Blended Multi-Modal Deep ConvNet Features for Diabetic Retinopathy Severity Prediction
Abstract
1. Introduction
- Effectiveness of the uni-modal feature representation is verified.
- A blended multi-modal feature representation approach is introduced
- Different pool-based approaches are proposed to blend deep features.
- A DNN architecture with dropout at the input layer is proposed to test the efficiency of the proposed uni-modal and blended multi-modal feature representations.
- APTOS 2019 benchmark dataset was used to compare the performance of the proposed approach with existing models.
2. Related Work
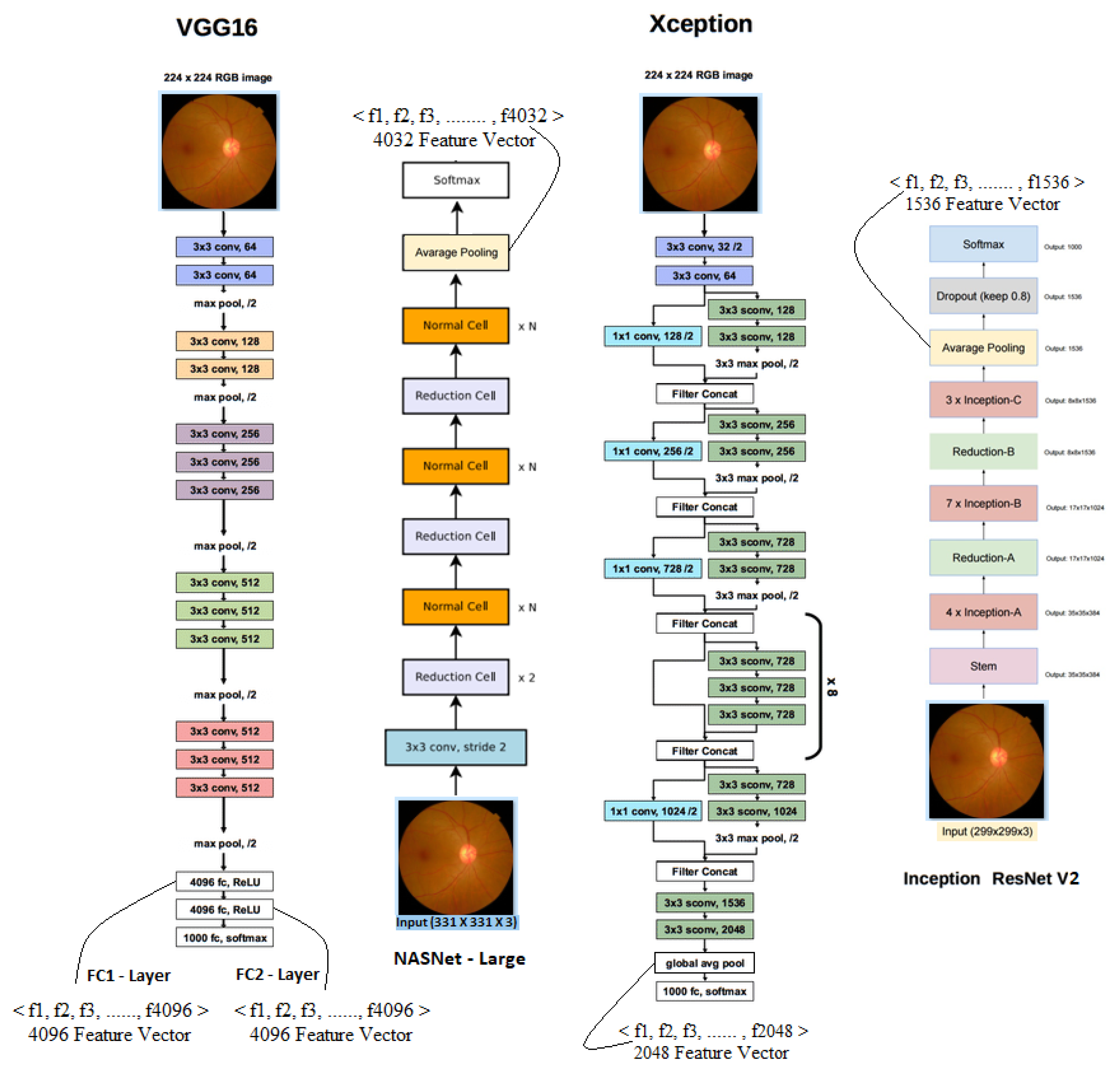
Deep ConvNets for Feature Extraction and Transfer Learning
- Visual Geometric Group (VGG 16): VGG16 is a deep ConvNet trained on 14 million images belonging to 1000 different classes and topped the leader board in ILSVR (ImageNet) challenge. In this architecture, 2 × 2 filters are used with stride 1 for convolution operation, and 2 × 2 filters with stride two and the same padding are used for max-pooling operation across the network. At the end of architecture, two fully connected dense layers of 4096 neurons are connected followed by soft-max layer.
- Neural Architecture Search Network (NASNet): This is a special kind of deep CNN which searches for a better architectural building block on small datasets such as CIFAR10 and transfers it to larger datasets such as ImageNet. It has a better regularization mechanism called scheduled drop path, which significantly improves generalization.
- Xception: Xception is another deep ConvNet architecture that supports depth-wise separable convolution operations and outperformed ResNet and InceptionV3 in the ILSVR challenge.
- Inception ResNetV2: This is popularly known as InceptionV4, as it combines two different architectures: InceptionV3 and ResNet152. It has both inception and residual connections, which boost the performance of the model.
3. Proposed Methodology
3.1. Feature Extraction
- Each of the first (fc1) and second (fc2) fully connected layers of VGG16 produces a feature vector of 4096 dimensions.
- The final global average pooling layer of NASNet, Xception, and InceptionResNetV2 gives feature vectors of size 4032, 2048, and 1536, respectively.
3.2. Uni-Modal Deep Feature Extraction
3.3. Blended (Multi-Modal) Deep Feature Extraction
Approaches to Blend Deep Features from Multiple ConvNets
| Algorithm 1: Blended multi-modal deep feature fusion based DR recognition task. |
| Input: Let and be the train and test datasets of fundus images, respectively, where and . represents ith color fundus image in the dataset and is the severity level of DR associated with . In the case of DR identification task, , whereas, in the case of DR severity classification, task . Output: for each Step 1: Preprocess each image in the dataset. Step 2: Feature Extraction For each preprocessed image three different features () are extracted. Features extracted from fc1 layer of VGG16 Features extracted from fc2 layer of VGG16 Features extracted from global avg pool layer of Xception Where and Step 3: Deep feature fusion Apply feature feature fusion on the deep features extracted from each image (1D max pooling) (1D max pooling) (Average Cross pooling) (Average Cross pooling) blended feature vector corresponding to Step 4: Model Training Training dataset is prepared using the blended features Train a deep neural network (DNN) using Step 5: Model evaluation Test dataset is prepared using the blended features Evaluate the performance of using the DNN trained in Step 4 |
3.4. Model Training and Evaluation
4. Experimental Results
4.1. Dataset Summary
4.2. Performance Measures
4.3. DR Identification and Severity Level Prediction
4.3.1. Task 1—DR Identification
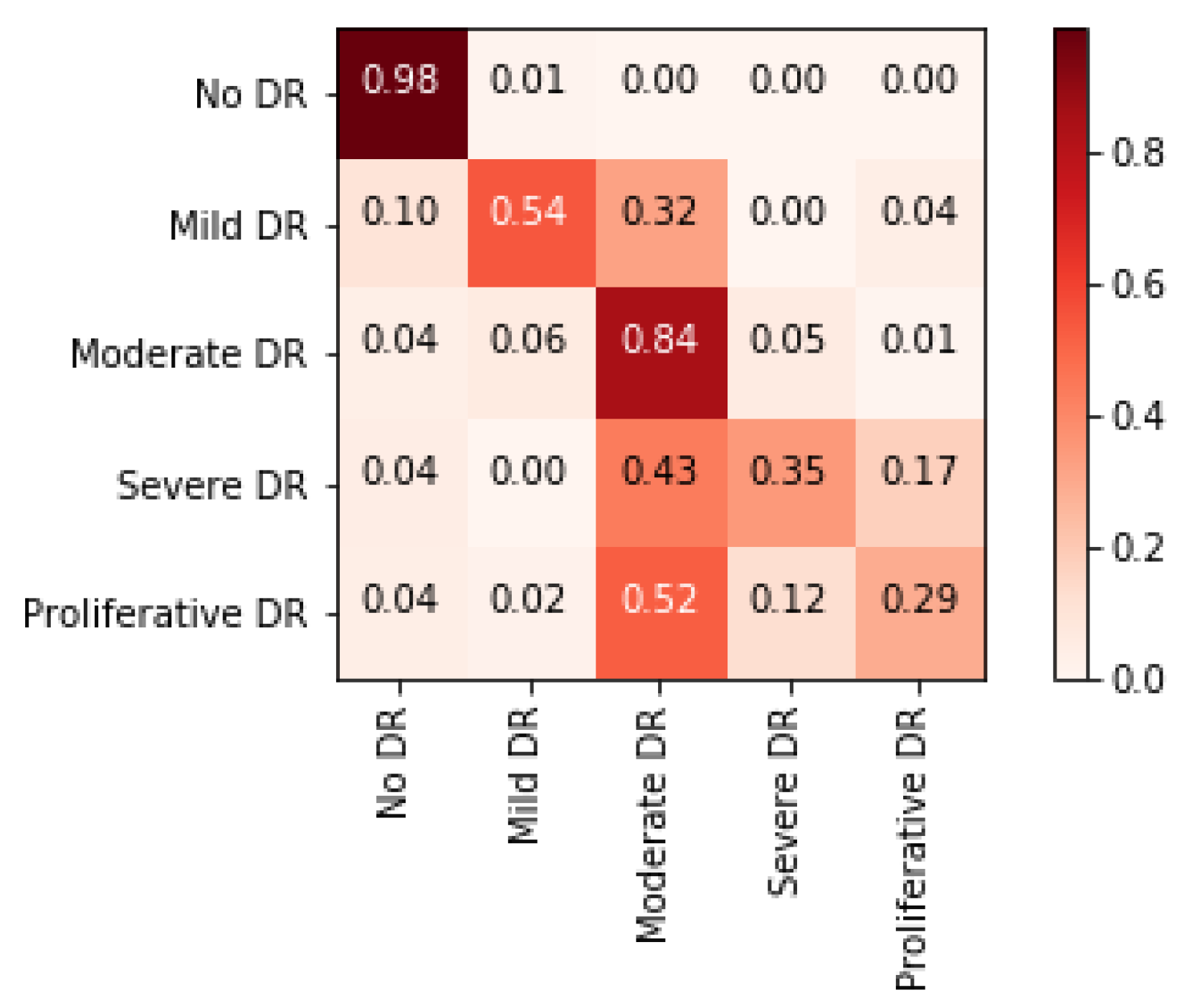
4.3.2. Task 2—Severity Level Prediction
4.4. Experimental Studies to Show the Representative Nature of Uni-Modal Features for Task 1
4.5. Experimental Studies to Show the Representative Nature of Uni-Modal Features for Task 2
4.6. Performance Evaluation of the Proposed Blended Multi-Modal Features
4.6.1. Blended Multi-Modal Deep Features for Task 1
4.6.2. Blended Multi-Modal Deep Features for Task 2
4.7. Comparison of Proposed Blended Feature Extraction with Existing Methods
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Cheung, N.; Rogers, S.L.; Donaghue, K.C.; Jenkins, A.J.; Tikellis, G.; Wong, T.Y. Retinal arteriolar dilation predicts retinopathy in adolescents with type 1 diabetes. Diabetes Care 2008, 31, 1842–1846. [Google Scholar] [CrossRef] [PubMed]
- Flaxman, S.; Bourne, R.; Resnikoff, S.; Ackland, P.; Braithwaite, T.; Cicinelli, M.; Das, A.; Jonas, J.; Keeffe, J.; Kempen, J.; et al. Global causes of blindness and distance vision impairment 1990-2020: A systematic review and meta-analysis. Lancet Glob. Health 2017, 5, e1221–e1234. [Google Scholar] [CrossRef]
- Gulshan, V.; Peng, L.; Coram, M.; Stumpe, M.C.; Wu, D.; Narayanaswamy, A.; Venugopalan, S.; Widner, K.; Madams, T.; Cuadros, J.; et al. Development and validation of a deep learning algorithm for detection of diabetic retinopathy in retinal fundus photographs. JAMA 2016, 316, 2402–2410. [Google Scholar] [CrossRef] [PubMed]
- Williams, R.; Airey, M.; Baxter, H.; Forrester, J.M.; Kennedy-Martin, T.; Girach, A. Epidemiology of diabetic retinopathy and macular oedema: A systematic review. Eye 2004, 18, 963–983. [Google Scholar] [CrossRef] [PubMed]
- Long, S.; Huang, X.; Chen, Z.; Pardhan, S.; Zheng, D. Automatic detection of hard exudates in color retinal images using dynamic threshold and SVM classification: Algorithm development and evaluation. BioMed Res. Int. 2019, 2019, 3926930. [Google Scholar] [CrossRef]
- Haloi, M.; Dandapat, S.; Sinha, R. A Gaussian scale space approach for exudates detection, classification and severity prediction. arXiv 2015, arXiv:1505.00737. [Google Scholar]
- Noushin, E.; Pourreza, M.; Masoudi, K.; Ghiasi Shirazi, E. Microaneurysm detection in fundus images using a two step convolution neural network. Biomed. Eng. Online 2019, 18, 67. [Google Scholar]
- Grinsven, M.; Ginneken, B.; Hoyng, C.; Theelen, T.; Sanchez, C. Fast convolution neural network training using selective data sampling. IEEE Trans. Med. Imaging 2016, 35, 1273–1284. [Google Scholar] [CrossRef]
- Haloi, M. Improved microaneurysm detection using deep neural networks. arXiv 2015, arXiv:1505.04424. [Google Scholar]
- Srivastava, R.; Duan, L.; Wong, D.W.; Liu, J.; Wong, T.Y. Detecting retinal microaneurysms and hemorrhages with robustness to the presence of blood vessels. Comput. Methods Programs Biomed. 2017, 138, 83–91. [Google Scholar] [CrossRef]
- Bodapati, J.D.; Veeranjaneyulu, N. Feature Extraction and Classification Using Deep Convolutional Neural Networks. J. Cyber Secur. Mobil. 2019, 8, 261–276. [Google Scholar] [CrossRef]
- Bodapati, J.D.; Veeranjaneyulu, N.; Shaik, S. Sentiment Analysis from Movie Reviews Using LSTMs. Ingénierie des Systèmes d’Information 2019, 24, 125–129. [Google Scholar] [CrossRef]
- Zhuo, P.; Zhu, Y.; Wu, W.; Shu, J.; Xia, T. Real-Time Fault Diagnosis for Gas Turbine Blade Based on Output-Hidden Feedback Elman Neural Network. J. Shanghai Jiaotong Univ. (Sci.) 2018, 23, 95–102. [Google Scholar] [CrossRef]
- Xia, T.; Song, Y.; Zheng, Y.; Pan, E.; Xi, L. An ensemble framework based on convolutional bi-directional LSTM with multiple time windows for remaining useful life estimation. Comput. Ind. 2020, 115, 103182. [Google Scholar] [CrossRef]
- Moreira, M.W.; Rodrigues, J.J.; Korotaev, V.; Al-Muhtadi, J.; Kumar, N. A comprehensive review on smart decision support systems for health care. IEEE Syst. J. 2019, 13, 3536–3545. [Google Scholar] [CrossRef]
- Gadekallu, T.R.; Khare, N.; Bhattacharya, S.; Singh, S.; Maddikunta, P.K.R.; Srivastava, G. Deep neural networks to predict diabetic retinopathy. J. Ambient Intell. Humaniz. Comput. 2020. [Google Scholar] [CrossRef]
- Patel, H.; Singh Rajput, D.; Thippa Reddy, G.; Iwendi, C.; Kashif Bashir, A.; Jo, O. A review on classification of imbalanced data for wireless sensor networks. Int. J. Distrib. Sens. Netw. 2020, 16, 1550147720916404. [Google Scholar] [CrossRef]
- Reddy, G.T.; Reddy, M.P.K.; Lakshmanna, K.; Rajput, D.S.; Kaluri, R.; Srivastava, G. Hybrid genetic algorithm and a fuzzy logic classifier for heart disease diagnosis. Evol. Intell. 2019, 13, 185–196. [Google Scholar] [CrossRef]
- Wu, L.; Fernandez-Loaiza, P.; Sauma, J.; Hernandez-Bogantes, E.; Masis, M. Classification of diabetic retinopathy and diabetic macular edema. World J. Diabetes 2013, 4, 290. [Google Scholar] [CrossRef]
- Akram, M.U.; Khalid, S.; Khan, S.A. Identification and classification of microaneurysms for early detection of diabetic retinopathy. Pattern Recognit. 2013, 46, 107–116. [Google Scholar] [CrossRef]
- Akram, M.U.; Khalid, S.; Tariq, A.; Khan, S.A.; Azam, F. Detection and classification of retinal lesions for grading of diabetic retinopathy. Comput. Biol. Med. 2014, 45, 161–171. [Google Scholar] [CrossRef] [PubMed]
- Casanova, R.; Saldana, S.; Chew, E.Y.; Danis, R.P.; Greven, C.M.; Ambrosius, W.T. Application of random forests methods to diabetic retinopathy classification analyses. PLoS ONE 2014, 9, e98587. [Google Scholar] [CrossRef] [PubMed]
- Verma, K.; Deep, P.; Ramakrishnan, A. Detection and classification of diabetic retinopathy using retinal images. In Proceedings of the 2011 Annual IEEE India Conference, Hyderabad, India, 16–18 December 2011; pp. 1–6. [Google Scholar]
- Welikala, R.; Dehmeshki, J.; Hoppe, A.; Tah, V.; Mann, S.; Williamson, T.H.; Barman, S. Automated detection of proliferative diabetic retinopathy using a modified line operator and dual classification. Comput. Methods Programs Biomed. 2014, 114, 247–261. [Google Scholar] [CrossRef] [PubMed]
- Welikala, R.A.; Fraz, M.M.; Dehmeshki, J.; Hoppe, A.; Tah, V.; Mann, S.; Williamson, T.H.; Barman, S.A. Genetic algorithm based feature selection combined with dual classification for the automated detection of proliferative diabetic retinopathy. Comput. Med. Imaging Graph. 2015, 43, 64–77. [Google Scholar] [CrossRef]
- Roychowdhury, S.; Koozekanani, D.D.; Parhi, K.K. DREAM: Diabetic retinopathy analysis using machine learning. IEEE J. Biomed. Health Inform. 2013, 18, 1717–1728. [Google Scholar] [CrossRef]
- Mookiah, M.R.K.; Acharya, U.R.; Martis, R.J.; Chua, C.K.; Lim, C.M.; Ng, E.; Laude, A. Evolutionary algorithm based classifier parameter tuning for automatic diabetic retinopathy grading: A hybrid feature extraction approach. Knowl. Based Syst. 2013, 39, 9–22. [Google Scholar] [CrossRef]
- Porter, L.F.; Saptarshi, N.; Fang, Y.; Rathi, S.; Den Hollander, A.I.; De Jong, E.K.; Clark, S.J.; Bishop, P.N.; Olsen, T.W.; Liloglou, T.; et al. Whole-genome methylation profiling of the retinal pigment epithelium of individuals with age-related macular degeneration reveals differential methylation of the SKI, GTF2H4, and TNXB genes. Clin. Epigenet. 2019, 11, 6. [Google Scholar] [CrossRef]
- Rahim, S.S.; Jayne, C.; Palade, V.; Shuttleworth, J. Automatic detection of microaneurysms in colour fundus images for diabetic retinopathy screening. Neural Comput. Appl. 2016, 27, 1149–1164. [Google Scholar] [CrossRef]
- Dutta, S.; Manideep, B.; Basha, S.M.; Caytiles, R.D.; Iyengar, N. Classification of diabetic retinopathy images by using deep learning models. Int. J. Grid Distrib. Comput. 2018, 11, 89–106. [Google Scholar] [CrossRef]
- Zeng, X.; Chen, H.; Luo, Y.; Ye, W. Automated diabetic retinopathy detection based on binocular Siamese-like convolutional neural network. IEEE Access 2019, 7, 30744–30753. [Google Scholar] [CrossRef]
- Mateen, M.; Wen, J.; Song, S.; Huang, Z. Fundus image classification using VGG-19 architecture with PCA and SVD. Symmetry 2019, 11, 1. [Google Scholar] [CrossRef]
- Reddy, G.T.; Reddy, M.P.K.; Lakshmanna, K.; Kaluri, R.; Rajput, D.S.; Srivastava, G.; Baker, T. Analysis of Dimensionality Reduction Techniques on Big Data. IEEE Access 2020, 8, 54776–54788. [Google Scholar] [CrossRef]
- Bhattacharya, S.; Kaluri, R.; Singh, S.; Alazab, M.; Tariq, U. A Novel PCA-Firefly based XGBoost classification model for Intrusion Detection in Networks using GPU. Electronics 2020, 9, 219. [Google Scholar] [CrossRef]
- Gadekallu, T.R.; Khare, N.; Bhattacharya, S.; Singh, S.; Reddy Maddikunta, P.K.; Ra, I.H.; Alazab, M. Early Detection of Diabetic Retinopathy Using PCA-Firefly Based Deep Learning Model. Electronics 2020, 9, 274. [Google Scholar] [CrossRef]
- Jindal, A.; Aujla, G.S.; Kumar, N.; Prodan, R.; Obaidat, M.S. DRUMS: Demand response management in a smart city using deep learning and SVR. In Proceedings of the 2018 IEEE Global Communications Conference (GLOBECOM), Abu Dhabi, UAE, 9–13 December 2018; pp. 1–6. [Google Scholar]
- Vinayakumar, R.; Alazab, M.; Srinivasan, S.; Pham, Q.V.; Padannayil, S.K.; Simran, K. A Visualized Botnet Detection System based Deep Learning for the Internet of Things Networks of Smart Cities. IEEE Trans. Ind. Appl. 2020. [Google Scholar] [CrossRef]
- Alazab, M.; Khan, S.; Krishnan, S.S.R.; Pham, Q.V.; Reddy, M.P.K.; Gadekallu, T.R. A Multidirectional LSTM Model for Predicting the Stability of a Smart Grid. IEEE Access 2020, 8, 85454–85463. [Google Scholar] [CrossRef]
- Simonyan, K.; Zisserman, A. Very deep convolutional networks for large-scale image recognition. arXiv 2014, arXiv:1409.1556. [Google Scholar]
- Chollet, F. Xception: Deep learning with depthwise separable convolutions. In Proceedings of the 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017; pp. 1251–1258. [Google Scholar]
- Zoph, B.; Vasudevan, V.; Shlens, J.; Le, Q.V. Learning transferable architectures for scalable image recognition. In Proceedings of the 2018 IEEE/CVF Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018; pp. 8697–8710. [Google Scholar]
- APTOS 2019. Available online: https://www.kaggle.com/c/aptos2019-blindness-detection (accessed on 30 December 2019).
- Gargeya, R.; Leng, T. Automated identification of diabetic retinopathy using deep learning. Ophthalmology 2017, 124, 962–969. [Google Scholar] [CrossRef]
- Kassani, S.H.; Kassani, P.H.; Khazaeinezhad, R.; Wesolowski, M.J.; Schneider, K.A.; Deters, R. Diabetic Retinopathy Classification Using a Modified Xception Architecture. In Proceedings of the 2019 IEEE International Symposium on Signal Processing and Information Technology (ISSPIT), Ajman, UAE, 10–12 December 2019; pp. 1–6. [Google Scholar]
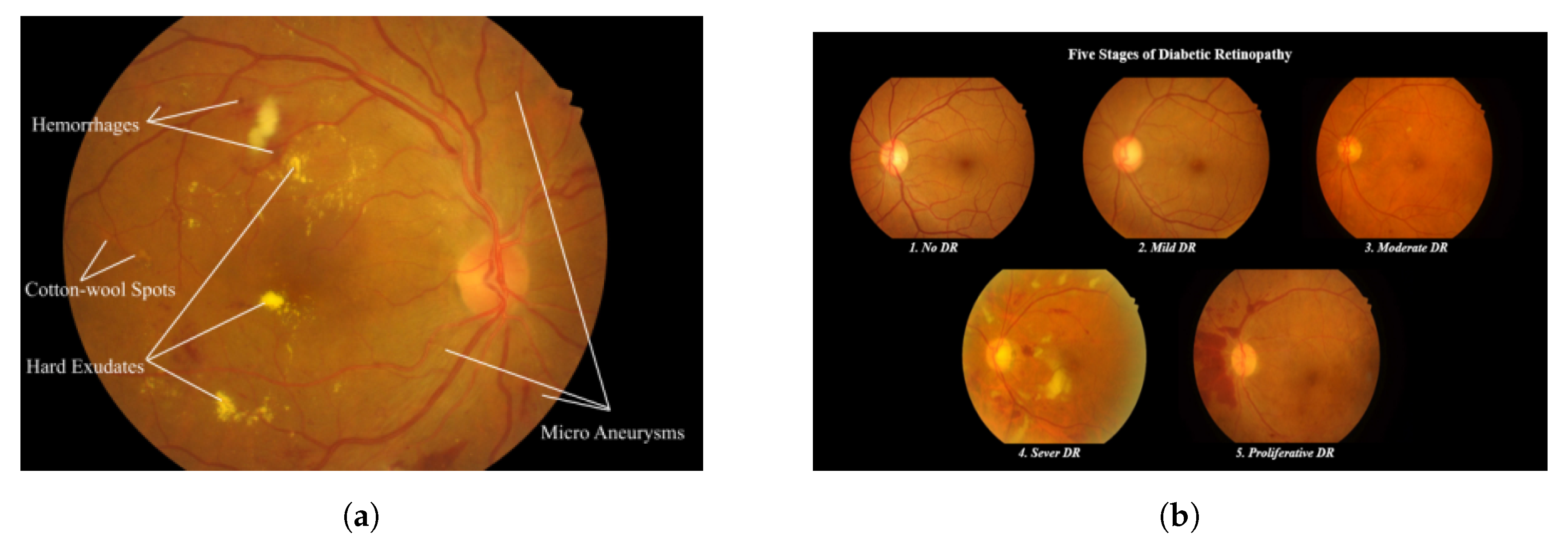




| Severity Level | # Samples |
|---|---|
| Class 0 (Normal) | 1805 |
| Class 1 (Mild Stage) | 370 |
| Class 2 (Moderate Stage) | 999 |
| Class 3 (Severe Stage) | 193 |
| Class 4 (Proliferative Stage) | 295 |
| Total | 3662 |
| Model | Accuracy | Precision | Recall | F1 Score | Kappa Statistic |
|---|---|---|---|---|---|
| Logistic Regression | 97.13 | 97 | 97 | 97 | 94.27 |
| KNN | 95.36 | 96 | 95 | 95 | 90.73 |
| Naive Bayes | 77.08 | 82 | 77 | 76 | 54.45 |
| Decision Tree | 91.27 | 91 | 91 | 91 | 82.52 |
| MLP | 96.45 | 97 | 96 | 96 | 92.91 |
| SVM (linear) | 96.58 | 97 | 97 | 97 | 93.17 |
| SVM (RBF) | 96.86 | 97 | 97 | 97 | 93.73 |
| DNN | 97.32 | 98 | 98 | 98 | 94.63 |
| Model | Accuracy | Precision | Recall | F1 Score | Kappa Statistic |
|---|---|---|---|---|---|
| Logistic Regression | 96.45 | 96 | 96 | 96 | 93 |
| KNN | 95.5 | 96 | 95 | 95 | 91 |
| Naive Bayes | 82.95 | 84 | 83 | 83 | 65.9 |
| Decision Tree | 87.59 | 88 | 88 | 88 | 75.17 |
| MLP | 96 | 96 | 96 | 96 | 91.89 |
| SVM (linear) | 96.18 | 96 | 96 | 96 | 92.36 |
| SVM (RBF) | 97.4 | 97 | 97 | 97 | 94.82 |
| DNN | 97.41 | 97 | 97 | 97 | 94.82 |
| Model | Accuracy | Precision | Recall | F1 Score | Kappa Statistic |
|---|---|---|---|---|---|
| VGG16-fc1 | 97.27 | 97 | 98 | 97 | 95.12 |
| VGG16-fc2 | 97.32 | 98 | 98 | 98 | 94.63 |
| NASNet | 97.14 | 97 | 97 | 97 | 94.27 |
| Xception | 97.41 | 97 | 97 | 97 | 94.82 |
| Inception ResNetV2 | 97.34 | 97 | 97 | 97 | 94.54 |
| Model | # Epochs | Loss | Accuracy |
|---|---|---|---|
| VGG16-fc1 | 65 | 0.0024 | 97.27 |
| VGG16-fc2 | 67 | 0.0139 | 97.32 |
| NASNet | 37 | 0.0310 | 97.14 |
| Xception | 16 | 0.0213 | 97.41 |
| Inception ResNet V2 | 19 | 0.0815 | 97.34 |
| Type of Uni-Modal Features | Accuracy | Precision | Recall | F1 Score | Kappa Statistic |
|---|---|---|---|---|---|
| VGG16-fc1 | 80.06 | 80 | 81 | 80 | 70.02 |
| VGG16-fc2 | 79.81 | 79 | 80 | 79 | 68.88 |
| NASNET | 76.4 | 75 | 76 | 75 | 63.87 |
| Xception | 78.99 | 78 | 79 | 78 | 67.67 |
| Inception ResNetV2 | 79.73 | 78 | 78 | 78 | 67.67 |
| Model | # Epochs | Loss | Accuracy |
|---|---|---|---|
| VGG16-fc1 | 76 | 0.3623 | 80.06 |
| VGG16-fc2 | 79 | 0.3986 | 79.81 |
| NASNet | 37 | 0.5612 | 76.39 |
| Xception | 23 | 0.4175 | 78.99 |
| Inception ResNet V2 | 89 | 0.382 | 79.73 |
| Modalities | Pooling | Accuracy | Kappa Statistic | Epochs | Loss |
|---|---|---|---|---|---|
| VGG16-fc1 and VGG16-fc2 | Max-pooling | 96.12 | 91.89 | 68 | 0.0352 |
| Avg-pooling | 97.39 | 94.61 | 51 | 0.0293 | |
| Sum-pooling | 95.5 | 91 | 64 | 0.0419 | |
| VGG16-fc1, VGG16-fc2 and Xception | Max-pooling | 96.85 | 92.6 | 69 | 0.0314 |
| Avg-pooling | 97.92 | 94.93 | 43 | 0.0201 | |
| Sum-pooling | 96.1 | 92.31 | 56 | 0.0396 |
| Modalities | Pooling | Accuracy | Kappa Statistic | Epochs | Loss |
|---|---|---|---|---|---|
| VGG16-fc1 and VGG16-fc2 | Maximum | 78.06 | 66.87 | 72 | 0.4176 |
| Average | 80.34 | 69.21 | 62 | 0.2987 | |
| Sum | 76.8 | 65.64 | 68 | 0.5693 | |
| VGG16-fc1, VGG16-fc2 and Xception | Maximum | 79.25 | 67.29 | 74 | 0.3986 |
| Average | 80.96 | 70.9 | 54 | 0.2619 | |
| Sum | 77.12 | 66.42 | 61 | 0.4782 |
| Model | Accuracy |
|---|---|
| DR detection using Deep Learning [43] | 57.2% |
| DR Classification Using Xception [44] | 79.59 |
| DR Classification Using InceptionV3 [44] | 78.72 |
| DR Classification Using MobileNet [44] | 79.01 |
| DR Classification Using ResNet50 [44] | 74.64 |
| Blended features + DNN (proposed) | 80.96 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bodapati, J.D.; Naralasetti, V.; Shareef, S.N.; Hakak, S.; Bilal, M.; Maddikunta, P.K.R.; Jo, O. Blended Multi-Modal Deep ConvNet Features for Diabetic Retinopathy Severity Prediction. Electronics 2020, 9, 914. https://doi.org/10.3390/electronics9060914
Bodapati JD, Naralasetti V, Shareef SN, Hakak S, Bilal M, Maddikunta PKR, Jo O. Blended Multi-Modal Deep ConvNet Features for Diabetic Retinopathy Severity Prediction. Electronics. 2020; 9(6):914. https://doi.org/10.3390/electronics9060914
Chicago/Turabian StyleBodapati, Jyostna Devi, Veeranjaneyulu Naralasetti, Shaik Nagur Shareef, Saqib Hakak, Muhammad Bilal, Praveen Kumar Reddy Maddikunta, and Ohyun Jo. 2020. "Blended Multi-Modal Deep ConvNet Features for Diabetic Retinopathy Severity Prediction" Electronics 9, no. 6: 914. https://doi.org/10.3390/electronics9060914
APA StyleBodapati, J. D., Naralasetti, V., Shareef, S. N., Hakak, S., Bilal, M., Maddikunta, P. K. R., & Jo, O. (2020). Blended Multi-Modal Deep ConvNet Features for Diabetic Retinopathy Severity Prediction. Electronics, 9(6), 914. https://doi.org/10.3390/electronics9060914







