Transcriptome Analysis Revealed Potential Neuro-Immune Interaction in Papillary Thyroid Carcinoma Tissues
Abstract
1. Introduction
2. Methods and Materials
2.1. Patient and Tissue Collection
2.2. RNA Sequencing Analysis
2.3. Gene Set Enrichment Analysis (GSEA)
2.4. Gene Set Variation Analysis (GSVA)
3. Results
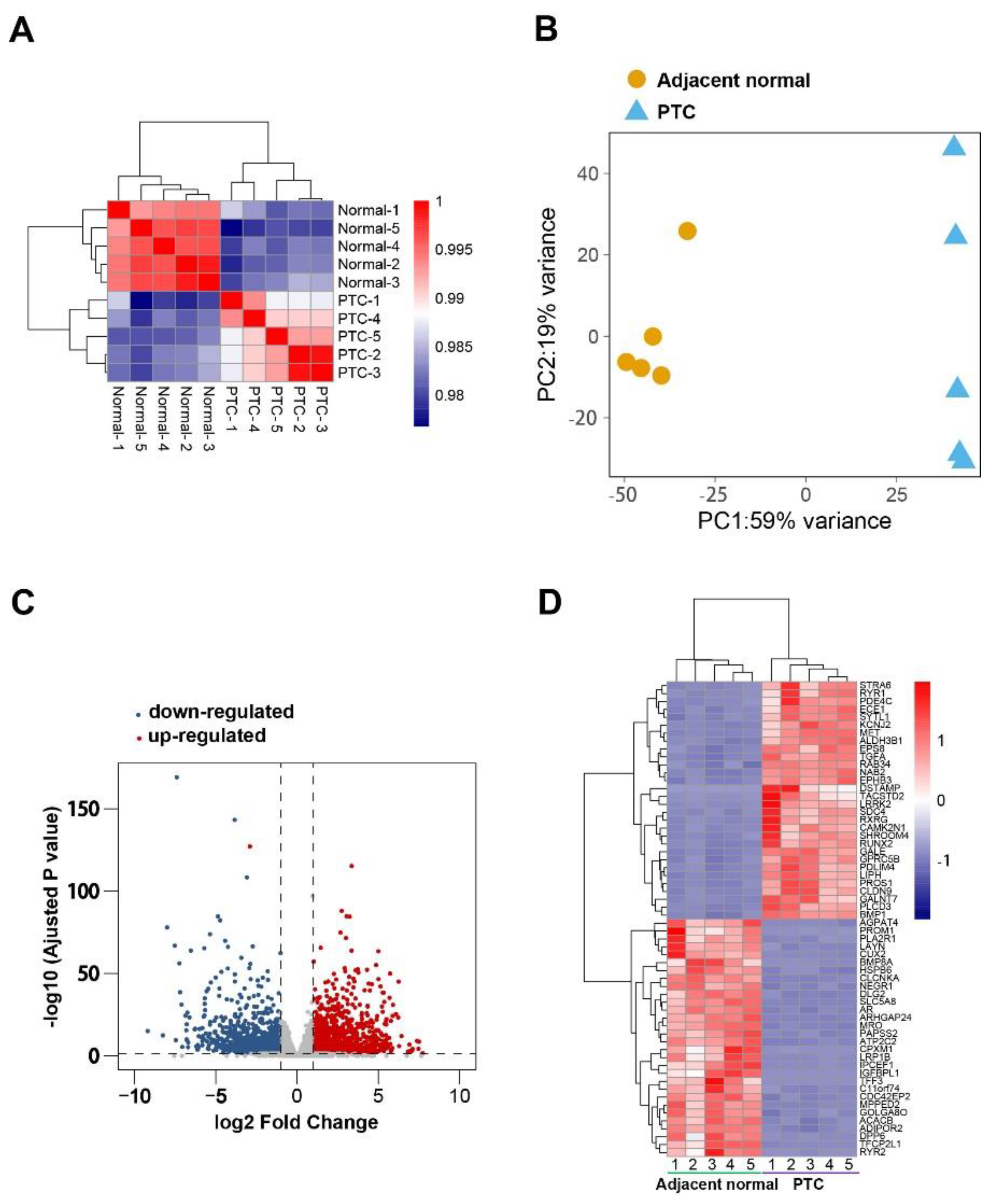
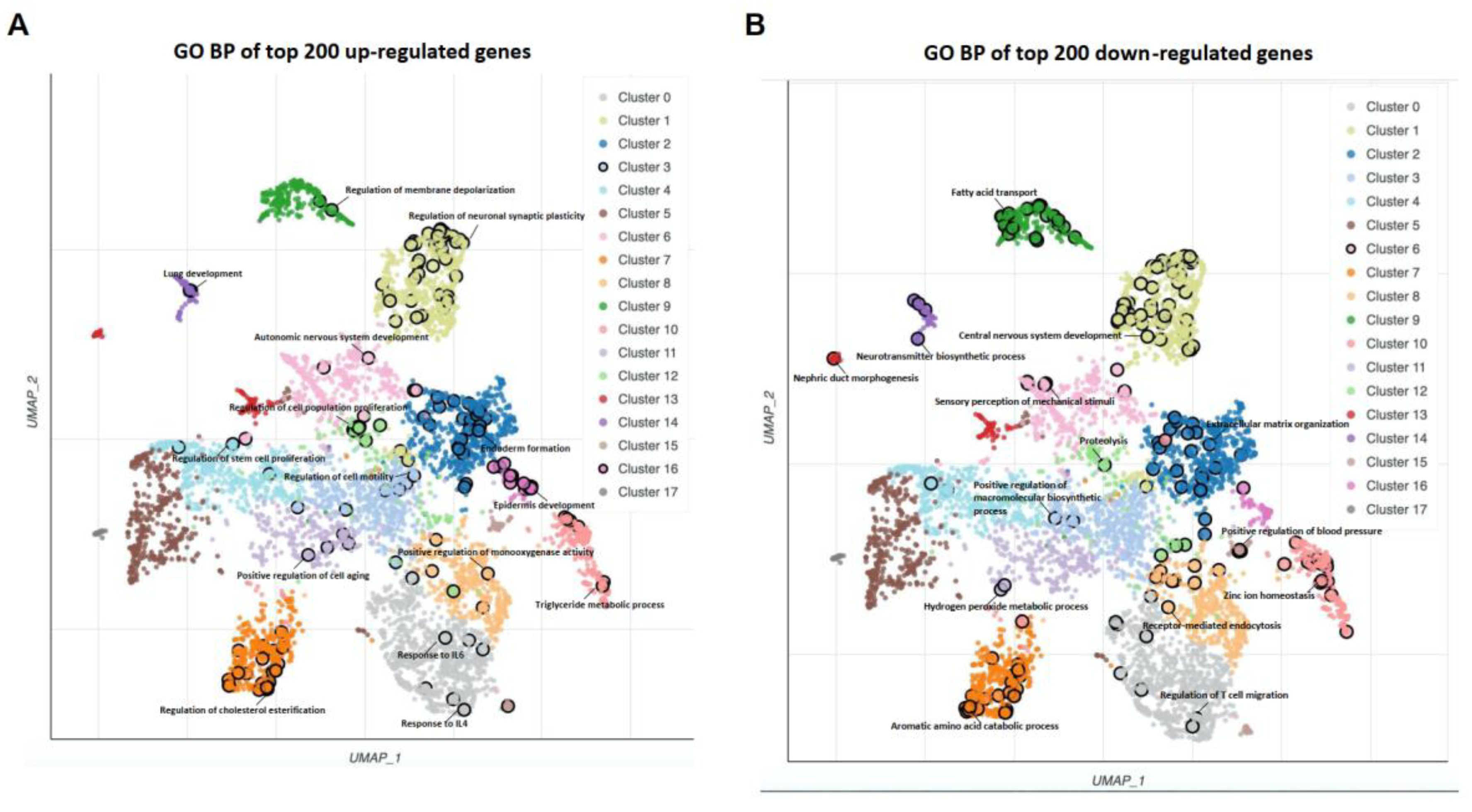
3.1. DEGs Identification and Gene Ontology Analysis
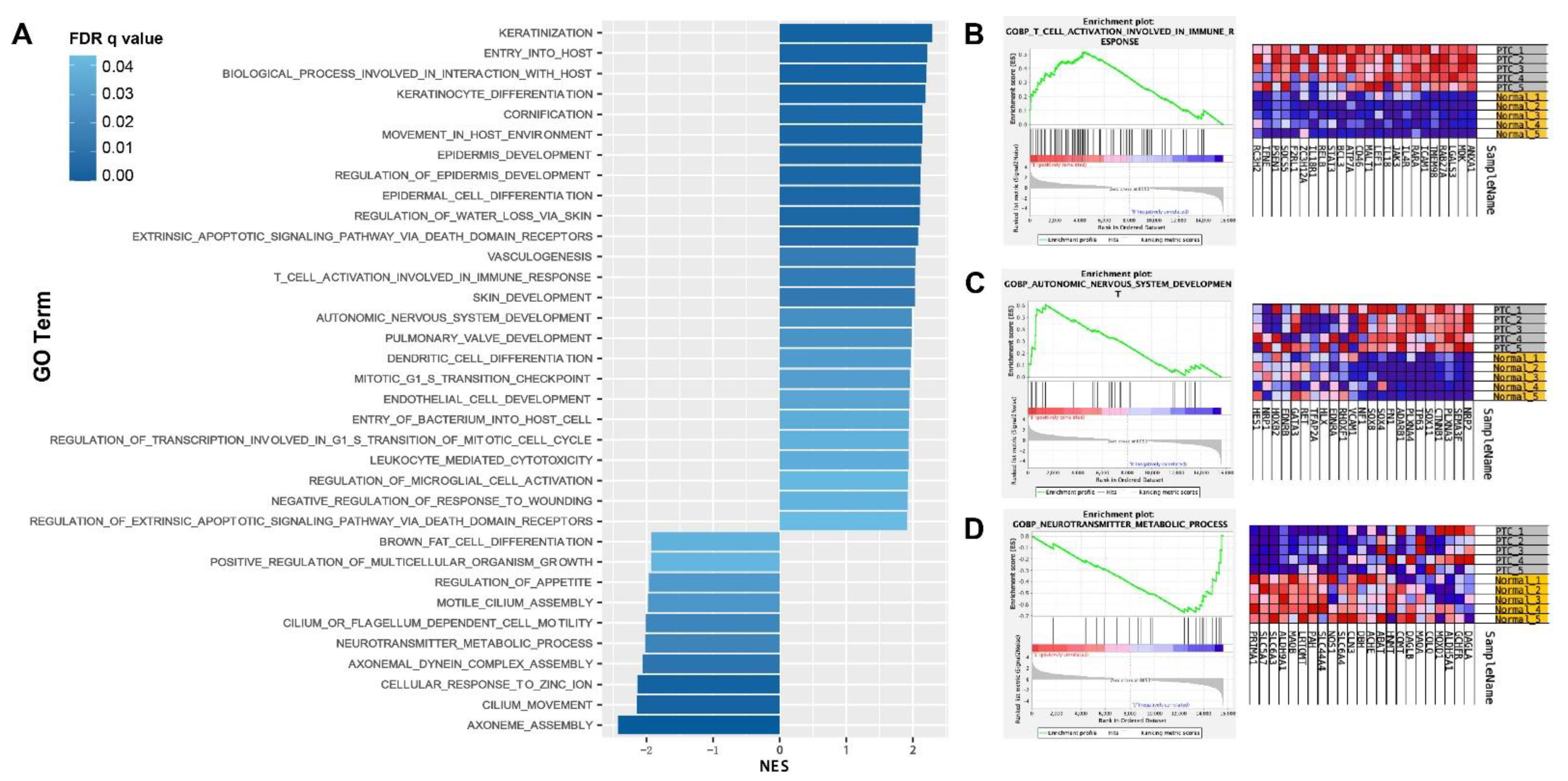
3.2. GSEA
3.3. GSVA of the Signatures of Tumor-Infiltrating Immune Cells and Tumor Microenvironment
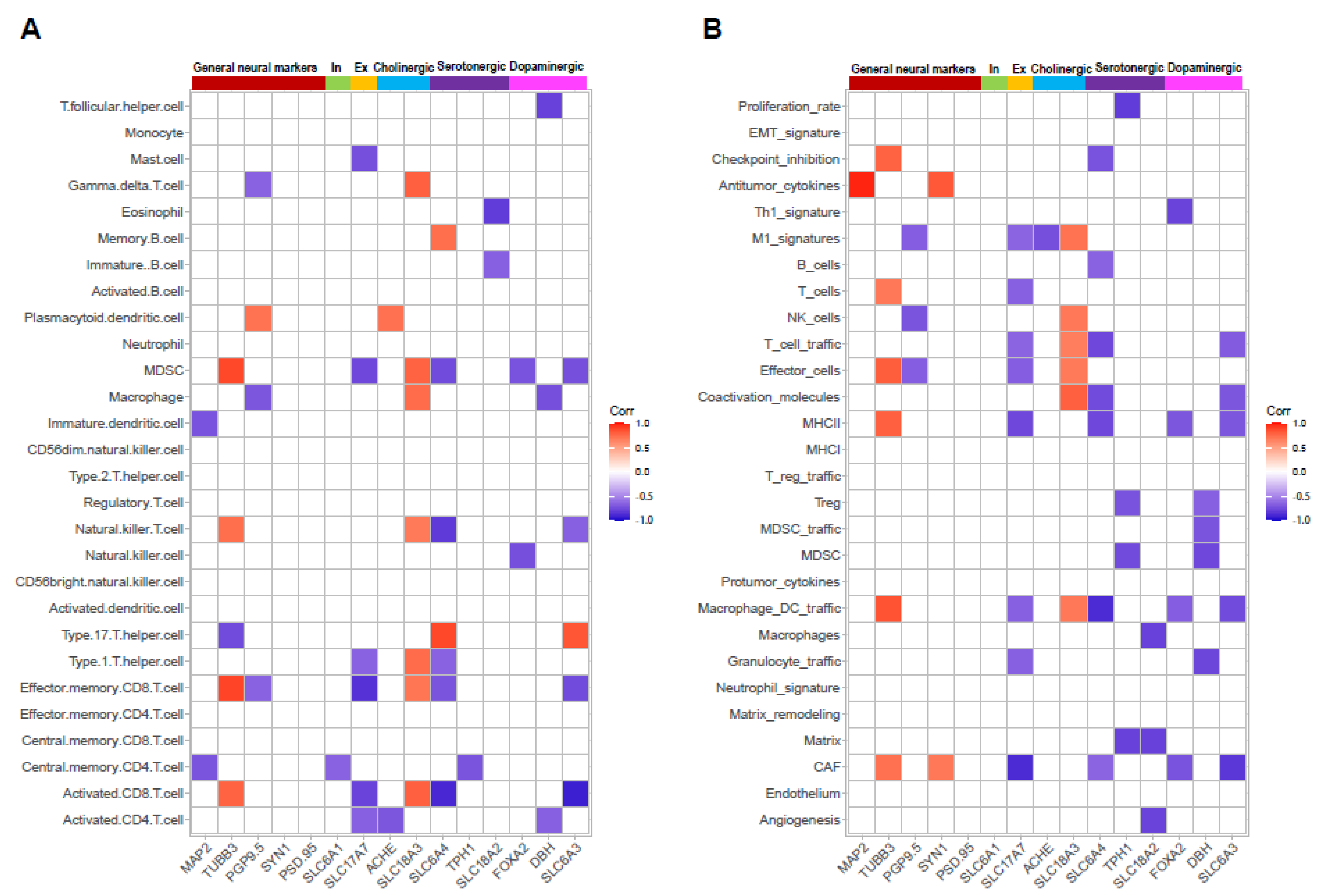
3.4. Correlation of Neuronal Markers and Immune Signatures
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Deng, Y.; Li, H.; Wang, M.; Li, N.; Tian, T.; Wu, Y.; Xu, P.; Yang, S.; Zhai, Z.; Zhou, L.; et al. Global Burden of Thyroid Cancer from 1990 to 2017. JAMA Netw. Open 2020, 3, e208759. [Google Scholar] [CrossRef] [PubMed]
- Thorsson, V.; Gibbs, D.L.; Brown, S.D.; Wolf, D.; Bortone, D.S.; Ou Yang, T.H.; Porta-Pardo, E.; Gao, G.F.; Plaisier, C.L.; Eddy, J.A.; et al. The Immune Landscape of Cancer. Immunity 2018, 48, 812–830. [Google Scholar] [CrossRef] [PubMed]
- Bao, X.; Shi, R.; Zhang, K.; Xin, S.; Li, X.; Zhao, Y.; Wang, Y. Immune Landscape of Invasive Ductal Carcinoma Tumor Microenvironment Identifies a Prognostic and Immunotherapeutically Relevant Gene Signature. Front. Oncol. 2019, 9, 903. [Google Scholar] [CrossRef] [PubMed]
- Bagaev, A.; Kotlov, N.; Nomie, K.; Svekolkin, V.; Gafurov, A.; Isaeva, O.; Osokin, N.; Kozlov, I.; Frenkel, F.; Gancharova, O.; et al. Conserved pan-cancer microenvironment subtypes predict response to immunotherapy. Cancer Cell 2021, 39, 845–865.e7. [Google Scholar] [CrossRef]
- Rowe, C.W.; Dill, T.; Griffin, N.; Jobling, P.; Faulkner, S.; Paul, J.W.; King, S.; Smith, R.; Hondermarck, H. Innervation of papillary thyroid cancer and its association with extra-thyroidal invasion. Sci. Rep. 2020, 10, 1539. [Google Scholar] [CrossRef]
- Cervantes-Villagrana, R.D.; Albores-Garcia, D.; Cervantes-Villagrana, A.R.; Garcia-Acevez, S.J. Tumor-induced neurogenesis and immune evasion as targets of innovative anti-cancer therapies. Signal Transduct. Target. Ther. 2020, 5, 99. [Google Scholar] [CrossRef]
- Gysler, S.M.; Drapkin, R. Tumor innervation: Peripheral nerves take control of the tumor microenvironment. J. Clin. Investig. 2021, 131, e147276. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Xie, Z.; Bailey, A.; Kuleshov, M.V.; Clarke, D.J.B.; Evangelista, J.E.; Jenkins, S.L.; Lachmann, A.; Wojciechowicz, M.L.; Kropiwnicki, E.; Jagodnik, K.M.; et al. Gene Set Knowledge Discovery with Enrichr. Curr. Protoc. 2021, 1, e90. [Google Scholar] [CrossRef] [PubMed]
- Traag, V.A.; Waltman, L.; van Eck, N.J. From Louvain to Leiden: Guaranteeing well-connected communities. Sci. Rep. 2019, 9, 5233. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef] [PubMed]
- Charoentong, P.; Finotello, F.; Angelova, M.; Mayer, C.; Efremova, M.; Rieder, D.; Hackl, H.; Trajanoski, Z. Pan-cancer Immunogenomic Analyses Reveal Genotype-Immunophenotype Relationships and Predictors of Response to Checkpoint Blockade. Cell Rep. 2017, 18, 248–262. [Google Scholar] [CrossRef] [PubMed]
- Ahmadi, A.; Balood, M.; Roversi, K.; Ahmadi, M.; Rafei, M.; Talbot, S. Teasing Out the Interplay Between Natural Killer Cells and Nociceptor Neurons. J. Vis. Exp. JoVE 2022, 30, e63800. [Google Scholar] [CrossRef]
- Regan, J.L.; Schumacher, D.; Staudte, S.; Steffen, A.; Lesche, R.; Toedling, J.; Jourdan, T.; Haybaeck, J.; Golob-Schwarzl, N.; Mumberg, D.; et al. Identification of a neural development gene expression signature in colon cancer stem cells reveals a role for EGR2 in tumorigenesis. iScience 2022, 25, 104498. [Google Scholar] [CrossRef]
- Xie, Z.; Li, X.; He, Y.; Wu, S.; Wang, S.; Sun, J.; He, Y.; Lun, Y.; Zhang, J. Immune Cell Confrontation in the Papillary Thyroid Carcinoma Microenvironment. Front. Endocrinol. 2020, 11, 570604. [Google Scholar] [CrossRef]
- Na, K.J.; Choi, H. Immune landscape of papillary thyroid cancer and immunotherapeutic implications. Endocr.-Relat. Cancer 2018, 25, 523–531. [Google Scholar] [CrossRef]
- Yang, Z.; Wei, X.; Pan, Y.; Xu, J.; Si, Y.; Min, Z.; Yu, B. A new risk factor indicator for papillary thyroid cancer based on immune infiltration. Cell Death Dis. 2021, 12, 51. [Google Scholar] [CrossRef]
- Cunha, L.L.; Morari, E.C.; Guihen, A.C.; Razolli, D.; Gerhard, R.; Nonogaki, S.; Soares, F.A.; Vassallo, J.; Ward, L.S. Infiltration of a mixture of immune cells may be related to good prognosis in patients with differentiated thyroid carcinoma. Clin. Endocrinol. 2012, 77, 918–925. [Google Scholar] [CrossRef] [PubMed]
- Xiang, Z.; Zhong, C.; Chang, A.; Ling, J.; Zhao, H.; Zhou, W.; Zhuo, X. Immune-related key gene CLDN10 correlates with lymph node metastasis but predicts favorable prognosis in papillary thyroid carcinoma. Aging 2020, 12, 2825–2839. [Google Scholar] [CrossRef] [PubMed]
- Krystel-Whittemore, M.; Dileepan, K.N.; Wood, J.G. Mast Cell: A Multi-Functional Master Cell. Front. Immunol. 2015, 6, 620. [Google Scholar] [CrossRef] [PubMed]
- Visciano, C.; Prevete, N.; Liotti, F.; Marone, G. Tumor-Associated Mast Cells in Thyroid Cancer. Int. J. Endocrinol. 2015, 2015, 705169. [Google Scholar] [CrossRef] [PubMed]
- Ferrari, S.M.; Fallahi, P.; Galdiero, M.R.; Ruffilli, I.; Elia, G.; Ragusa, F.; Paparo, S.R.; Patrizio, A.; Mazzi, V.; Varricchi, G.; et al. Immune and Inflammatory Cells in Thyroid Cancer Microenvironment. Int. J. Mol. Sci. 2019, 20, 4413. [Google Scholar] [CrossRef]
- Melillo, R.M.; Guarino, V.; Avilla, E.; Galdiero, M.R.; Liotti, F.; Prevete, N.; Rossi, F.W.; Basolo, F.; Ugolini, C.; de Paulis, A.; et al. Mast cells have a protumorigenic role in human thyroid cancer. Oncogene 2010, 29, 6203–6215. [Google Scholar] [CrossRef]
- Wen, S.; Qu, N.; Ma, B.; Wang, X.; Luo, Y.; Xu, W.; Jiang, H.; Zhang, Y.; Wang, Y.; Ji, Q. Cancer-Associated Fibroblasts Positively Correlate with Dedifferentiation and Aggressiveness of Thyroid Cancer. OncoTargets Ther. 2021, 14, 1205–1217. [Google Scholar] [CrossRef]
- Minna, E.; Brich, S.; Todoerti, K.; Pilotti, S.; Collini, P.; Bonaldi, E.; Romeo, P.; Cecco, L.; Dugo, M.; Perrone, F.; et al. Cancer Associated Fibroblasts and Senescent Thyroid Cells in the Invasive Front of Thyroid Carcinoma. Cancers 2020, 12, 112. [Google Scholar] [CrossRef]
- Zimring, J.C.; Kapp, L.M.; Yamada, M.; Wess, J.; Kapp, J.A. Regulation of CD8+ cytolytic T lymphocyte differentiation by a cholinergic pathway. J. Neuroimmunol. 2005, 164, 66–75. [Google Scholar] [CrossRef]
- Fujii, T.; Mashimo, M.; Moriwaki, Y.; Misawa, H.; Ono, S.; Horiguchi, K.; Kawashima, K. Expression and Function of the Cholinergic System in Immune Cells. Front. Immunol. 2017, 8, 1085. [Google Scholar] [CrossRef]
- Qiao, G.; Bucsek, M.J.; Winder, N.M.; Chen, M.; Giridharan, T.; Olejniczak, S.H.; Hylander, B.L.; Repasky, E.A. beta-Adrenergic signaling blocks murine CD8(+) T-cell metabolic reprogramming during activation: A mechanism for immunosuppression by adrenergic stress. Cancer Immunol. Immunother. CII 2019, 68, 11–22. [Google Scholar] [CrossRef] [PubMed]
- Estrada, L.D.; Agac, D.; Farrar, J.D. Sympathetic neural signaling via the beta2-adrenergic receptor suppresses T-cell receptor-mediated human and mouse CD8(+) T-cell effector function. Eur. J. Immunol. 2016, 46, 1948–1958. [Google Scholar] [CrossRef] [PubMed]

| Sample ID | Gender | Age | TNM Staging | Clinical Stage |
|---|---|---|---|---|
| 1 | Female | 39 | T1bN0M0 | I |
| 2 | Female | 22 | T2N1aM0 | I |
| 3 | Male | 55 | T1aN1aM0 | III |
| 4 | Female | 24 | T1bN1aM0 | I |
| 5 | Male | 43 | T1bN1aM0 | I |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hu, H.; Chen, Q.; Zheng, S.; Du, S.; Ding, S.; Lun, Y. Transcriptome Analysis Revealed Potential Neuro-Immune Interaction in Papillary Thyroid Carcinoma Tissues. Diseases 2023, 11, 9. https://doi.org/10.3390/diseases11010009
Hu H, Chen Q, Zheng S, Du S, Ding S, Lun Y. Transcriptome Analysis Revealed Potential Neuro-Immune Interaction in Papillary Thyroid Carcinoma Tissues. Diseases. 2023; 11(1):9. https://doi.org/10.3390/diseases11010009
Chicago/Turabian StyleHu, Haibei, Qiang Chen, Siqi Zheng, Shan Du, Siqin Ding, and Yongzhi Lun. 2023. "Transcriptome Analysis Revealed Potential Neuro-Immune Interaction in Papillary Thyroid Carcinoma Tissues" Diseases 11, no. 1: 9. https://doi.org/10.3390/diseases11010009
APA StyleHu, H., Chen, Q., Zheng, S., Du, S., Ding, S., & Lun, Y. (2023). Transcriptome Analysis Revealed Potential Neuro-Immune Interaction in Papillary Thyroid Carcinoma Tissues. Diseases, 11(1), 9. https://doi.org/10.3390/diseases11010009





