacteria. Br. J. Nutr. 2011, 106, 408–416. [Google Scholar] [CrossRef] [PubMed]
- Fahey, J.W.; Talalay, P.; Kensler, T.W. Notes from the Field: “Green” Chemoprevention as Frugal Medicine. Cancer Prev. Res. (Phila. Pa.) 2012, 5, 179–188. [Google Scholar] [CrossRef] [PubMed]
- Riso, P.; Martini, D.; Visioli, F.; Martinetti, A.; Porrini, M. Effect of broccoli intake on markers related to oxidative stress and cancer risk in healthy smokers and nonsmokers. Nutr. Cancer 2009, 61, 232–237. [Google Scholar] [CrossRef] [PubMed]
- Riso, P.; Martini, D.; Møller, P.; Loft, S.; Bonacina, G.; Moro, M.; Porrini, M. DNA damage and repair activity after broccoli intake in young healthy smokers. Mutagenesis 2010, 25, 595–602. [Google Scholar] [CrossRef] [PubMed]
- Riso, P.; Del Bo’, C.; Vendrame, S.; Brusamolino, A.; Martini, D.; Bonacina, G.; Porrini, M. Modulation of plasma antioxidant levels, glutathione S-transferase activity and DNA damage in smokers following a single portion of broccoli: A pilot study. J. Sci. Food Agric. 2014, 94, 522–528. [Google Scholar] [CrossRef] [PubMed]
- Greenwald, P.; Clifford, C.; Milner, J. Diet and cancer prevention. Eur. J. Cancer 2001, 37, 948–965. [Google Scholar] [CrossRef]
- Spitz, M.R.; Duphorne, C.M.; Detry, M.A.; Pillow, P.C.; Amos, C.I.; Lei, L.; de Andrade, M.; Gu, X.; Hong, W.K.; Wu, X. Dietary intake of isothiocyanates: Evidence of a joint effect with glutathione S-transferase polymorphisms in lung cancer risk. Cancer Epidemiol. Biomark. Prev. Publ. Am. Assoc. Cancer Res. Cosponsored Am. Soc. Prev. Oncol. 2000, 9, 1017–1020. [Google Scholar]
- London, S.J.; Yuan, J.-M.; Chung, F.-L.; Gao, Y.-T.; Coetzee, G.A.; Ross, R.K.; Mimi, C.Y. Isothiocyanates, glutathione S-transferase M1 and T1 polymorphisms, and lung-cancer risk: A prospective study of men in Shanghai, China. Lancet 2000, 356, 724–729. [Google Scholar] [CrossRef]
- Fimognari, C.; Turrini, E.; Ferruzzi, L.; Lenzi, M.; Hrelia, P. Natural isothiocyanates: Genotoxic potential versus chemoprevention. Mutat. Res. Mutat. Res. 2012, 750, 107–131. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Li, J.; Tang, L. Cancer-preventive isothiocyanates: Dichotomous modulators of oxidative stress. Free Radic. Biol. Med. 2005, 38, 70–77. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Li, W.; Su, Z.; Kong, A.-N.T. The complexity of the Nrf2 pathway: Beyond the antioxidant response. J. Nutr. Biochem. 2015, 26, 1401–1413. [Google Scholar] [CrossRef] [PubMed]
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).