4.1. Analysis of Findings
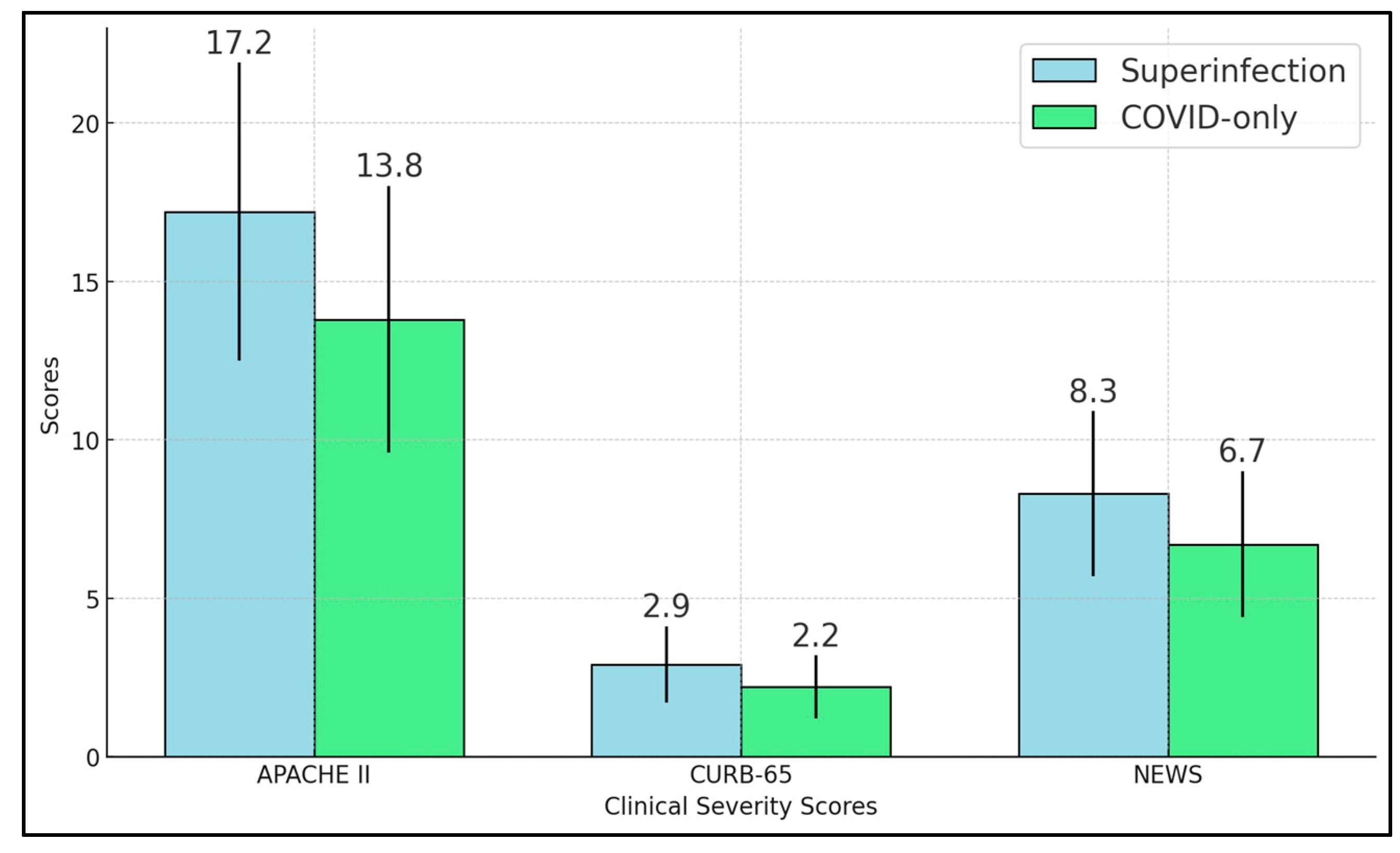
Our study shows that bacterial superinfections significantly affect the clinical symptoms and microbiological data of hospitalized COVID-19 patients. The superinfection group displayed marked elevations in inflammatory markers, including procalcitonin, neutrophil-to-lymphocyte ratio, and SII, aligning with the anticipated immune response to bacterial pathogens [
28,
29]. Severity scores—APACHE II, CURB-65, and NEWS—were all significantly higher in these patients, underscoring how secondary infections intensify physiological stress and contribute to more complicated clinical courses [
30].
Microbiological data confirm that
Klebsiella pneumoniae and
Pseudomonas aeruginosa are principal culprits in these secondary pneumonias, with Staphylococcus aureus also playing a notable role. The detection of multidrug-resistant strains in roughly one-third of isolates further underscores the difficulties in crafting effective empirical antibiotic regimens. These findings suggest that early and repeated respiratory sampling, such as BAL in high-risk cases, is instrumental for targeted therapy. Blood culture positivity, though less common, indicates higher severity and possible systemic spread [
31].
Clinically, superinfection strongly correlates with increased ICU admissions, mechanical ventilation, and longer hospital stays [
32]. Although mortality differences did not always reach statistical significance, the trend toward worse outcomes is evident. Taken together, the data imply that vigilant monitoring for bacterial superinfection is essential in COVID-19 management. Strategies such as robust antimicrobial stewardship, rapid identification of MDR organisms, and prompt de-escalation of antibiotics based on culture results could help balance the need for broad coverage with the risk of driving resistance.
Other similar studies investigated the prevalence and impact of infections in hospitalized COVID-19 patients. Garcia-Vidal et al. [
33] reported a low incidence of community-acquired coinfections at 3.1% and a 4.7% rate of hospital-acquired superinfections among 989 patients, with superinfections mainly due to
Pseudomonas aeruginosa and
Escherichia coli occurring approximately 10.6 days post-admission, and an overall mortality of 9.8%. In a similar vein, Scott et al. [
34] found a lower rate of bacterial infections at 8% among COVID-19 patients compared to 13% in pre-pandemic pneumonia patients, with most infections being nosocomial, and a higher mortality rate of 15% versus 9% in non-COVID-19 patients. This highlights the critical need for vigilant infection control and management in COVID-19 hospital settings, given the significant impact of nosocomial infections and the heightened mortality associated with such complications.
Yoon et al. [
35] identified bacterial superinfections in 30% of the 106 patients studied, with a median onset of 13 days post-ICU admission. This cohort displayed substantially worse outcomes, such as fewer ventilator-free days (median 0 days compared to 19 for those without superinfections) and prolonged ICU (32 days vs. 11 days) and hospital stays (39 days vs. 18 days), although ICU and in-hospital mortality rates did not differ significantly between groups. Similarly, Al-Ali et al. [
36] reported superinfections in 17.4% of their 230 patients, occurring approximately 17.6 days after starting antibiotics. These patients experienced longer ICU stays (27.1 days vs. 7.1 days), more complications (92.5% vs. 52.6%), and higher ICU mortality rates (45% vs. 22.1%). Both studies underline the severe challenges posed by superinfections in the management of critically ill COVID-19 patients, emphasizing the need for diligent antibiotic stewardship and monitoring to mitigate these risks and improve patient outcomes.
Søvik et al. [
37] found that 43% of their 155 mechanically ventilated patients developed superinfections, predominantly pneumonia, and these were significantly associated with dexamethasone treatment (66% vs. 32%,
p < 0.0001), autoimmune diseases, and longer ICU stays (26 vs. 17 days,
p < 0.001). They also noted a higher incidence of invasive fungal infections specifically in patients treated with dexamethasone. Despite these complications, 90-day survival rates did not differ significantly between patients with and without superinfections. In a similar manner, De Francesco et al. [
38] reported that 66.3% of the 713 SARS-CoV-2-positive ICU patients developed bacterial and/or fungal superinfections, a significantly higher rate compared to 30% in COVID-19-negative ICU patients. They also identified obesity, length of ICU stay, and duration of mechanical ventilation as risk factors for acquiring superinfections, which were associated with higher mortality rates (45.8% vs. 26.2%,
p < 0.0001).
Similarly, Moreno-Torres et al. [
39] found that 8.5% of 1594 hospitalized patients developed bacterial infections, with urinary tract infections, bacteremia, and pneumonia being the most common types. Independent predictors of these infections included older age, neurological disease, prior immunosuppression, and ICU admission. Although patients with bacterial infections experienced more severe ARDS and had a higher mortality rate (25% vs. 6.7%), bacterial infections themselves were not independently associated with increased mortality when other factors such as age and underlying conditions were considered. In a similar manner, Hughes et al. [
40] observed a low incidence of bacterial and fungal coinfections among 836 hospitalized patients during the initial COVID-19 wave in the UK, with only 3.2% showing early confirmed bacterial infections. Their study highlighted that significant bacterial or fungal infections, particularly in the early stages of hospitalization, were uncommon, suggesting that the baseline severity of COVID-19 rather than secondary infections primarily influenced patient outcomes. Both studies underscore the complexity of managing COVID-19 in the hospital setting, pointing to the need for careful consideration of antibiotic use and close monitoring of at-risk patient subgroups.
In their study, Nag and Kaur [
41] reported that up to half of COVID-19-related deaths could be linked to superinfections, with ventilator-associated pneumonia (VAP) being the most prevalent, followed by bacteremia and urinary tract infections. In a similar manner, Seitz et al. [
42] found that among 117 critically ill COVID-19 patients, 55% developed a superinfection, including a 65.2% rate of VAP, and 13.6% experienced fungal infections (5.9% candidemia and 7.7% COVID-19-associated pulmonary aspergillosis). Both studies underscored that patients with superinfections had prolonged hospital stays and higher mortality, particularly in cases involving multidrug-resistant pathogens or fungal complications. These findings pointed to the urgent need for enhanced surveillance, judicious use of antimicrobials, and further prospective research to guide effective stewardship strategies.
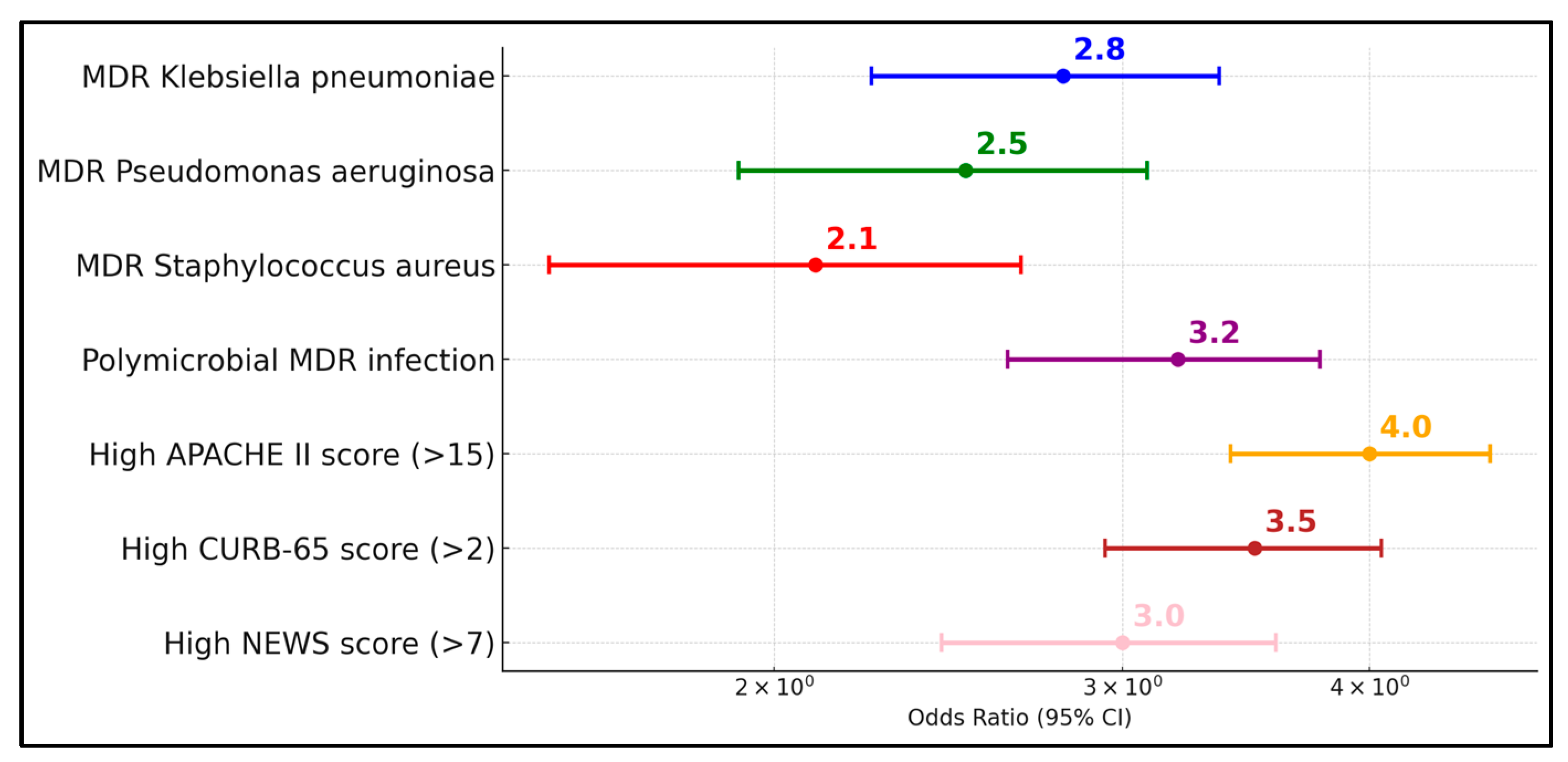
The observed high rates of resistance, including ESBL and fluoroquinolone resistance, necessitate urgent revisions in antimicrobial stewardship programs. Particularly concerning is the correlation between the time from symptom onset to hospital admission and the severity of infection, which suggests that delayed medical intervention may contribute to the advancement of these resistant infections. The high rates of MDR and the presence of ESBLs significantly influence treatment decisions, underscoring the urgent need for enhanced antimicrobial stewardship and targeted therapy. This study’s findings demonstrate a clear association between delayed hospital admission and increased severity of bacterial superinfections, suggesting that earlier intervention could mitigate the progression to severe outcomes.
Recent insights into AMR show an alarming increase in MDR and XDR pathogens, particularly in
Enterobacteriaceae,
Pseudomonas aeruginosa, and
Staphylococcus aureus. Genomic surveillance indicates that resistance genes are rapidly spreading via plasmids, threatening last-resort antibiotics like carbapenems and colistin. The COVID-19 pandemic has worsened AMR due to antibiotic overuse and disrupted monitoring, significantly increasing cases of COVID-19-associated bacterial pneumonia, especially with temperature > 38 °C, LDH > 250 U/L, and d-dimer > 1000 ng/mL [
42]. New mechanisms such as biofilm-mediated resistance complicate treatment further. Experts stress the need for rapid diagnostics, phage therapy, and antimicrobial stewardship to combat the growing threat of AMR, projected to cause 10 million deaths annually by 2050 if left unaddressed [
43,
44].
This study’s findings highlight critical implications for antimicrobial stewardship, underscoring the need for targeted antibiotic therapy based on local antibiogram data to combat the high prevalence of multidrug-resistant organisms like
Klebsiella pneumoniae and
Pseudomonas aeruginosa. Rapid diagnostic techniques, such as BAL, are essential for quickly identifying pathogens and tailoring antibiotic treatments, particularly in managing severe and polymicrobial infections. Additionally, the data support rigorous updates to hospital antimicrobial policies and enhanced educational programs for prescribers to optimize antibiotic use, reduce the spread of resistance, and improve patient outcomes in COVID-19 cases complicated by secondary bacterial infections [
41].
4.2. Study Limitations
Several limitations warrant discussion. First, our study is retrospective and single-center, restricting the generalizability of the findings to other populations with varied microbial landscapes and infection control practices. Second, microbiological testing was clinician-driven, meaning some patients with superinfection might have gone undetected if sampling was not performed. Third, while our cohort size of 141 patients provided meaningful insights, certain subgroup analyses (e.g., ICU outcomes, multidrug resistance patterns) had limited statistical power. Fourth, we did not detail specific treatment regimens, including antivirals, corticosteroids, and antibiotic choices, which may differ between physicians and impact patient trajectories. There is also an explicit limitation by excluding BAL-only diagnoses. Also, the mortality rate was low; therefore, it was not feasible to run a separate analysis for mortality risk. Finally, the observational design prevents causal inferences, although strong associations point to the deleterious effect of secondary bacterial infections in COVID-19. Prospective, multicenter studies with standardized protocols for sampling, antimicrobial therapy, and severity scoring are needed to substantiate these findings and guide best practices.