Abstract
With the increasing richness of medical images and clinical data, abundant data support is provided for multimodal chest disease diagnosis methods. However, traditional multimodal fusion methods are often relatively simple, leading to insufficient exploitation of crossmodal complementary advantages. At the same time, existing multimodal chest disease diagnosis methods usually focus on two modalities, and their scalability is poor when extended to three or more modalities. Moreover, in practical clinical scenarios, missing modality problems often arise due to equipment limitations or incomplete data acquisition. To address these issues, this paper proposes a novel multimodal chest disease classification model, MDFormer. This model designs a crossmodal attention fusion mechanism, MFAttention, and combines it with the Transformer architecture to construct a multimodal fusion module, MFTrans, which effectively integrates medical imaging, clinical text, and vital signs data. When extended to multiple modalities, MFTrans significantly reduces model parameters. At the same time, this paper also proposes a two-stage masked enhancement classification and contrastive learning training framework, MECCL, which significantly improves the model’s robustness and transferability. Experimental results show that MDFormer achieves a classification precision of 0.8 on the MIMIC dataset, and when 50% of the modality data are missing, the AUC can reach 85% of that of the complete data, outperforming models that did not use two-stage training.
1. Introduction
Chest diseases, including pneumonia, lung cancer, and tuberculosis, are among the types of diseases with high incidence and mortality rates worldwide. According to the World Health Organization (WHO), six out of the top ten global causes of death in 2021 were chest diseases [1], posing significant challenges to global public health. Due to the subtle early symptoms of these chest diseases, many patients are diagnosed at advanced stages, missing the optimal treatment window. Therefore, early and accurate diagnosis is crucial for improving patient survival rates and quality of life. Traditional diagnostic methods for chest diseases primarily rely on medical imaging (such as X-rays, CT scans, etc.) or clinical data (such as medical history, symptoms, and signs). However, information from a single modality is often insufficient to provide a comprehensive diagnostic basis. Numerous medical studies have repeatedly shown that the lack of access to clinical and laboratory data during image interpretation can lead to a decline in diagnostic performance by doctors [2,3]. Furthermore, in a survey of radiologists, the majority (87%) of doctors stated that clinical information significantly impacts their interpretation [4]. As a result, the fusion and analysis of multimodal medical data have garnered extensive attention and research in the current medical field [5,6].
However, existing multimodal fusion methods still face many challenges in practical applications. First, data from different modalities are heterogeneous; for example, medical images are high-dimensional spatial data, clinical texts are discrete symbolic data, and vital sign time-series are continuous time data. Effectively aligning and merging these heterogeneous data is a problem that needs to be addressed. Traditional methods often use simple weighted averaging or concatenation techniques, which fail to capture the complex relationships between modalities and may lead to the loss of fine-grained semantic information. Second, some Transformer-based multimodal fusion methods have poor scalability, and when expanded to multiple modalities, the model complexity increases significantly. Additionally, in practical clinical settings, due to equipment limitations, incomplete data collection, and other reasons, complete modal data are often unavailable. This requires chest disease diagnosis models to be robust enough to handle incomplete data. However, existing research has paid little attention to the missing modality problem and lacks effective solutions.
To address the challenges of multimodal fusion difficulties, poor model scalability, and insufficient adaptability to the missing modality problem, this study proposes a Transformer-based multimodal fusion module, MFTrans, and builds a multimodal model, MDFormer, for chest disease classification on this basis. Additionally, we design an efficient two-stage training framework, MECCL. This approach not only improves classification accuracy but also effectively reduces the model’s parameter count, enhances scalability, and allows for flexible handling of various modality combinations. More importantly, the model demonstrates excellent robustness in the presence of missing modality. The main contributions of this paper are summarized as follows:
- We extend the self-attention mechanism to use different modalities as queries, simultaneously learning both intra-modality and inter-modality correlations. By combining group convolutions to extract local information, we design a lightweight fusion module MFTrans based on a multi-head attention mechanism. This approach effectively enhances fusion performance while maintaining low computational overhead.
- By integrating multimodal data such as time-series vital signs, chest X-ray images, and patient diagnostic reports, we propose a Transformer-based multimodal chest disease classification model, MDFormer. This model processes data from different modalities through modality-specific encoders, implements multimodal feature interaction and fusion in MFTrans, and ultimately achieves accurate chest disease diagnosis.
- This study combines modality masking and contrastive learning to propose a two-stage Mask-Enhanced Classification and Contrastive Learning training framework, MECCL. To further balance the optimization process of different task objectives in the two-stage training, we introduce a Sigmoid dynamic balance loss weight, enabling adaptive adjustment between loss terms. This framework allows the model to maintain effective diagnostic and classification performance even when some modality data are missing, thereby enhancing the model’s robustness and training stability.
The structure of the rest of this paper is arranged as follows: Section 2 provides a review of related work, including deep learning-based chest disease diagnosis, medical multimodal disease diagnosis, and Transformer-based multimodal data fusion methods. Section 3 details the construction process and preprocessing methods of the dataset used, the proposed improved Transformer-based multimodal fusion model, and the two-stage training framework employed. Section 4 presents the experimental setup, demonstrates comparative experimental results with current state-of-the-art methods, and provides an in-depth analysis and discussion of the results. Finally, Section 5 summarizes the research work presented in this paper and discusses future research directions.
2. Related Works
2.1. Deep Learning-Based Chest Disease Diagnosis
Accurate diagnosis and classification of chest diseases are crucial for early detection, personalized treatment, and improving prognosis. In recent years, deep learning technologies have made significant progress in medical image analysis. For example, Asuntha et al. improved the accuracy of lung cancer detection and classification by combining various image feature extraction methods with a fuzzy particle swarm optimization algorithm (FPSO) [7]. Schroeder et al. successfully predicted COPD by combining X-ray images and pulmonary function data using a CNN model, achieving an AUC of 0.814 [8]. Pham et al. improved the efficiency of pneumonia diagnosis by combining CNN and ViT models, outperforming traditional models [9]. Hussein et al. proposed a hybrid architecture that significantly improved the classification accuracy of lung diseases by combining SVM and CNN [10]. Mann et al. evaluated the performance of various deep learning models and found that DenseNet121 and ResNet50 performed exceptionally well in diagnosing diseases such as emphysema [11]. Chandrashekar et al. evaluated the application of transfer learning in COVID-19 diagnosis, with the MobileNet model performing best, achieving an accuracy of 98% [12]. Hayat et al. proposed a hybrid deep learning model that combines EfficientNetV2 and Vision Transformer (ViT) for breast cancer histopathological image classification. Experiments conducted on the BreakHis dataset showed that the model achieved an accuracy of 99.83% in binary classification and 98.10% in multi-class classification, demonstrating its superior performance in breast cancer detection [13].
In addition, lung sounds are also an important basis for diagnosing chest diseases. Tariq et al. achieved approximately 97% accuracy in lung sound classification by analyzing lung sound spectrogram features using a CNN model [14]. Pham et al. proposed a CNN-MoE-based framework that outperformed existing methods in lung disease detection [15]. Lal et al. proposed an algorithm combining VGGish with BiGRU, improving the accuracy of lung sound recognition [16].
Currently, in the diagnosis of chest diseases, despite the availability of various modal data (such as images, physiological signals, clinical texts, etc.), the utilization of these data is often limited to single-modal or simple combinatorial analyses, failing to fully explore the potential correlations between multimodal information. This single-modal or shallow multimodal analysis approach limits the comprehensiveness and accuracy of chest disease diagnosis.
2.2. Medical Multimodal Disease Diagnosis
Deep neural networks, with their powerful ability to learn hierarchical representations, are particularly well suited for multimodal learning problems. However, how to effectively combine the marginal and joint representations of heterogeneous modalities remains a core challenge in multimodal fusion [6]. Multimodal fusion can occur at different stages, primarily including data-level fusion, feature-level fusion, and decision-level fusion [6]. Data-level fusion involves concatenating the raw data from different modalities into a single vector, which is then treated as a single-modal input for processing by deep learning models. Feature-level fusion is performed after learning the marginal representations of each modality, followed by their integration. Decision-level fusion involves independently modeling multiple sub-models and combining them with ensemble strategies to arrive at a final decision.
Data-level fusion has the advantage of ease of implementation, as it does not require the construction of complex unimodal feature extraction frameworks [17]. The EHR-KnowGen model proposed by Niu et al., by integrating multimodal data from electronic health records, improves the quality of disease diagnosis [18]. Glicksberg et al. used the pre-trained language model BlueBERT to analyze emergency department patient data, achieving AUCs of 0.9014 and 0.6475, although this approach may lose some accuracy when converting numerical data into text [19]. When input features have structural information, such as genomic data or time-series data, RNNs [20] or CNNs can effectively process the concatenated inputs. For example, the multitask learning method proposed by Bichindaritz et al., combining Cox loss and ordinal loss tasks, uses genomic data to predict patient survival risk and improves model performance [20]. Despite the simplicity and frequent use of data-level fusion in multimodal learning, its limitations are also evident. Directly modeling joint representations makes it difficult to extract useful marginal representations of modalities, especially when the features between modalities only become apparent in high-level abstractions. Furthermore, data-level fusion is suitable for cases where the modality distributions are similar, but when there are significant modality differences, such as between images and molecular data, it may not perform well.
In feature-level fusion, different types of deep learning networks are used to learn the marginal representations of each modality, and these features are then integrated through a fusion layer to learn a joint representation. This method is suitable for tasks that require the preservation of modality-specific features [17]. Yan et al. integrated pathological images and electronic medical record data, achieving an accuracy of 92.9%. Additionally [21], Lee et al. combined fundus images and clinical risk factors to predict cardiovascular diseases, with an AUC of 0.781 [22]. The MMD framework proposed by Cui et al., by handling missing data, significantly improved the accuracy and robustness of brain cancer survival prediction [23]. Liu et al. proposed the self-attention-based SFusion module, which can automatically learn the correlations between multimodal data and generate shared representations, thereby enhancing model performance [24]. In 2024, Al et al. introduced an AI-based multimodal breast cancer diagnostic framework, achieving a classification accuracy of 97.73% [25].
In decision-level fusion, the key is to synthesize the decisions of unimodal models rather than directly merging the data or features [26]. The most straightforward aggregation method is to take the average of the results from different models, assuming that each sub-model contributes equally. The LVH-fusion model developed by Soto et al. surpassed traditional methods in predicting left ventricular hypertrophy [27]. El et al. evaluated deep convolutional neural network models, and decision-level fusion outperformed unimodal methods in classifying eye diseases [28]. Since different modalities contribute unequally to prediction performance, using a weighted aggregation method can better adjust the contribution of each sub-model. Liu et al. integrated genomic and imaging data to predict breast cancer molecular subtypes, using weighted fusion to improve prediction accuracy [29]. The 3D convolutional neural network framework proposed by Saikia et al., combining CT and PET images, significantly improved cancer prediction accuracy [30]. Meta-learning methods can learn the complex relationships between different sub-models and integrate prediction results through classifiers, improving the final accuracy. For example, Reda et al. combined clinical biomarkers and imaging data and used a meta-learning approach to enhance early prostate cancer diagnosis accuracy, achieving a diagnostic accuracy of 94.4% [31]. Decision-level fusion is suitable for situations where signals are not complementary or when data are missing, as it can still provide effective predictive power. In contrast, data-level and feature-level fusion may not be adaptable when data is missing.
2.3. Transformer-Based Multimodal Data Fusion Methods
In 2017, Vaswani et al. proposed the Transformer model, which is based on the multi-head attention mechanism and constructs a sequence-to-sequence encoder–decoder structure, providing a new solution for multimodal data fusion [32]. Compared to traditional convolutional neural networks and recurrent neural networks, the Transformer reduces inductive bias [33], allowing it to learn higher-quality feature representations from multimodal inputs. The consistency of the self-attention mechanism across different modalities enables the Transformer to flexibly represent multimodal clinical data. Common Transformer-based multimodal data fusion methods are shown in the Figure 1.
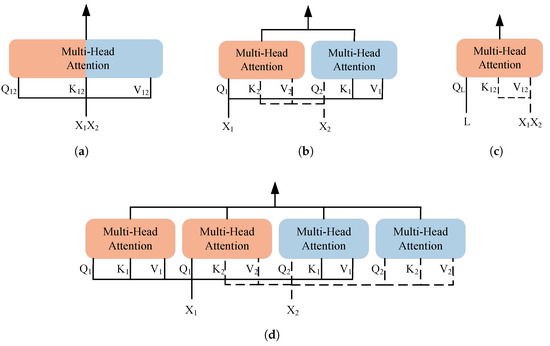
Figure 1.
Transformer-based multimodal fusion methods. (a) SelfAttention. (b) CrossAttention. (c) LearnableQAttention. (d) Cross-Self-Attention.
Simply concatenating the data or features of two modalities is referred to as SelfAttention. This method enables modality interaction by directly merging information from two modalities. Its advantages lie in efficiency and simplicity, avoiding complex modality interaction models, and it has been proven effective in multimodal pretraining and some downstream tasks [34,35,36,37].
CrossAttention is the most commonly used multimodal fusion method today, which calculates attention across the representations of different modalities. This method has been widely applied in multimodal medical data fusion [38]. However, CrossAttention faces the issue of parameter explosion when extended to multiple modalities [39], and it cannot fully capture the self-attention within each modality, potentially leading to information loss. In addition, combining CrossAttention with SelfAttention forms the Cross-Self-Attention mechanism [40]. This method not only preserves the internal information of each modality but also promotes effective interaction between modalities, though it still faces scalability issues. To mitigate the parameter explosion problem, some methods treat one modality as the primary modality and reduce the influence of the other modalities [41]. In addition, Flamingo and its medical adaptation Med-Flamingo adopt decoder-based architectures with CrossAttention for vision-language fusion, achieving strong few-shot performance [42,43].
Furthermore, LearnableQAttention captures the complex relationships between modalities by introducing trainable queries, increasing the model’s adaptability and flexibility when it comes to multimodal inputs [44,45]. However, similar to CrossAttention, this method also overlooks the correlations within each modality.
3. Data and Methodology
3.1. Dataset Construction
In this study, we constructed a multimodal, multi-label dataset for chest disease classification by combining MIMIC-CXR-JPG [46] and MIMIC-IV-ED [47], as illustrated in Figure 2. MIMIC-CXR-JPG contains chest X-ray images (in JPEG format), corresponding radiology reports, and disease label information. MIMIC-IV-ED provides time-series vital sign data of emergency department patients. First, we retained one chest X-ray image for each radiology report from the MIMIC-CXR-JPG dataset. At this stage, the data contained 65,379 patients and 227,827 reports. Then, for each chest X-ray image, we retrieved the corresponding emergency department stay record for the patient. In the edstays table, the intime and outtime columns record the time span of hospitalization, while the StudyDate and StudyTime columns in the metadata table indicate the time the chest X-ray image was taken. If the image’s capture time falls within the span of an emergency department stay, the image is associated with the corresponding stay-id; otherwise, we continue searching other hospitalization records to ensure the integrity and accuracy of the data. During the integration process, only 61,854 images matched with a stay-id, while the remaining images might have been taken after the patient left the emergency department. The final MIMIC multimodal dataset contains 61,854 chest X-ray images, corresponding radiology reports, 7 time-series vital signs, and 14 structured labels. The final dataset exhibits class imbalance in its label distribution. To alleviate this issue, we incorporated class weights during training to reduce the impact of label imbalance on model performance.
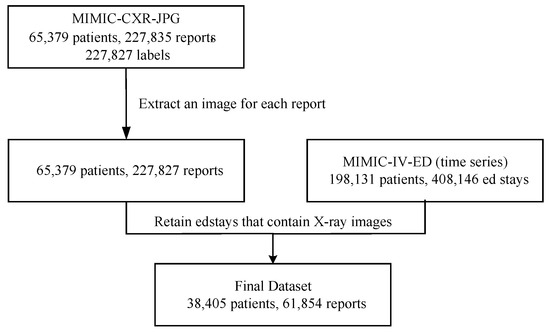
Figure 2.
Dataset construction workflow.
3.2. Data Preprocessing
In text preprocessing, we first used regular expressions to remove punctuation and special characters to retain the core content of the text. In addition, in the MIMIC-CXR-JPG dataset, radiology reports mainly consist of two sections: ‘Findings’ and ‘Impression’. The ‘Findings’ section mainly includes descriptive statements about the patient’s images, while the ’Impression’ section mainly contains brief diagnostic information. Considering that the labels in the MIMIC-CXR-JPG dataset are automatically extracted from the reports using the CheXpert method, we remove the ‘Impression’ part of the radiology reports during preprocessing (82.4% of reports in the original dataset contain the ’Impression’ section). Directly using the ’Impression’ information may lead to label leakage. Additionally, to ensure the completeness of the text expression, we removed short phrases consisting of fewer than three words.
For image preprocessing, we implemented several measures to enhance the model’s generalization ability and robustness. First, we resized the images to the specified dimensions through random cropping, ensuring image diversity, and used cubic interpolation to preserve image quality. Second, random horizontal flipping was applied to increase data diversity, allowing the model to learn richer information from different perspectives. Finally, normalization was performed to ensure pixel values fell within a reasonable range, improving the stability of training.
For time-series data, we conducted feature selection and outlier handling to improve data quality. First, we removed the feature “rhythm” due to its high missing rate. Next, we transformed the “pain” feature by converting non-numeric data into numeric values and standardizing different recording methods. Additionally, outliers in temperature data were corrected, specifically fixing the misrecording between Celsius and Fahrenheit. When handling missing values, we applied a neighboring value imputation method within the same stay-id to ensure data integrity and usability.
3.3. Multimodal Chest Disease Diagnosis Model
3.3.1. Overview
In the diagnosis of chest diseases, a patient’s health condition often involves multiple interrelated diseases. For instance, certain chest diseases (such as Chronic Obstructive Pulmonary Disease, COPD) may coexist with other diseases (such as lung infections or cardiovascular diseases). Therefore, multi-class classification methods cannot effectively capture the complexity of medical scenarios, and multimodal multi-label classification becomes a more clinically relevant approach for modeling. Specifically, let the set of modalities be denoted as , where m is the total number of modalities. Then, each patient’s data can be represented as a multimodal sample , where represents the feature of the m-th modality for the i-th sample, and is the feature space of the m-th modality. For example, this could include imaging data, laboratory test data, gene expression data, and clinical text. Data from different modalities each carry important medical information and have strong complementarity. The final prediction result is a set of associated disease labels , where C is the total number of potential labels, which may include categories like pneumonia, tuberculosis, pleural effusion, heart failure, and others. Our goal is to learn a mapping function from the given dataset by optimizing the binary cross-entropy loss (BCE), such that is as close as possible to the true label .
In related work, we emphasize the importance of multimodal fusion and the challenges of extending models to multiple modalities. To address these two challenges, we have developed a multimodal multi-label chest disease classification model, MDFormer, as shown in Figure 3. The model consists of three parts: a modality-specific encoder , a fusion module , and a classifier . First, each modality is processed through its own encoder to learn the marginal representation of each modality, all of which are then jointly input into the fusion module to obtain a multimodal fused representation. Finally, the fused representation is subjected to multi-label classification to generate the multi-label disease classification results. MDFormer can be configured with different modality-specific encoders according to the use case, enabling plug-and-play adaptation to different modalities.
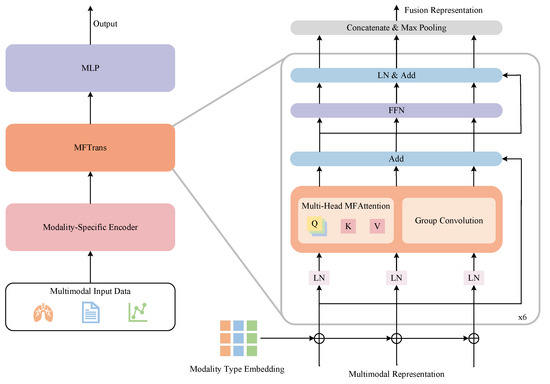
Figure 3.
MDFormer for multimodal chest disease classification.
3.3.2. Modality-Specific Encoders
In the implementation of MDFormer, we use BioViL to initialize our image and text encoders, which are responsible for extracting the input features from images and texts. BioViL is a self-supervised vision-language model proposed by Microsoft Health in 2022 [48], specifically adapted to the chest X-ray domain. The inputs to the image and text encoders include an image (in this study, the image size is uniformly set to , with representing the number of channels), and a text , where is the text sequence length, and is the word embedding dimension. The specific encoding process is as follows:
Here, is the mapping operation applied to the image features to ensure they align with the dimensions of the text features. The image features are generated by the image encoder , and the text features are generated by the text encoder , where represents the length of the image features. These modality representations are then input into the multimodal fusion module to further learn both global and local crossmodal associations.
We use LightTS as the time-series encoder to model the time-series features. LightTS is a lightweight multivariate time-series model based on the MLP architecture, which improves efficiency and modeling capabilities through sampling strategies [49]. Specifically, for the vital sign time-series data , where is the length of the time-series and represents the number of variables, the vital sign time-series data are processed by the time-series encoder and mapping module to obtain the time-series representation . The encoding process is as follows:
Here, is the projection layer, which ensures the time-series representation’s dimension is compatible with the subsequent modalities. Through this design, MDFormer is able to efficiently learn from the vital sign data and provide rich time-series features for the subsequent multimodal fusion module.
3.3.3. MFTrans
Before performing multimodal fusion, we first apply each modality-specific encoder independently to extract high-dimensional feature representations for each modality. Specifically, for the j-th modality data of the i-th patient, after being processed by the encoder, we obtain the feature representation , where . To further enhance the distinguishability of features from different modalities, we introduce modality type embeddings, denoted as , to the representation of each modality. Modality type embeddings assign a learnable vector to each modality, helping the model understand the specific attributes of that modality while strengthening the interactions between different modalities. Then, we perform an addition operation on and , followed by normalization via LayerNorm to stabilize the training process, resulting in . The specific process is as follows:
Based on this, we design a Multimodal Fusion Attention (MFAttention) mechanism for crossmodal information fusion, as shown in Figure 4. In MFAttention, there are three independent query mapping layers , where the features of each modality are passed through their respective query mappings to obtain the corresponding query matrices . Additionally, MFAttention includes a key mapping layer and a value mapping layer . The concatenated representation of the three modalities is passed through the key mapping layer and the value mapping layer to obtain the key matrix and the value matrix . The specific computation process is as follows:
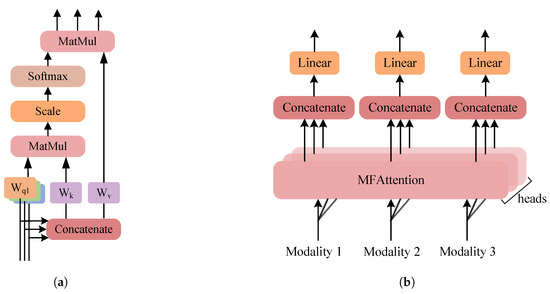
Figure 4.
Multimodal fusion attention. (a) MFAttention. (b) MultiHead-MFAttention.
Subsequently, based on scaled dot-product attention, the attention output for each modality is calculated, where represents the features after multimodal interaction. To further enhance the expression of local information, we introduce group convolution [50] on each modality feature, obtaining the convolution output . Group convolution is an efficient feature extraction method that divides the features into several groups and performs convolution operations independently on each group, reducing computational cost and enhancing the model’s ability to perceive local features. In this study, we set the number of groups to 12, with a kernel size of 3, stride of 1, and padding of 1, ensuring the input and output have the same shape. After that, we add the outputs from MFAttention , the group convolution , and the residual connection to obtain . Next, we input the three fused modality features into a shared Feedforward Neural Network (FFN) module to obtain the representation . We then stack 6 layers of the fusion layer described above to form the fusion module MFTrans, as shown on the right side of Figure 3. Finally, the outputs of the three modalities are concatenated and aggregated through max-pooling to obtain the final output of MFTrans, :
In summary, MFAttention primarily models global modality interactions and is suitable for long-term dependency modeling across modalities, while group convolution enhances the perception of local features, helping to capture fine-grained modality-specific features. Together, they complement each other. This combination allows MDFormer to effectively fuse multimodal information at different scales, improving its ability to understand clinical data.
3.3.4. Classification Head
In MDFormer, our classification head is used for multi-label chest disease classification tasks. Given the fused multimodal feature vector , we first perform feature mapping through two fully connected layers, where the first layer uses a ReLU activation function to introduce non-linear transformations. Finally, we use a Sigmoid activation function to generate the class probability distribution, obtaining the final prediction vector , where C is the number of classes. We optimize the model using binary cross-entropy loss, as shown below:
Here, N is the batch size, C is the total number of classes, represents the true label of sample i for class j, and is the predicted probability output by the Sigmoid function.
3.4. Two-Stage Training Framework
3.4.1. Overview
In the process of actual clinical data collection, missing modality is an unavoidable problem. We aim to build a multimodal multi-label classification model that can handle missing modality. Let the set of modalities for a sample be , and the actual available subset of modalities be . Then, the input data for the i-th patient becomes , meaning the input data includes only the modalities observed in . At this point, the mapping that the model needs to learn becomes .
To address the above problem, we design a two-stage Masked Enhanced Classification and Contrastive Learning training framework, MECCL, to improve the model’s classification performance in the case of missing modality while maintaining the stability of its performance when all modalities are available. The core idea of this training framework is to simulate missing modality during training so that the model learns how to classify effectively with incomplete data. The overall structure of the framework is shown in Figure 5. To ensure the efficiency of model training and fully utilize the representations learned in the first stage, we only adjust the parameters of the model fusion module MFTrans and the classification head in the second stage, while freezing the modality-specific encoders. The parameters adjusted in the second stage account for only 20% of the original model size. Four losses are jointly optimized: the classification loss for complete modalities , the classification loss for modality masks , the modality mask contrastive learning loss , and the KL divergence loss .
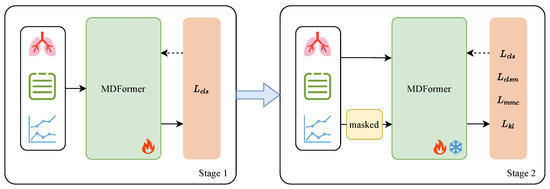
Figure 5.
MECCl two-stage training framework.
3.4.2. Modality Masked Classification
Based on the first stage, we introduce a random dynamic masking enhancement strategy in the second stage to improve the model’s performance on missing modalities while maintaining stability for complete modalities. Specifically, in each training iteration, we apply a random mask to the input sample , meaning that we randomly mask one or two modalities to generate a masked sample . This process simulates the incomplete data scenarios often encountered in real clinical applications. The dynamic masking strategy forces the model to learn in the presence of missing modalities, thereby enhancing its adaptability to missing modalities.
Subsequently, for the complete modality input and the masked modality input , two forward passes are performed, yielding prediction results based on the complete dataset and prediction results based on the masked dataset . Then, the binary cross-entropy classification losses are computed for each:
Through , we constrain the classification ability on the complete modality input, ensuring the model maintains high classification accuracy when modalities are intact. By introducing the random dynamic masking enhancement strategy, directly addresses the classification task on masked modalities, allowing the model to make accurate predictions even with missing modalities. Since the mask is random, the model must learn to predict under different modality combinations, enhancing the information complementarity between modalities and preventing the model from over-relying on any specific modality.
3.4.3. Modality Masked Contrastive Learning
In order to enable the model to effectively represent disease information even in the case of missing modality, we introduce Multimodal Masked Contrastive Learning, as shown in the Figure 6. This method enhances the consistency of representations between modalities through contrastive learning, thereby alleviating the issue of incomplete information caused by modality missing. Contrastive Learning is a self-supervised learning method that aims to bring the representations of positive samples closer together while separating those of negative samples. A commonly used loss function is the InfoNCE loss:
where N is the batch size, is the similarity between vectors and , and is the temperature coefficient, used to control the smoothness of the Softmax distribution.
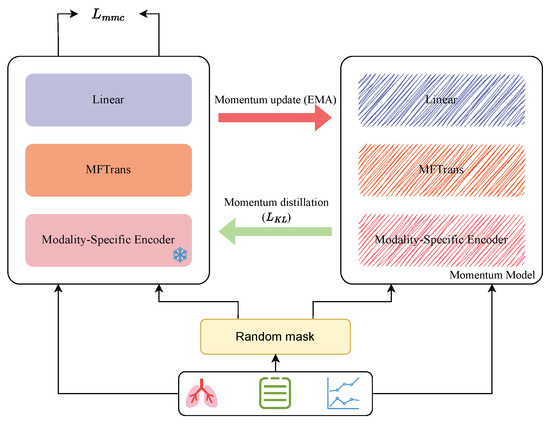
Figure 6.
Multimodal masked contrastive learning.
In Multimodal Masked Contrastive Learning, we treat the complete modality data and the corresponding masked modality data as positive sample pairs, while other samples in a batch are considered negative samples. The goal of the model is to maximize the similarity between these positive samples while minimizing their similarity to negative samples. Specifically, for complete modality input sample and masked modality input sample , we first pass them through modality-specific encoders for feature extraction to obtain preliminary modality features. These preliminary representations are then processed by the fusion module MFTrans to produce the fused representations and . To further optimize the contrastive learning of features, we add a linear transformation layer and a normalization layer for feature transformation. After this processing, we obtain two normalized representations and . The specific formulas are as follows:
where represents a fully connected layer used to map the fused representations to the feature space of contrastive learning, and refers to L2 normalization to enhance the stability of similarity calculation. We then perform contrastive learning on these two normalized representations and compute the Multimodal Masked Contrastive Learning loss :
This objective encourages the model to learn crossmodal relationships and maximize the similarity between representations of complete data and modality-masked data. Furthermore, in contrastive learning, the choice of negative samples is crucial to the model’s training. However, in real-world applications, due to the uncertainty of the missing modality problem, negative samples may contain examples semantically similar to positive samples, leading to false negatives, where the model misjudges the sample as negative. This problem is referred to as the false negative problem. In the process of multimodal masked contrastive learning, a batch may still contain relevant positive samples, but the one-hot labels used in contrastive learning will penalize all negative samples indiscriminately, introducing noise during training. To alleviate this issue, we introduce pseudo-targets generated by a momentum model for learning. The momentum model is a continuously evolving teacher model, obtained by the Exponential Moving Average (EMA) of MDFormer.
where is the momentum update coefficient. We then pass the complete modality input sample and modality-masked input sample into the momentum model to obtain the corresponding momentum representations and .
where is the momentum representation of the complete modality sample, and is the momentum representation of the modality-masked sample. We then generate additional predictions through the momentum model and introduce the KL divergence loss to constrain the distribution consistency between MDFormer and the momentum model:
where and represent the probability distributions of the similarity between complete modality samples and modality-masked samples after Softmax normalization, generated by MDFormer. and are the probability distributions generated by the momentum model. calculates the KL divergence between two probability distributions, encouraging the model’s prediction distribution to match the momentum model’s distribution as closely as possible.
3.4.4. Dynamic Balancing of Loss Weights
To better balance different loss functions in the second-stage training, we leverage the non-linear adjustment properties of the Sigmoid function to dynamically adjust the loss weights based on the number of training epochs. This allows for the allocation of the most appropriate optimization objectives at different stages of training. Specifically, in the first 5 epochs of training, we use the Sigmoid function to gradually increase the loss weight for and decrease the loss weight for . Meanwhile, the weights and for and are kept constant at 1. This ensures training stability, allowing the model to more stably adapt to the modality mask early in the second-stage training and establish a more robust representational foundation. The specific weight adjustment formulas are as follows:
Here, represents the current training epoch, is the midpoint of the adjustment, i.e., half of the total adjustment epochs, and is the total number of adjustment epochs, set to 5 epochs. This means that the loss weights are dynamically adjusted in the first 5 epochs, after which the loss weights remain constant. k controls the steepness of the Sigmoid curve, governing the rate of weight adjustment; it is set to 10, where a larger k results in a steeper adjustment process, and a smaller k results in a gentler one. Finally, combining the four loss terms, the second-stage loss function adopts the following weighted form:
This method allows the model to better focus on modality mask contrastive learning during the early stages of training, thereby learning the effective relationship between full modality samples and modality mask samples. As training progresses, the model gradually shifts more attention to the modality mask classification task, ensuring that its classification capability in the case of missing modalities is fully improved.
4. Results and Discussion
4.1. Experimental Setup
This paper compares the performance of different types of models on the MIMIC dataset, divided into three categories for analysis. The first category consists of unimodal models, including DenseNet121 [51], BioLinkBert [52], and Informer [53]; the second category includes bimodal models such as MedFuse [54], ALBEF [55], and MedCLIP [56]; and the third category includes trimodal models, mainly ConcantateFusion and MedFusePlus.
Among these, ConcantateFusion is a simple multimodal fusion model. Its encoder structure is identical to that of MDFormer, but in the fusion stage, the representations of the three modalities are directly concatenated to generate a unified feature representation, which is then used for classification tasks. MedFusePlus is an extended version of MedFuse, incorporating BioLinkBert as the text encoder, and further extends multimodal learning from clinical time-series data and chest X-ray images into a trimodal model. Additionally, we compared various Transformer-based multimodal fusion methods, including Self-Attention, Cross-Attention, LearnableQAttn, and Cross-Self-Attention. The experiments use precision, recall, F1 score, ROC curve, and AUC value as evaluation metrics.
The replication of all baseline models follows the configuration from the original papers or official code, with a consistent classification head design across all models. In the experiments, 10% of the samples were kept for evaluation. In the second-stage masked test set, 50% of the data were randomly masked, with one or two modalities masked at a time to simulate missing modality scenarios. Our method used the AdamW optimizer for training, with the following parameter settings: , , and , a maximum learning rate of , and cosine learning rate decay. The batch size was set to 256, and for models exceeding memory capacity, it was adjusted to 128. The training process was stopped early after 15 epochs, with a weight decay coefficient set to 0.05 and a learning rate warmup ratio of 0.03.
4.2. Comparative Experiment
The comparison of the experiments mainly includes two aspects: first, the classification performance comparison of different models on the same dataset; second, exploring the impact of different multimodal data fusion strategies on classification performance under the same encoder. The best and second-best values of the experimental results are marked in bold and underlined, respectively, for an intuitive comparison of the model’s performance.
“I” represents chest X-ray image data, “T” represents radiology report data, “TS” represents vital signs time-series data, and “✓” indicates the modality used during training. Table 1 presents the classification performance of different models on the MIMIC dataset. It can be seen that MDFormer achieved the best results across all metrics. Specifically, MDFormer reached an F1 score of 0.7966, which is a 3% improvement over the second-best model, ALBEF, confirming its effectiveness in multimodal fusion. Notably, among all the unimodal models, BioLinkBert achieved an AUC of 0.8164, significantly higher than DenseNet121 and Informer. This indicates that the text modality performs better than chest X-ray images and vital signs time-series data in the chest disease diagnosis task. This may be because radiology reports provide a more intuitive summary of the condition, not only highlighting key points from the image data but also incorporating the doctor’s professional knowledge. Additionally, we observed that multimodal fusion methods generally outperform unimodal models. For example, MedFuse and ConcatenateFusion significantly surpass the unimodal baselines. Furthermore, ALBEF, which uses both radiology reports and chest X-ray images, outperforms ConcatenateFusion and MedFusePlus, indicating that simple concatenation or LSTM-based fusion strategies cannot fully leverage multimodal information, while more advanced crossmodal interaction mechanisms can further improve classification performance.

Table 1.
Results of MDFormer and baseline models.
To further assess the practicality of each model in clinical settings, we compare their average inference time on the test set. As shown in Table 1, although MDFormer achieves the best overall performance across all evaluation metrics, its inference time (26.63 ms) remains within a reasonable range. In fact, it is comparable to or even faster than some less accurate multimodal models, such as MedFuse (31.71 ms) and MedCLIP (29.70 ms). This demonstrates that MDFormer not only provides high accuracy but also maintains efficient inference speed, making it suitable for real-time or time-sensitive medical applications. In addition, we performed 100 bootstrap resamplings on the test set. MDFormer achieved an average F1 score of 0.7920 (95% CI: 0.7796–0.8044) and an AUC of 0.9011 (95% CI: 0.8948–0.9074), while ALBEF achieved an average F1 score of 0.7720 (95% CI: 0.7596–0.7844) and an AUC of 0.8860 (95% CI: 0.8798–0.8922). Furthermore, using the same data splits, we conducted paired Wilcoxon signed-rank tests to examine the differences in F1 score and AUC between the two methods. The results showed statistically significant improvements (F1: p = 0.0047; AUC: p = 0.0031), further validating the stability and reliability of our approach. MDFormer achieves the best results across all metrics mainly due to its use of the MFAttention and group convolution feature extraction, which effectively capture the correlations between different modalities and integrate both local and global features.
From the above experimental data, the following conclusions can be drawn: Unimodal models generally perform poorly due to the lack of crossmodal complementary information, limiting the model’s discriminative ability. Simple multimodal fusion methods fail to fully utilize the complementary advantages of modalities and do not adequately explore the synergistic information between modalities. MDFormer achieves optimal results on various datasets thanks to its multimodal attention mechanism and the adaptive fusion module, MFTrans, which can flexibly handle different multimodal data.
Figure 7 shows the ROC curves of MDFormer and baseline models on the MIMIC dataset. The x-axis represents the false positive rate (FPR), and the y-axis represents the true positive rate (TPR). The values in parentheses in the legend represent the model’s AUC score. It is clearly visible that the ROC curve of MDFormer is above all the baseline models, further confirming its advantage in the chest disease classification task. Especially compared to MedFusionPlus and ALBEF, MDFormer shows a significant improvement in TPR in the low-FPR region, indicating stronger discriminatory ability.
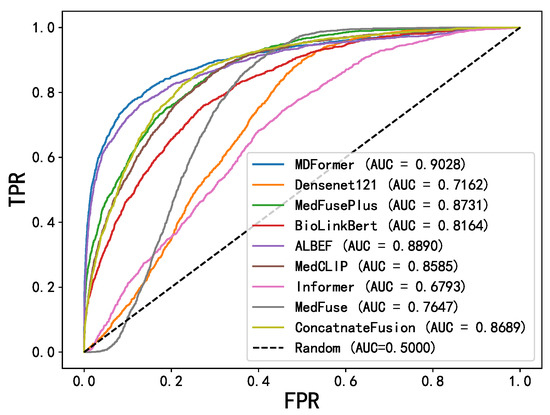
Figure 7.
ROC curves.
Additionally, we used different Transformer-based multimodal fusion methods in combination with a fixed encoder for comparative analysis, and the experimental results are shown in Table 2. To visually demonstrate the relationship between performance and parameter count for different fusion methods on the MIMIC dataset, we use a bubble chart for visualization, as shown in Figure 8. In this chart, the x-axis represents AUC, the y-axis represents F1, and the size of the bubbles corresponds to the model’s parameter count. First, MFTrans achieved the best performance on all key metrics. At the same time, MFTrans uses only 36M parameters, while CrossAttention and Cross-Self-Attention require 3.1 times and 4.6 times the number of parameters as MFTrans, respectively. This indicates that, although CrossAttention-related methods have certain performance advantages, they come with a large parameter overhead and poor scalability, whereas MFTrans achieves optimal performance while maintaining a lower parameter count. Furthermore, SelfAttention and LearnableQAttn have smaller parameter counts but significantly lag behind MFTrans in all performance metrics.

Table 2.
Results of different fusion methods.
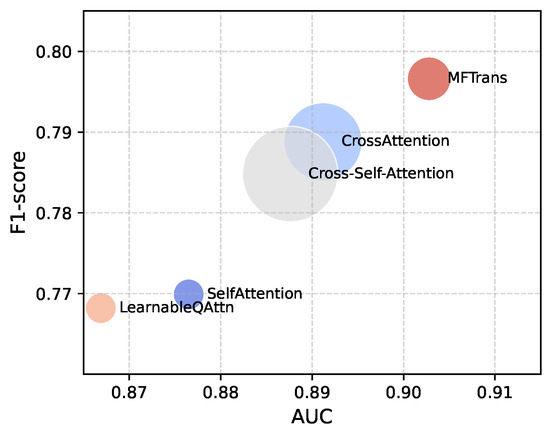
Figure 8.
Bubble chart of parameters and performance for different multimodal fusion methods.
Finally, we present the hyperparameter tuning experiments for the fusion module MFTrans, focusing on evaluating the impact of different numbers of fusion layers and grouped convolution groups on model performance. The experimental results are shown in the Figure 9. As illustrated in the left panel, the model’s F1 score steadily improves as the number of fusion layers increases from two to six, with a particularly notable gain between two and four layers. This suggests that increasing the number of fusion layers helps to more effectively model multimodal information. However, when the number of layers increases to eight, the performance gain saturates and even slightly declines, possibly due to redundant information or overfitting introduced by the overly deep structure. In the right panel, the number of grouped convolutions has a limited effect on performance when it exceeds two. Specifically, from 4 to 12 groups, the F1 score remains around 0.796, but performance slightly drops when the group number exceeds 16. These results indicate that moderately increasing the number of convolution groups can reduce the number of parameters, but an excessive number may harm model performance. Considering both performance and computational cost, the optimal configuration for the fusion module is six fusion layers and 12 convolution groups.
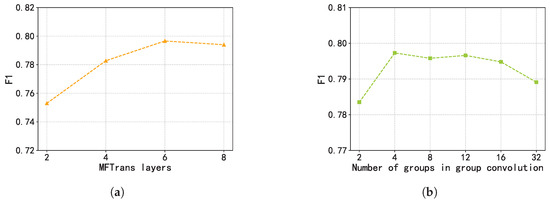
Figure 9.
Hyperparameter tuning experiments of MFTrans. (a) MFTrans layers. (b) Number of groups in group convolution.
4.3. Ablation Study
MDFormer uses MFTrans to jointly interpret multimodal information, effectively capturing both global and local features. To evaluate the actual contribution of each component of MFTrans to the model’s performance, we conducted ablation experiments on the MIMIC dataset, comparing the following MDFormer variants:
- w/o MFAttention: Removes the MFAttention, relying only on the self-attention mechanisms of individual modalities for information extraction in the fusion module.
- w/o Group Convolution: Removes the group convolution bypass.
- w/o MFTrans: Completely removes the MFTrans fusion module.
Table 3 present the results of the ablation experiments. It can be observed that, on both datasets, the classification performance of MDFormer slightly decreases when the group convolution bypass is removed, indicating that group convolution effectively extracts local information from each modality and contributes to the model’s classification performance. When MFAttention is removed, the model’s metrics are significantly lower than those of the full model, suggesting that MFAttention, compared to the self-attention of individual modalities, better captures the correlations between modalities, thus enhancing the feature fusion capability. Additionally, combining MFAttention with group convolution further improves the model’s performance, indicating that both components complement each other in the multimodal information fusion process. Among all the ablation experiments, removing MFTrans had the most significant impact, with a noticeable drop in model performance, further confirming the crucial role of the fusion module in enhancing the model’s predictive ability.

Table 3.
The results of the ablation experiments.
4.4. Two-Stage Training Experiment
To comprehensively evaluate the effectiveness of the MECCL two-stage training method, we compared the classification performance differences of the MDFormer model before and after the two-stage training on both the full modality dataset and the modality-masked dataset. The experimental results are shown in Table 4. The changes in ROC before and after the two-stage training are shown in Figure 10.

Table 4.
MDFormer two-stage training results.
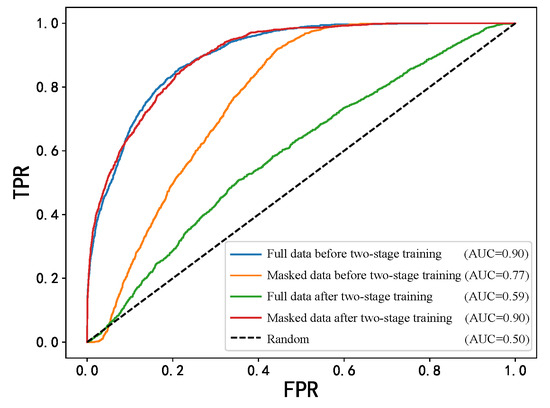
Figure 10.
ROC curves before and after two-stage training.
As can be seen from the table, on the MIMIC dataset, the F1 score of the one-stage model on the original test set is 0.7966, while it drops to only 0.4441 on the masked test set. In contrast, after the MECCL two-stage training, although the F1 score on the original test set remains nearly unchanged at 0.7923, its performance on the masked test set significantly improves to 0.6675, which is about a 50% increase compared to the one-stage model, with an AUC improvement of 29.6%. These results indicate that the two-stage training effectively enhanced the model’s classification ability in the presence of modality missing data.
Meanwhile, the two-stage model’s classification performance on the full modality test set remains almost unchanged, indicating that the MECCL training strategy does not negatively affect the model’s performance with complete modalities. Additionally, the two-stage training further optimized the model’s performance on the modality-masked dataset while freezing modality-specific encoders. This suggests that the model structure of MDFormer has already learned to represent multimodal data in the first-stage training and has extracted robust feature representations. Through modality-masked classification and modality-masked contrastive learning, the model can better identify the important features of existing modalities, and by comparing the representations of modality-missing and complete data, it further aids the model in recovering complete labels from missing data. This training strategy not only improved the model’s classification performance on modality-missing data but also enhanced its generalization ability, allowing it to more flexibly handle data with different modality combinations.
4.5. Visualization Analysis
We performed PCA dimensionality reduction and visualization on the final representations obtained by MDFormer for all cases in the test set, as shown in Figure 11. The reduced-dimensional results reveal a clear clustering of samples from different categories in the two-dimensional space, indicating that MDFormer effectively integrates multimodal medical data.
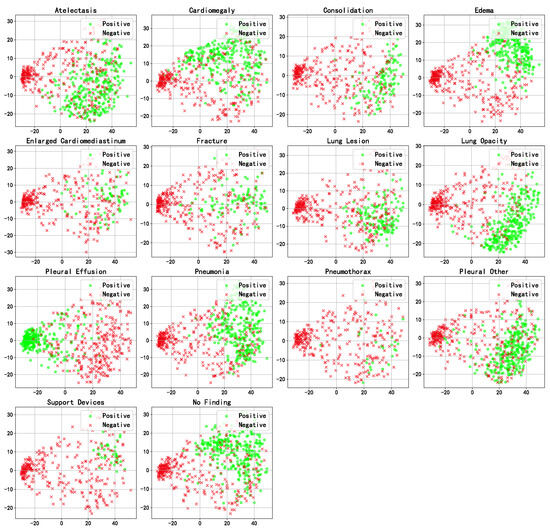
Figure 11.
PCA visualization of the fused representations of samples in the test set.
5. Discussion
Beyond technical advancements, we also deeply recognize the significant importance of applying MDFormer in real clinical environments. In emergency departments or resource-limited medical settings, the ability to make accurate decisions quickly based on incomplete multimodal data is critical for clinical decision-making. Our experimental results show that MDFormer demonstrates strong robustness even under conditions of modality missingness, making it an ideal choice for such application scenarios. Especially in the context of telemedicine or remote regions with scarce medical resources, MDFormer shows great potential for practical deployment. To further enhance MDFormer’s adaptability to real-world clinical settings, we plan to expand our experiments to more comprehensive multimodal datasets (such as CheXpert) and incorporate textual perturbation analysis involving low-quality or non-standard radiology reports. This will allow us to systematically assess the model’s stability and practicality when faced with cross-institutional and heterogeneous data. Future research will also focus on the seamless integration of MDFormer into existing clinical workflows, such as Electronic Health Record (EHR) systems, targeting key tasks like early disease detection and critical case triage. This includes addressing the challenges posed by asynchronous or real-time multimodal data input. Specifically, MDFormer is expected to serve as an intelligent decision support tool within EHR systems, enabling real-time analysis of patient data from various modalities to assist clinicians in identifying early disease signals more accurately, providing diagnostic recommendations, or automatically assessing the urgency of clinical cases. Furthermore, we will explore the potential of MDFormer in disease progression prediction and personalized treatment planning. To meet the practical needs of telemedicine under edge-computing resource constraints, we also plan to conduct research on model lightweighting and deployment optimization. In summary, these efforts will lay a solid foundation for advancing MDFormer from research into clinical application. Future work will primarily focus on the following directions:
- Future work may consider integrating more data modalities, such as genomic data, pathology data, and laboratory test results, to build a more comprehensive multimodal fusion framework for more accurate diagnosis.
- It could also validate the model’s generalization ability on a broader range of multimodal datasets and systematically evaluate its robustness under low-quality or non-standard input conditions.
- Additionally, self-supervised or semi-supervised learning methods can be adopted to train multimodal medical encoders with strong representation abilities, better adapting to the needs of downstream classification tasks.
- Such work can explore lightweight deployment techniques such as model quantization, pruning, and knowledge distillation to enable real-time, resource-efficient inference on edge devices, which is essential for mobile healthcare and remote diagnostic scenarios.
6. Conclusions
This paper first combines the attention mechanism and grouped convolutions to propose a multimodal fusion attention mechanism called MFAttention. MFAttention can not only mine valuable features within each individual modality but also capture crossmodal global semantic information, further improving the effectiveness of multimodal data fusion. Based on this, the paper further constructs a multimodal fusion module, MFTrans, which effectively integrates heterogeneous data from medical images, clinical texts, and vital sign time-series. This process not only enhances the model’s expressive power but also significantly improves the model’s accuracy and stability in disease diagnosis. Experimental comparisons with baseline models on the MIMIC dataset demonstrate that MDFormer performs well in chest disease classification tasks. Additionally, ablation experiments validate the effectiveness of the various modules in MDFormer.
Next, considering the issue of missing modality in practical applications, this paper proposes an innovative two-stage Mask-enhanced Classification and Contrastive Learning training framework, MECCL. Specifically, the second-stage training simultaneously optimizes the classification loss of complete modality data and the classification loss of masked data. By combining contrastive learning, it maximizes the representation consistency between the two, thereby improving the model’s robustness and generalization ability. Furthermore, to balance various losses, the paper introduces a Sigmoid function to construct a dynamic balancing loss weight. The experimental results show that the proposed model not only achieves excellent classification performance when multiple modalities are complete but also maintains good stability and diagnostic effectiveness when facing missing modalities. We will release our code at https://github.com/thulxl/MFTrans upon publication to support reproducibility (accessed on 6 May 2025).
Author Contributions
Conceptualization, X.L., F.P., H.S., S.C., C.L., and T.L.; methodology, X.L., F.P., and H.S.; formal analysis, X.L., F.P., H.S., and S.C.; validation, X.L.; data curation, X.L.; writing—original draft preparation, X.L. and S.C.; writing—review and editing, F.P., H.S., C.L., and T.L.; visualization, X.L. and S.C.; project administration, F.P., H.S., C.L., and T.L. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Data Availability Statement
The datasets used in this work are freely available on public websites: the MIMIC-CXR-JPG dataset (https://physionet.org/content/mimic-cxr-jpg/2.1.0/, accessed on 6 May 2025) and the MIMIC-ED dataset (https://physionet.org/content/mimic-iv-ed/2.2/, accessed on 6 May 2025).
Conflicts of Interest
The authors declare no conflicts of interest.
References
- World Health Organization. Global Health Estimates 2021, 2021. [In press]. Available online: https://www.who.int/data/global-health-estimates (accessed on 6 May 2025).
- Leslie, A.; Jones, A.; Goddard, P. The influence of clinical information on the reporting of CT by radiologists. Br. J. Radiol. 2000, 73, 1052–1055. [Google Scholar] [CrossRef] [PubMed]
- Cohen, M.D. Accuracy of information on imaging requisitions: Does it matter? J. Am. Coll. Radiol. 2007, 4, 617–621. [Google Scholar] [CrossRef] [PubMed]
- Boonn, W.W.; Langlotz, C.P. Radiologist use of and perceived need for patient data access. J. Digit. Imaging 2009, 22, 357–362. [Google Scholar] [CrossRef]
- Li, Y.; Wu, F.X.; Ngom, A. A review on machine learning principles for multi-view biological data integration. Brief. Bioinform. 2018, 19, 325–340. [Google Scholar] [CrossRef]
- Baltrušaitis, T.; Ahuja, C.; Morency, L.P. Multimodal machine learning: A survey and taxonomy. IEEE Trans. Pattern Anal. Mach. Intell. 2018, 41, 423–443. [Google Scholar] [CrossRef]
- Asuntha, A.; Srinivasan, A. Deep learning for lung Cancer detection and classification. Multimed. Tools Appl. 2020, 79, 7731–7762. [Google Scholar] [CrossRef]
- Schroeder, J.D.; Bigolin Lanfredi, R.; Li, T.; Chan, J.; Vachet, C.; Paine III, R.; Srikumar, V.; Tasdizen, T. Prediction of obstructive lung disease from chest radiographs via deep learning trained on pulmonary function data. Int. J. Chronic Obstr. Pulm. Dis. 2020, 15, 3455–3466. [Google Scholar] [CrossRef]
- Pham, T.A.; Hoang, V.D. Chest X-ray image classification using transfer learning and hyperparameter customization for lung disease diagnosis. J. Inf. Telecommun. 2024, 8, 587–601. [Google Scholar] [CrossRef]
- Hussein, F.; Mughaid, A.; AlZu’bi, S.; El-Salhi, S.M.; Abuhaija, B.; Abualigah, L.; Gandomi, A.H. Hybrid clahe-cnn deep neural networks for classifying lung diseases from x-ray acquisitions. Electronics 2022, 11, 3075. [Google Scholar] [CrossRef]
- Mann, M.; Badoni, R.P.; Soni, H.; Al-Shehri, M.; Kaushik, A.C.; Wei, D.Q. Utilization of deep convolutional neural networks for accurate chest X-ray diagnosis and disease detection. Interdiscip. Sci. Comput. Life Sci. 2023, 15, 374–392. [Google Scholar] [CrossRef]
- Chandrashekar Uppin, G.G. Assessing the Efficacy of Transfer Learning in Chest X-ray Image Classification for Respiratory Disease Diagnosis: Focus on COVID-19. Lung Opacity, Viral Pneumonia 2024, 10, 11–20. [Google Scholar]
- Hayat, M.; Ahmad, N.; Nasir, A.; Tariq, Z.A. Hybrid Deep Learning EfficientNetV2 and Vision Transformer (EffNetV2-ViT) Model for Breast Cancer Histopathological Image Classification. IEEE Access 2024, 12, 184119–184131. [Google Scholar] [CrossRef]
- Tariq, Z.; Shah, S.K.; Lee, Y. Lung disease classification using deep convolutional neural network. In Proceedings of the 2019 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), San Diego, CA, USA, 18–21 November 2019; pp. 732–735. [Google Scholar]
- Pham, L.; Phan, H.; Palaniappan, R.; Mertins, A.; McLoughlin, I. CNN-MoE based framework for classification of respiratory anomalies and lung disease detection. IEEE J. Biomed. Health Inform. 2021, 25, 2938–2947. [Google Scholar] [CrossRef] [PubMed]
- Lal, K.N. A lung sound recognition model to diagnoses the respiratory diseases by using transfer learning. Multimed. Tools Appl. 2023, 82, 36615–36631. [Google Scholar] [CrossRef]
- Stahlschmidt, S.R.; Ulfenborg, B.; Synnergren, J. Multimodal deep learning for biomedical data fusion: A review. Brief. Bioinform. 2022, 23, bbab569. [Google Scholar] [CrossRef]
- Niu, S.; Ma, J.; Bai, L.; Wang, Z.; Guo, L.; Yang, X. EHR-KnowGen: Knowledge-enhanced multimodal learning for disease diagnosis generation. Inf. Fusion 2024, 102, 102069. [Google Scholar] [CrossRef]
- Glicksberg, B.S.; Timsina, P.; Patel, D.; Sawant, A.; Vaid, A.; Raut, G.; Charney, A.W.; Apakama, D.; Carr, B.G.; Freeman, R.; et al. Evaluating the accuracy of a state-of-the-art large language model for prediction of admissions from the emergency room. J. Am. Med. Inform. Assoc. 2024, 31, 1921–1928. [Google Scholar] [CrossRef]
- Bichindaritz, I.; Liu, G.; Bartlett, C. Integrative survival analysis of breast cancer with gene expression and DNA methylation data. Bioinformatics 2021, 37, 2601–2608. [Google Scholar] [CrossRef]
- Yan, R.; Zhang, F.; Rao, X.; Lv, Z.; Li, J.; Zhang, L.; Liang, S.; Li, Y.; Ren, F.; Zheng, C.; et al. Richer fusion network for breast cancer classification based on multimodal data. BMC Med. Inform. Decis. Mak. 2021, 21, 1–15. [Google Scholar] [CrossRef]
- Lee, Y.C.; Cha, J.; Shim, I.; Park, W.Y.; Kang, S.W.; Lim, D.H.; Won, H.H. Multimodal deep learning of fundus abnormalities and traditional risk factors for cardiovascular risk prediction. NPJ Digit. Med. 2023, 6, 14. [Google Scholar] [CrossRef]
- Cui, C.; Liu, H.; Liu, Q.; Deng, R.; Asad, Z.; Wang, Y.; Zhao, S.; Yang, H.; Landman, B.A.; Huo, Y. Survival prediction of brain cancer with incomplete radiology, pathology, genomic, and demographic data. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Singapore, 18–22 September 2022; Lecture Notes in Computer Science. Springer: Cham, Switzerland, 2022; pp. 626–635. [Google Scholar]
- Liu, Z.; Wei, J.; Li, R.; Zhou, J. SFusion: Self-attention based n-to-one multimodal fusion block. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Singapore, 18–22 September 2022; Lecture Notes in Computer Science. Springer: Cham, Switzerland, 2023; pp. 159–169. [Google Scholar]
- Al-Tam, R.M.; Al-Hejri, A.M.; Alshamrani, S.S.; Al-antari, M.A.; Narangale, S.M. Multimodal breast cancer hybrid explainable computer-aided diagnosis using medical mammograms and ultrasound Images. Biocybern. Biomed. Eng. 2024, 44, 731–758. [Google Scholar] [CrossRef]
- Ramachandram, D.; Taylor, G.W. Deep multimodal learning: A survey on recent advances and trends. IEEE Signal Process. Mag. 2017, 34, 96–108. [Google Scholar] [CrossRef]
- Soto, J.T.; Weston Hughes, J.; Sanchez, P.A.; Perez, M.; Ouyang, D.; Ashley, E.A. Multimodal deep learning enhances diagnostic precision in left ventricular hypertrophy. Eur. Heart-J. Digit. Health 2022, 3, 380–389. [Google Scholar] [CrossRef]
- El-Ateif, S.; Idri, A. Eye diseases diagnosis using deep learning and multimodal medical eye imaging. Multimed. Tools Appl. 2024, 83, 30773–30818. [Google Scholar] [CrossRef]
- Liu, T.; Huang, J.; Liao, T.; Pu, R.; Liu, S.; Peng, Y. A hybrid deep learning model for predicting molecular subtypes of human breast cancer using multimodal data. Irbm 2022, 43, 62–74. [Google Scholar] [CrossRef]
- Saikia, M.J.; Kuanar, S.; Mahapatra, D.; Faghani, S. Multi-modal ensemble deep learning in head and neck cancer HPV sub-typing. Bioengineering 2023, 11, 13. [Google Scholar] [CrossRef]
- Reda, I.; Khalil, A.; Elmogy, M.; Abou El-Fetouh, A.; Shalaby, A.; Abou El-Ghar, M.; Elmaghraby, A.; Ghazal, M.; El-Baz, A. Deep learning role in early diagnosis of prostate cancer. Technol. Cancer Res. Treat. 2018, 17, 1533034618775530. [Google Scholar] [CrossRef]
- Vaswani, A.; Shazeer, N.; Parmar, N.; Uszkoreit, J.; Jones, L.; Gomez, A.N.; Kaiser, Ł.; Polosukhin, I. Attention is all you need. Adv. Neural Inf. Process. Syst. 2017, 30, 5998–6008. [Google Scholar]
- Nguyen, D.K.; Assran, M.; Jain, U.; Oswald, M.R.; Snoek, C.G.; Chen, X. An image is worth more than 16x16 patches: Exploring transformers on individual pixels. arXiv 2024, arXiv:2406.09415. [Google Scholar]
- Chen, Y.C.; Li, L.; Yu, L.; El Kholy, A.; Ahmed, F.; Gan, Z.; Cheng, Y.; Liu, J. Uniter: Universal image-text representation learning. In Proceedings of the European Conference on Computer Vision, Virtual, 23–28 August 2020; Springer: Cham, Switzerland, 2020; pp. 104–120. [Google Scholar]
- Kim, W.; Son, B.; Kim, I. Vilt: Vision-and-language transformer without convolution or region supervision. In Proceedings of the International Conference on Machine Learning, Virtual, 18–24 July 2021; pp. 5583–5594. [Google Scholar]
- Singh, A.; Hu, R.; Goswami, V.; Couairon, G.; Galuba, W.; Rohrbach, M.; Kiela, D. Flava: A foundational language and vision alignment model. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, New Orleans, LA, USA, 18–24 June 2022; pp. 15638–15650. [Google Scholar]
- Wang, J.; Yang, Z.; Hu, X.; Li, L.; Lin, K.; Gan, Z.; Liu, Z.; Liu, C.; Wang, L. Git: A generative image-to-text transformer for vision and language. arXiv 2022, arXiv:2205.14100. [Google Scholar]
- Liu, S.; Wang, X.; Hou, Y.; Li, G.; Wang, H.; Xu, H.; Xiang, Y.; Tang, B. Multimodal data matters: Language model pre-training over structured and unstructured electronic health records. IEEE J. Biomed. Health Inform. 2022, 27, 504–514. [Google Scholar] [CrossRef] [PubMed]
- Tsai, Y.H.H.; Bai, S.; Liang, P.P.; Kolter, J.Z.; Morency, L.P.; Salakhutdinov, R. Multimodal transformer for unaligned multimodal language sequences. In Proceedings of the Conference Association for Computational Linguistics, Florence, Italy, 28 July–2 August 2019; Volume 2019, p. 6558. [Google Scholar]
- Zhou, H.Y.; Yu, Y.; Wang, C.; Zhang, S.; Gao, Y.; Pan, J.; Shao, J.; Lu, G.; Zhang, K.; Li, W. A transformer-based representation-learning model with unified processing of multimodal input for clinical diagnostics. Nat. Biomed. Eng. 2023, 7, 743–755. [Google Scholar] [CrossRef] [PubMed]
- Xu, T.; Chen, W.; Wang, P.; Wang, F.; Li, H.; Jin, R. Cdtrans: Cross-domain transformer for unsupervised domain adaptation. arXiv 2021, arXiv:2109.06165. [Google Scholar]
- Alayrac, J.B.; Donahue, J.; Luc, P.; Miech, A.; Barr, I.; Hasson, Y.; Lenc, K.; Mensch, A.; Millican, K.; Reynolds, M.; et al. Flamingo: A visual language model for few-shot learning. Adv. Neural Inf. Process. Syst. 2022, 35, 23716–23736. [Google Scholar]
- Moor, M.; Huang, Q.; Wu, S.; Yasunaga, M.; Dalmia, Y.; Leskovec, J.; Zakka, C.; Reis, E.P.; Rajpurkar, P. Med-flamingo: A multimodal medical few-shot learner. In Proceedings of the Machine Learning for Health (ML4H), New Orleans, LA, USA, 10 December 2023; pp. 353–367. [Google Scholar]
- Li, J.; Li, D.; Savarese, S.; Hoi, S. Blip-2: Bootstrapping language-image pre-training with frozen image encoders and large language models. In Proceedings of the International conference on machine learning, Honolulu, HI, USA, 23–29 July 2023; pp. 19730–19742. [Google Scholar]
- Bai, J.; Bai, S.; Yang, S.; Wang, S.; Tan, S.; Wang, P.; Lin, J.; Zhou, C.; Zhou, J. Qwen-VL: A Versatile Vision-Language Model for Understanding, Localization, Text Reading, and Beyond. arXiv 2023, arXiv:2308.12966. [Google Scholar] [CrossRef]
- Johnson, A.E.; Pollard, T.J.; Greenbaum, N.R.; Lungren, M.P.; Deng, C.y.; Peng, Y.; Lu, Z.; Mark, R.G.; Berkowitz, S.J.; Horng, S. MIMIC-CXR-JPG, a large publicly available database of labeled chest radiographs. arXiv 2019, arXiv:1901.07042. [Google Scholar]
- Johnson, A.; Bulgarelli, L.; Pollard, T.; Celi, L.A.; Mark, R.; Horng, S. MIMIC-IV-ED (Version 2.2); PhysioNet: Boston, MA, USA, 2023. [Google Scholar] [CrossRef]
- Boecking, B.; Usuyama, N.; Bannur, S.; Castro, D.C.; Schwaighofer, A.; Hyland, S.; Wetscherek, M.; Naumann, T.; Nori, A.; Alvarez-Valle, J.; et al. Making the most of text semantics to improve biomedical vision–language processing. In Proceedings of the European conference on computer vision, Tel Aviv, Israel, 23–27 October; Springer: Cham, Switzerland, 2022; pp. 1–21. [Google Scholar]
- Zhang, T.; Zhang, Y.; Cao, W.; Bian, J.; Yi, X.; Zheng, S.; Li, J. Less is more: Fast multivariate time series forecasting with light sampling-oriented mlp structures. arXiv 2022, arXiv:2207.01186. [Google Scholar]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. ImageNet classification with deep convolutional neural networks. Commun. ACM 2017, 60, 84–90. [Google Scholar] [CrossRef]
- Huang, G.; Liu, Z.; Van Der Maaten, L.; Weinberger, K.Q. Densely connected convolutional networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, US, 21–26 July 2017; pp. 4700–4708. [Google Scholar]
- Yasunaga, M.; Leskovec, J.; Liang, P. Linkbert: Pretraining language models with document links. arXiv 2022, arXiv:2203.15827. [Google Scholar]
- Zhou, H.; Zhang, S.; Peng, J.; Zhang, S.; Li, J.; Xiong, H.; Zhang, W. Informer: Beyond efficient transformer for long sequence time-series forecasting. In Proceedings of the AAAI Conference on Artificial Intelligence, Virtual, 2–9 February 2021; Volume 35, pp. 11106–11115. [Google Scholar]
- Hayat, N.; Geras, K.J.; Shamout, F.E. MedFuse: Multi-modal fusion with clinical time-series data and chest X-ray images. In Proceedings of the Machine Learning for Healthcare Conference, Virtual, 28 November 2022; pp. 479–503. [Google Scholar]
- Li, J.; Selvaraju, R.; Gotmare, A.; Joty, S.; Xiong, C.; Hoi, S.C.H. Align before fuse: Vision and language representation learning with momentum distillation. Adv. Neural Inf. Process. Syst. 2021, 34, 9694–9705. [Google Scholar]
- Wang, Z.; Wu, Z.; Agarwal, D.; Sun, J. Medclip: Contrastive learning from unpaired medical images and text. In Proceedings of the Conference on Empirical Methods in Natural Language Processing. Conference on Empirical Methods in Natural Language Processing, Abu Dhabi, United Arab Emirates, 7–11 December 2022; Volume 2022, p. 3876. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).