1. Introduction
High-Performance Computing (HPC) is essential for modern scientific research. Fields such as physics, genomics, and economics rely on HPC to drive their research forward. During the COVID-19 pandemic, HPC applications in genomics played a vital role, including genome assembly, which provided crucial information on genome-wide mutation patterns of the virus. This has been instrumental in characterizing different concern variants [
1].
HPC is crucial for many industries and society, but the need for large-scale data collection and processing comes with costs. The current approach to HPC system design is no longer feasible for the exascale era, where computers must be able to perform over
calculations per second. Energy efficiency is a critical factor in ensuring the sustainability of future exascale HPC systems. The most advanced and energy-efficient HPC systems utilize accelerators integrated with general-purpose processors to achieve the optimal energy consumption per computation [
2,
3,
4]. Developing a robust European HPC supply chain with independent components and technologies is crucial for achieving strategic autonomy and digital sovereignty in Europe [
5]. To address the exascale computing needs and digital sovereignty, the European Union (EU) has been supporting the strategic European Processor Initiative (EPI) project [
6,
7] to develop critical components and make a successful transition towards exascale while fostering a world-class, competitive, and innovative supercomputing ecosystem across Europe.
One promising approach for achieving exascale computing is using novel ARM HPC processor architectures. The latest Top500 list [
4] demonstrates the potential of ARM-based architectures in supercomputing. The Fugaku supercomputer, featuring ARM-based A64FX processors from Fujitsu, is ranked second on the TOP500 list with
Petaflops/s of peak performance in June 2023. New ARM HPC architectures are expected to surpass this performance in the future.
Single Instruction, Multiple Data (SIMD) in Central Processing Units (CPUs) is crucial for achieving high performance with improved power efficiency. By utilizing the power of parallelism, SIMD allows for faster processing of large amounts of data, making it an essential component for future scientific computing with a higher ratio of FLOPS per watt [
8]. To facilitate the deployment of SIMD on various CPU architectures, we have utilized SIMD Everywhere (SIMDe) in our research. SIMDe automatically transcodes previously vectorized source code to other supported instruction set architectures, allowing for efficient porting of code without sacrificing functionality [
9]. Furthermore, if the hardware supports the native implementation, there is no performance penalty (e.g., SSE/AVX will run at full speed on x86 and ARM NEON).
In this paper, we optimized and ported GASM (Genome ASseMbler), a genome assembly pipeline for ARM CPUs. The GASM pipeline includes minimap2, miniasm, and Racon, with the latter being the most suitable for hardware acceleration due to the implementation of Partial Order Alignment (POA) algorithm using SSE4, AVX512 for CPUs, and CUDA for Graphical Processing Units (GPUs). Racon, also known as rapid consensus, is a consensus module that allows for constructing high-accuracy genome sequences without needing an error-correction step [
10]. We will compare the performance gains achieved by rewriting the Racon code using intrinsics and autovectorization with both
g++ and
armclang++ compilers. The key contributions of this paper are as follows:
We updated the Racon source code to utilize two intrinsic types: Scalable Vector Extension (SVE) intrinsics, which were manually written, and NEON intrinsics, which were generated using SIMDe.
We compared the performance of auto-vectorized versus nonvectorized instructions on the g++ and armclang++ compilers and compared the percentages of emulated instructions relative to the total number of instructions in cases of auto-vectorized versus manually vectorized code and on different register widths in SVE.
We also compared the performance gain of manually vectorized versus nonvectorized instructions on the armclang++ and gcc compiler.
Finally, we present the design of the CPU dispatcher included in the Racon consensus module, which enables the automatic selection of the fastest instruction set supported by the utilized CPU.
The remainder of this paper is organized as follows. In
Section 2, we present an overview of relevant literature and previous work within the realm of ARM-based High-Performance Computing (HPC) and genome assembly.
Section 3 provides background information and motivation for this study, emphasizing the current state of the art in these domains. It focuses on ARM intrinsics, specifically ARM NEON and ARM SVE, and their significance in optimizing genome assembly algorithms. Additionally, we introduce the GASM pipeline and its key components, notably highlighting the Racon module, which is the prime focus of our optimization endeavors.
Section 4 delves into the specifics of our code optimization process, detailing the utilization of NEON and SVE and elucidating the performance improvements achieved through these optimization techniques. Furthermore, this section features a comparison between hand-ported and auto-vectorized code.
Section 5 elaborates on our approach to devising a CPU dispatcher capable of automatically selecting the optimal instruction set for a given architecture. This feature facilitates efficient and adaptable execution of the GASM pipeline.
Section 6 discusses the measured results and provides insights derived from these outcomes. Finally,
Section 7 concludes the paper and outlines potential avenues for future work in this domain.
2. Related Work
There has been a growing interest in using HPC systems for genome assembly in recent years due to their ability to provide highly accurate analysis while meeting scalability and reliability targets [
11]. For example, Angelova et al. [
12] introduced the SnakeCube, a de novo genome assembly pipeline that can be run on an isolated environment and scaled to HPC domains. The pipeline was tested and benchmarked using the Zorba HPC cluster, which consists of 328 cores and 2.3 TB of memory from the Institute of Marine Biology, Biotechnology, and Aquaculture [
13]. In addition, Castrignano et al. [
14] described the HPC system CINECA, which offers streamlined access for bioinformatics researchers. Using CINECA, investigators have significantly reduced the computing times for their research projects, even when dealing with large amounts of data. Overall, these studies demonstrate the potential of HPC systems for genome assembly and highlight the importance of utilizing such resources in this field.
In a recent study, Jacob et al. [
15] demonstrated using SIMD in genome sequencing by implementing the Smith-Waterman algorithm on Intel Pentium III and 4 processors provided by the T. S. Santhanam Computing Centre, Tamil Nadu, India. Although this approach resulted in significant speedups, the resulting code was not portable and would not be compatible with modern 64-bit processors. Gao et al. [
16] proposed a SIMD-based library for POA using adaptive banded dynamic programming. As POA is also used in long-read sequencing, the results of this study are relevant to the current field. Their algorithm, which utilized SIMD parallelization, achieved speeds that were 2.7–9.5 times faster than SPOA (SIMD POA) on NanoSim emulated sequences.
Previous studies have focused on genome assembly, high-performance computing, and SIMD techniques. However, none have addressed using ARM-based HPC systems and SIMD for parallel genome assembly. This gap in the literature will be addressed in our study.
3. Background
This section provides background on ARM-based HPC systems and their potential for improving computing resources and power efficiency. We also discuss the current state of the art in genome assembly and explore ways to accelerate these algorithms. By considering these factors, we aim to provide a comprehensive overview of the potential of ARM-based systems in genome assembly.
3.1. Arm-Based HPC
ARM-based processors are a viable option for HPC systems, offering performance comparable to high-end x86-based processors. One example of an ARM-based CPU designed explicitly for HPC is the Marvell ThunderX2 (TX2), which has been deployed on several systems offering many cores with high memory bandwidth. However, its short 128-bit vector size makes it less suitable for applications requiring intensive computation [
17]. Despite this limitation, its high memory bandwidth makes it well-suited for memory-intensive applications [
18].
The Fugaku system, built by Fujitsu at the Riken Center for Computational Science in Kobe, Japan, deployed the new A64FX CPU which represents an improvement over the TX2. The A64FX offers impressive capabilities, including a peak memory bandwidth of 1 TB/s and a 512-bit vector size based on SVE. These improvements helped Fugaku achieve the top ranking on the TOP500 list until 2022 [
19,
20]. In addition, the system’s Neoverse V1 architecture enables the construction of exascale-class CPUs for artificial intelligence and other HPC applications, providing state-of-the-art power and performance efficiency.
The Mont-Blanc project is another example of an ARM-based HPC system that focuses on low-power technology. This project is pioneering the development of energy-efficient HPC systems using embedded and mobile devices. It has achieved an impressive exascale-level power efficiency of 50 Gflops/Watt at the processor level [
21].
3.2. Genome Assembling
Genomics studies an organism’s complete genetic material and genome through DNA sequencing methods and bioinformatics. This field aims to sequence, assemble, and analyze the structure and function of genomes to gain a better understanding of biological processes [
22]. DNA sequencing involves determining the precise order of nucleotides in a DNA molecule by breaking it down into smaller segments and sequencing those segments to obtain reads. This information can be used to map and compare DNA sequences within a cell, leading to new insights into biological processes.
Current DNA sequencing technology is limited in its ability to read entire genomes as a continuous sequence, producing only short reads. To overcome this limitation, sequence assembly algorithms merge these fragments to reconstruct the original DNA sequence [
23]. There are two main approaches to genome assembly: (1) de novo assembly, which is used for genomes that do not have a previously sequenced reference, and (2) comparative assembly, which uses existing sequences as a reference point during the assembly process [
24].
In this study, we explore the use of de novo genome assembly from long uncorrected reads to overcome the challenges posed by high error rates in long reads. Through genome polishing, we aim to improve annotation quality and enhance the performance of advanced scaffolding technologies. However, we acknowledge that the polishing process is computationally demanding and time-consuming [
25]. We focus on developing efficient and effective methods for genome polishing to address these limitations.
3.3. Accelerating Genome Assembling
The acceleration of the genome assembly process has been a widely researched topic, with numerous studies focused on improving the speed and accuracy of this complex task. One notable example is the work of DiGenova et al. [
26], who proposed the WNGAN algorithm for de novo assembly of human genomes and utilized SIMD intrinsics to accelerate the computation of the POA graph.
The SWPepNovo algorithm proposed by Li et al. [
27] leverages parallel computing techniques for efficient de novo peptide sequencing. This demonstrates the ongoing efforts to improve genome assembly technology and its applications in various fields. In their large-scale protein analysis, the SW26010 HPC system was used with asynchronous task transfers and optimization strategies to reduce communication costs between management and computing elements. Using these techniques and balancing the workload between computing units, they achieved a ten-fold speedup compared to the unoptimized version. These technological advancements are crucial for improving the accuracy and efficiency of genome assembly.
In [
28], the authors demonstrated the potential for improving the performance of the Smith-Waterman sequence alignment algorithm using SIMD vector extensions in ARM processors. By leveraging the benefits of vector-agnostic code and implementing SVE, they achieved an impressive 18 times speedup over the baseline NEON implementation and 54 times speedup over a scalar CPU implementation. These results highlight the potential for using SVE in ARM processors to accelerate computational biology applications.
Long-read genome sequencing offers highly contiguous genome assemblies but often comes with a high error rate (>5%) that requires resource-intensive error-correction steps. To address resource limitation, Li et al. [
29] introduced a new assembly approach called
miniasm, which can generate long reads without the need for error correction but at the cost of an increased error rate. However, this approach has the downside of a 10-fold higher error rate than other methods [
30]. To address the challenges of
miniasm, Vaser et al. [
10] developed a new module called Racon (rapid consensus) that can be used in combination with miniasm to achieve efficient genomic sequencing with high accuracy without the need for error correction. Additionally, the authors implemented SIMD intrinsics in the Racon module to reduce its time complexity. This approach is effective in generating high-quality genome assemblies with long reads.
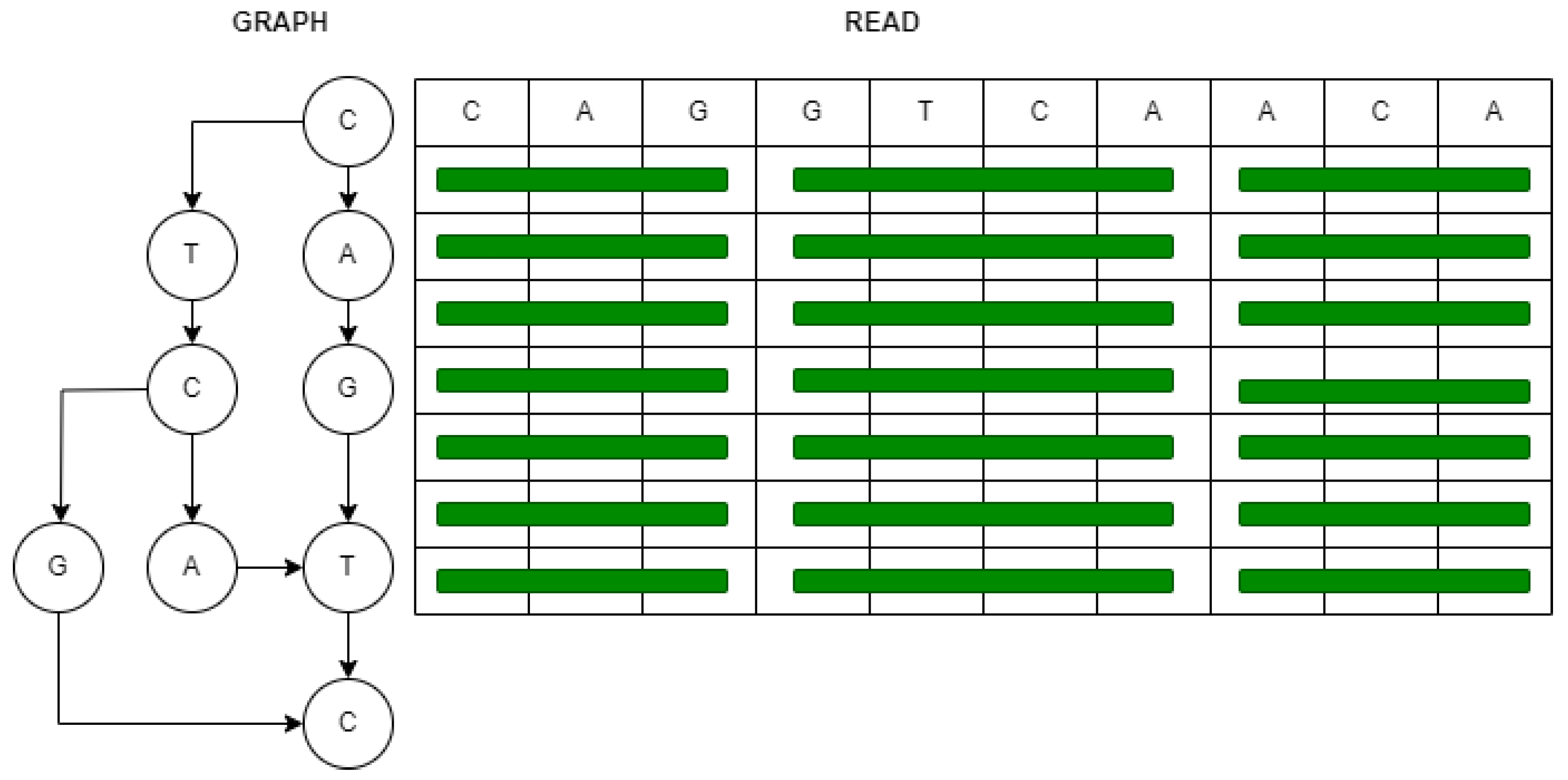
Figure 1 illustrates the utilization of SIMD intrinsics for the vectorization of genome sequence reads. In this depiction, two dimensions are employed: the
read dimension signifies SIMD vectors aligned parallel to the query sequence, while the
graph dimension represents a directed acyclic graph. In this graph, nodes correspond to bases for input sequences, and edges indicate whether two bases are neighbors in any sequence [
10].
3.4. ARM Intrinsics
This section provides an in-depth analysis of the ARM NEON and ARM SVE intrinsics, examining their capabilities and limitations. By exploring these features, we aim to comprehensively understand how these intrinsics can optimize code performance on ARM architecture.
3.4.1. ARM NEON
CPUs with SIMD instruction sets, such as NEON instructions in the ARM Cortex-A8, can significantly enhance parallel processing performance in low-power environments. These instructions, designed specifically for multimedia processing, allow concurrent processing of multiple data streams, improving overall application performance. However, it is important to note that using NEON in applications without significant parallelism may not be beneficial, as it can lead to trade-offs regarding power consumption, execution time, and code size [
31].
3.4.2. ARM SVE
One effective strategy for increasing the computing speed of HPC systems is the introduction of multiple levels of parallelism, including data-level parallelism through code vectorization. Vector processors, in contrast to scalar processors, support SIMD operations using vectors rather than individual elements. The ARM SVE architecture, in particular, offers a unique advantage with its variable-length vector registers, allowing the code to adapt to the current vector length at runtime through Vector Length Agnostic (VLA) programming. This allows for a wide range of vector lengths, from 128 to 2048 bits in 128-bit increments [
32].
The ARM C Language Extensions (ACLE) provide access to the features of SIMD extensions in the form of special built-in functions handled by the compiler. These functions, known as SVE intrinsics, allow efficient use of the SVE hardware in C/C++ programs. However, it should be noted that the variable size of SVE ACLE data types (referred to as
sizeless structs) may prevent their use in certain contexts, such as members of structures or classes, as static variables, and as arguments to the
sizeof function [
33].
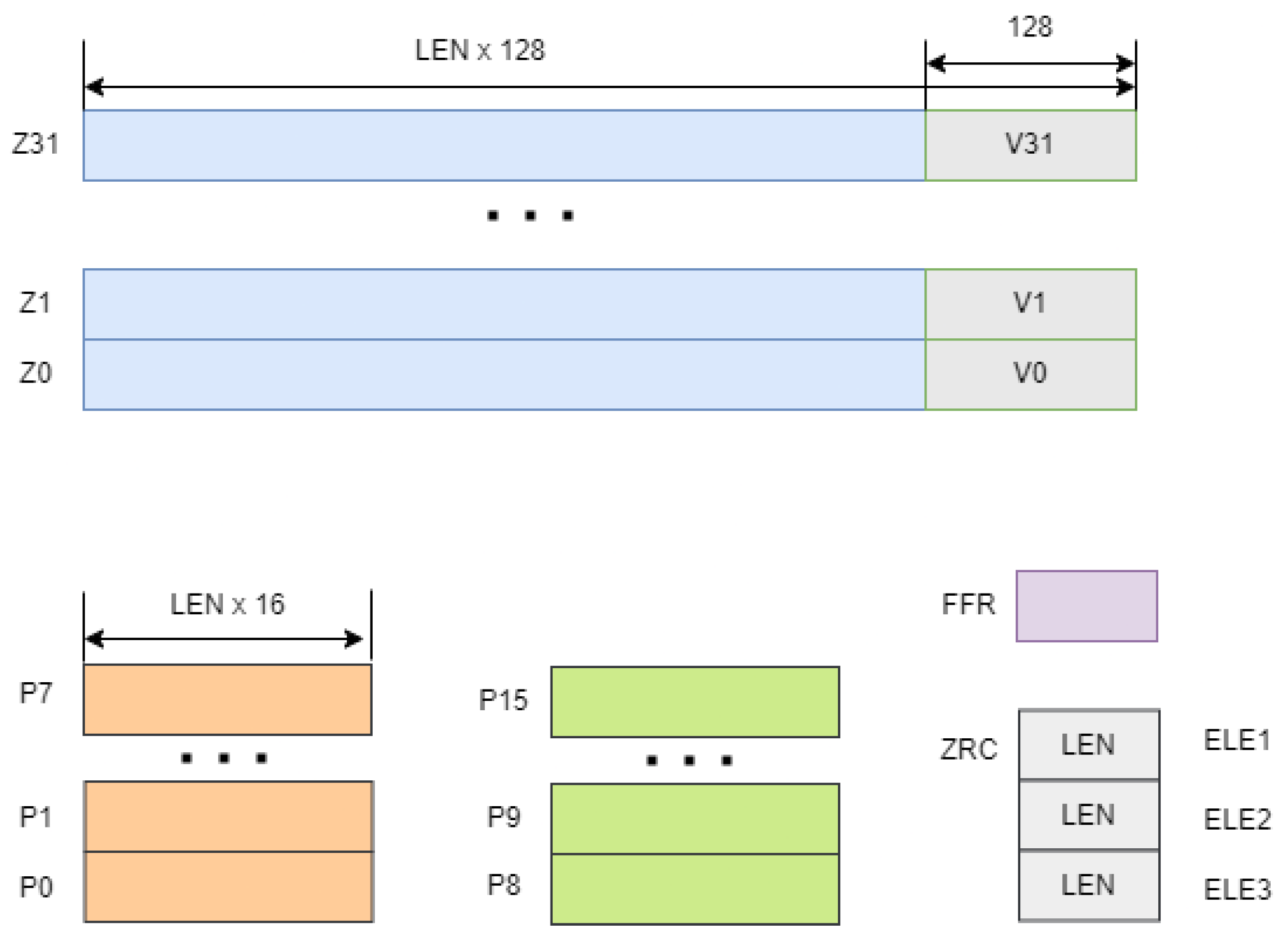
Figure 2 illustrates the SVE vector (
Z) and predicate (
P) registers. The SVE architecture includes 32 vector registers, ranging from
to
, with a size of LEN
bits. The supported vector length,
, is unique for each machine and is implicitly used in the instruction set, which is stored in the system registry. In addition, each vector register contains a corresponding NEON SIMD register in its lower 128 bits, making NEON a subset of SVE. Predicate registers, denoted from
to
, specify the elements that will be operated on in the vector registers [
34].
3.5. GASM
GASM (Genome ASseMbly) is a rapid de novo assembly pipeline consisting of several widely used tools:
minimap2 [
35]—a general-purpose alignment program for mapping DNA or large mRNA sequences against an extensive reference database.
miniasm [
29]—de novo assembler that maps and assembles Single Molecule Real-Time (SMRT) and Oxford Nanopore Technologies (ONT) reads without an error-correction stage.
Racon [
10]—platform-independent consensus module for long and erroneous reads used as an error-correction tool. The goal of Racon is to generate genomic consensus, which is of similar or better quality compared to the output generated by assembly methods, which employ both error-correction and consensus steps while providing a speedup of several times compared to those methods.
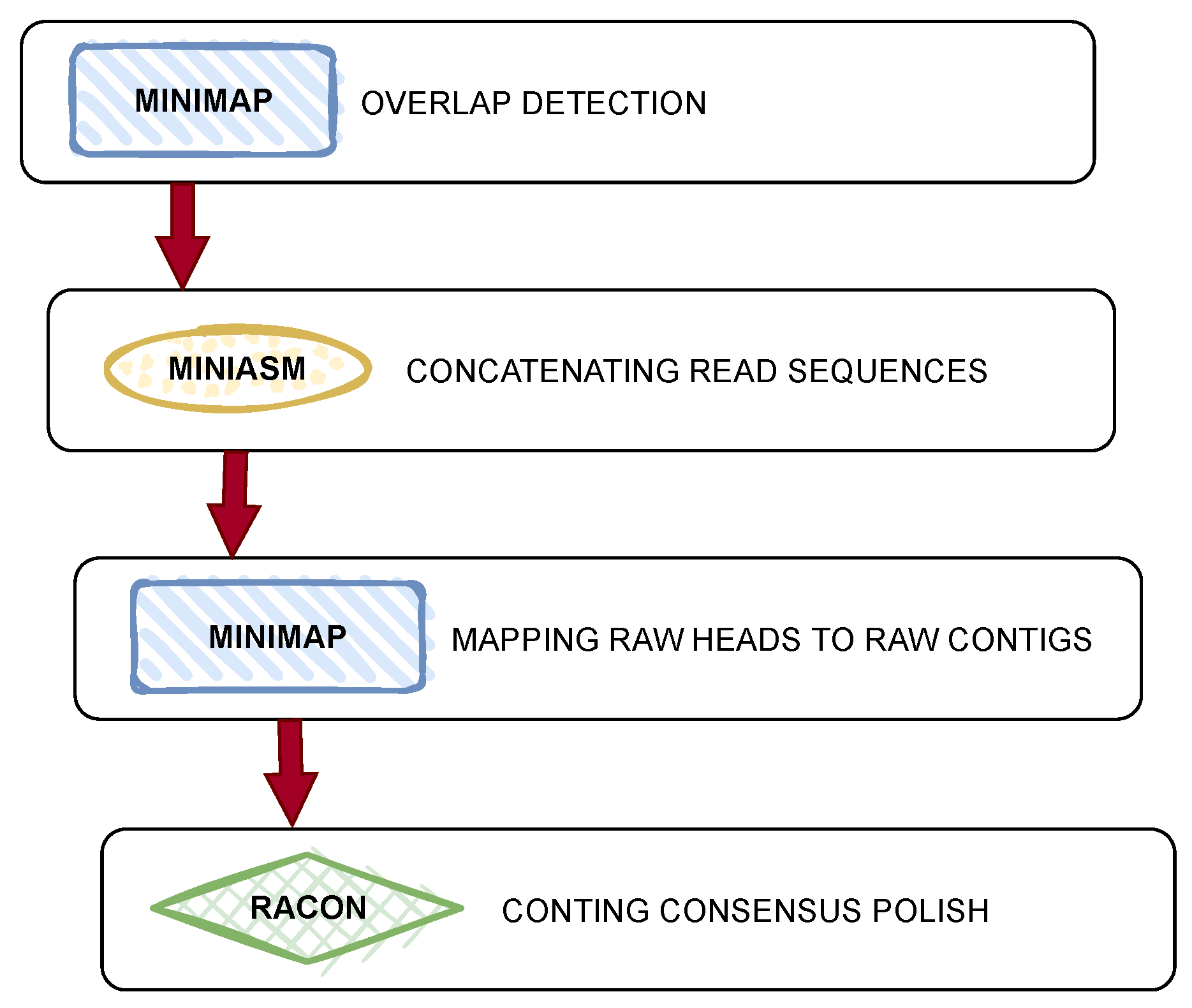
As shown in
Figure 3, the GASM pipeline consists of four key steps. In the first step, minimap2 is used to detect overlaps in the sequence. Then, miniasm concatenates the read sequences to form contigs. In the third step, minimap2 maps the raw heads of the sequences to the raw contigs. Finally, Racon is used to polish the contig consensus and generate a high-quality genomic consensus. This process allows for rapid de novo assembly of SMRT and ONT reads without error correction.
All experiments were conducted using the Racon module, developed from scratch by the University of Zagreb, and depend only on portable open-source third-party libraries. The module is written in C/C++ and can be compiled for any operating system with a C/C++ compiler. In addition, Racon employs the POA algorithm, which has several implementations in the SPOA library, including Single Instruction Single Data (SISD), SIMD in SSE4.1 and AVX2, and a CUDA implementation for General-Purpose GPU (GPGPU) processing.
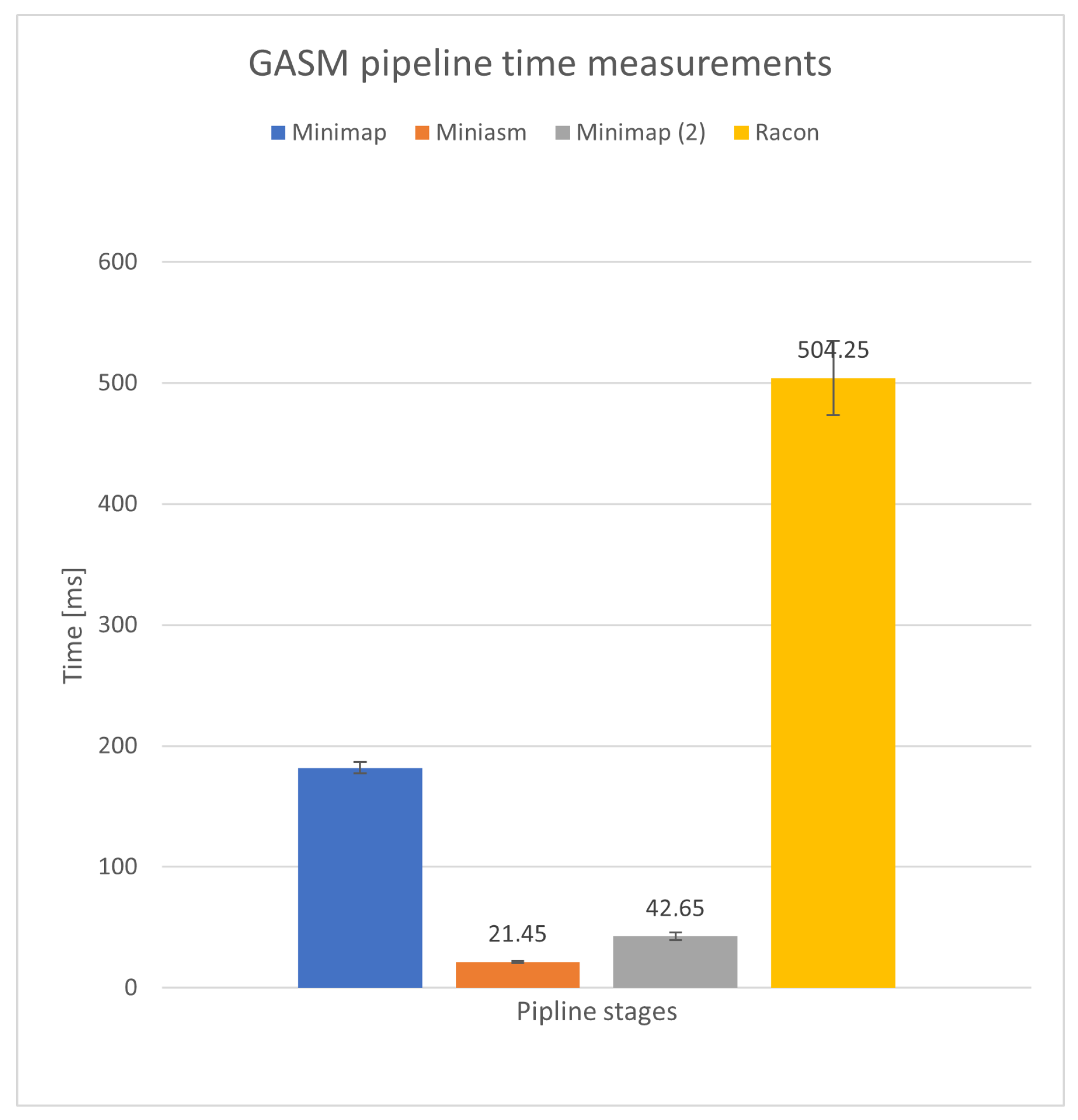
Our decision to focus on Racon in our experiments was based on several factors. First, profiling showed that the majority (on average
) of time in the pipeline was spent on the polishing step in Racon, indicating its potential impact on overall performance.
Figure 4 shows the average execution in each phase of the GASM pipeline measured over
iterations on a
Tiny MB dataset. The longest measured times were in the Racon step with average execution of
ms. Second, Racon utilizes the POA algorithm, which is implemented in the SPOA library and is easily parallelizable. This allows for more flexibility in experimentation and optimization. Additionally, the SPOA library has several implementations of the POA algorithm, including nonvectorized and vectorized versions for different instruction sets. This is more convenient than the parallelization options available in Minimap2, which only offers SSE2 and SSE4.1 implementations of SIMD instructions.
The combination of Racon and miniasm has been shown to generate consensus genomes with a quality comparable to or better than state-of-the-art methods but with significantly faster processing times. In this study, we successfully adapted Racon for ARM CPUs and tested its performance using both SISD and SIMD implementations. Our results demonstrate the potential of this approach for improving the efficiency and speed of genome assembly.
All experiments were conducted on an Ubuntu-based system with two six-core Intel Xeon E5645 CPUs at 2.4 GHz.
3.6. Porting GASM to ARM Using Emulator
For porting GASM to the ARM architecture, we used the armclang++ compiler to recompile the entire suite. We utilized the Racon SISD implementation for autovectorization with both NEON and SVE. All tests in this section were conducted on the armie emulator.
Racon is designed to be run as a multi-threaded program, which is how it was tested and evaluated. Additionally, since multithreading does not impact vectorization, speed improvements achieved by implementing multiple threads can be applied to nonvectorized and vectorized implementations. The configuration of Racon used in our testing is shown in
Table 1. This configuration is standard, except for the number of threads.
We have used two different genomic problems as input:
Tiny: standard Racon test problem, uncompressed reads size: MB
Medium: Escherichia coli str. K-12 substr. MG1655 (E. coli), uncompressed reads size: MB
Using two datasets with varying read sizes allows for a deeper understanding of the algorithm’s scaling properties regarding execution time. Additionally, since some experiments are run on the armie emulator, the size of the datasets is limited to ensure reasonable execution times. This results in the workload being compute-bound in our experiments. However, if larger datasets were used, the workload would be memory-bound.
When possible, utilizing both multithreading and SIMD techniques also significantly improves performance [
36]. Therefore, our experiments utilized 24 threads to achieve the best performance.
Table 2 describes all parameters of the datasets.
Table 3 presents the compiler and application-specific flags used in tests.
3.7. Porting GASM to ARM NEON
To evaluate the impact of ARM vectorization on the GASM pipeline, we initially focused on the ARM NEON SIMD architecture extension, as the ARM cores on the ARM Neoverse N1 platform do not support SVE instructions and only provide SVE emulation. By utilizing the NEON extension, we could measure the real-time performance benefits of vectorization without the added overhead of emulation. Although SVE is not an extension of NEON, the hardware logic for SVE overlays the NEON hardware implementation, allowing for some comparison between the two.
Implementing Racon’s POA using SIMD has allowed us to utilize the SIMDe library to port the SSE4.1 implementation to NEON intrinsics automatically. We then evaluated the performance of the recompiled application using two compilers (
g++ and
armclang++), with the latter being used with the flag
-march=armv8-a+simd -O3. The evaluation was performed on two datasets (
Tiny and
Medium), and the execution times are presented in
Table 4.
In the first case, where the intrinsics were set to None, the compiler automatically optimized the code by utilizing the intrinsics. In the second case, we utilized custom functions in the code specific to the ARM NEON intrinsics. From the results, the armclang++ compiler does a good job at optimizing the code on its own, unlike g++, where the execution times are significantly higher with automatic code optimization.
The speedups achieved are shown in
Table 5. The most significant improvement was observed on the
Medium dataset using the
g++ compiler, while the least improvement was seen on the same dataset with the
armclang++ compiler. This is because
armclang++ is better at optimizing the code. Thus, manual optimization did not yield significant improvements.
Utilizing SIMDe
To vectorize code and transcode the instruction set to another supported architecture, we utilized SIMDe. With SIMDe, we could automatically port the SSE4.1 implementation to NEON intrinsics without any changes to the source code. SIMDe identifies the equivalent function, such as _mm_add_ps from SSE, and implements it using vaddq_ps on the NEON. If there is no equivalent function, SIMDe will attempt to find the best-suited function, focusing on maintaining high execution speed. Implementing SIMDe at the source level was straightforward; we simply defined the SIMDE_ENABLE_NATIVE_ALIASES macro and included SIMDe headers simde/x86/sse4.1.h instead of smmintrin.h headers.
3.8. Porting to ARM SVE
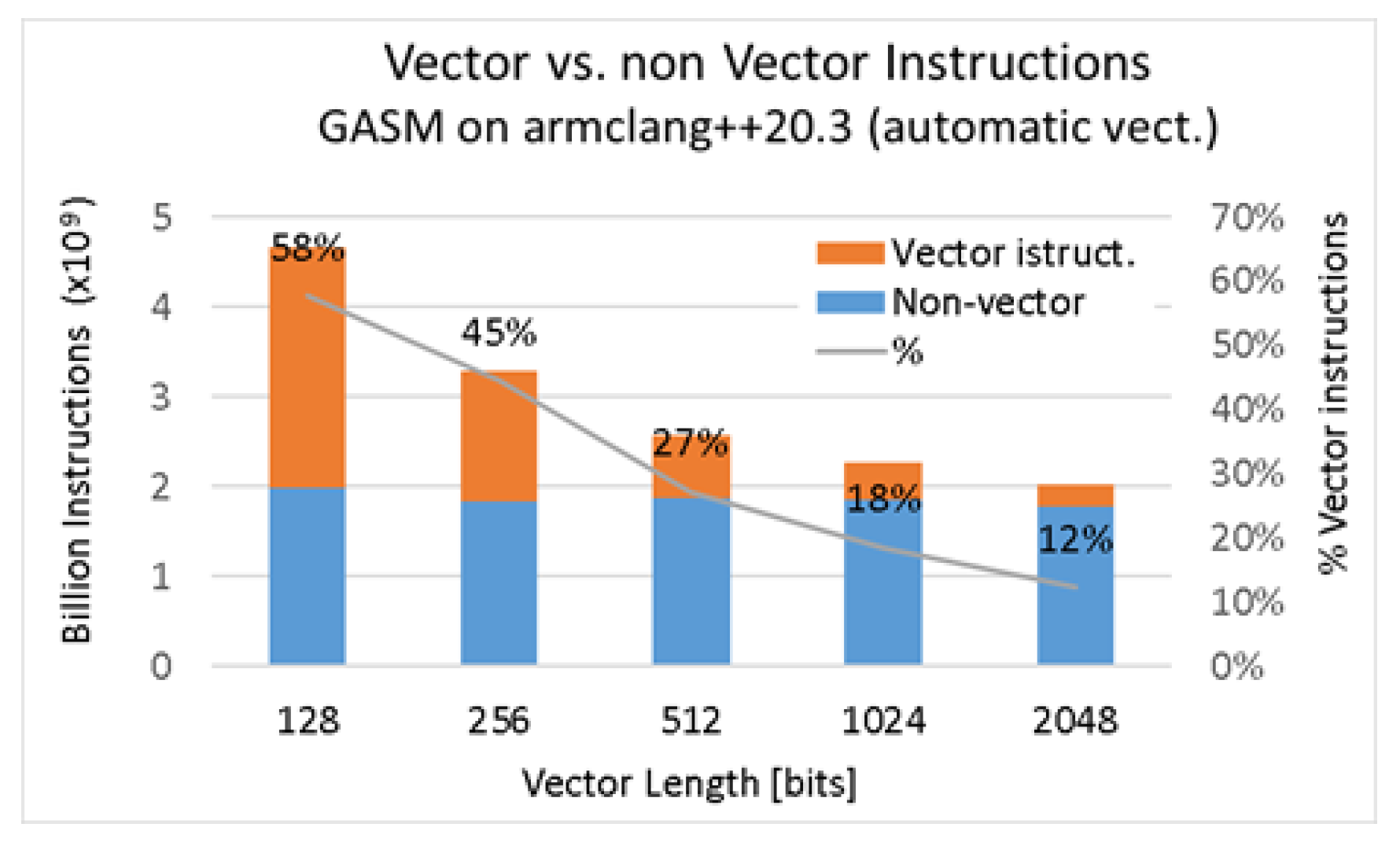
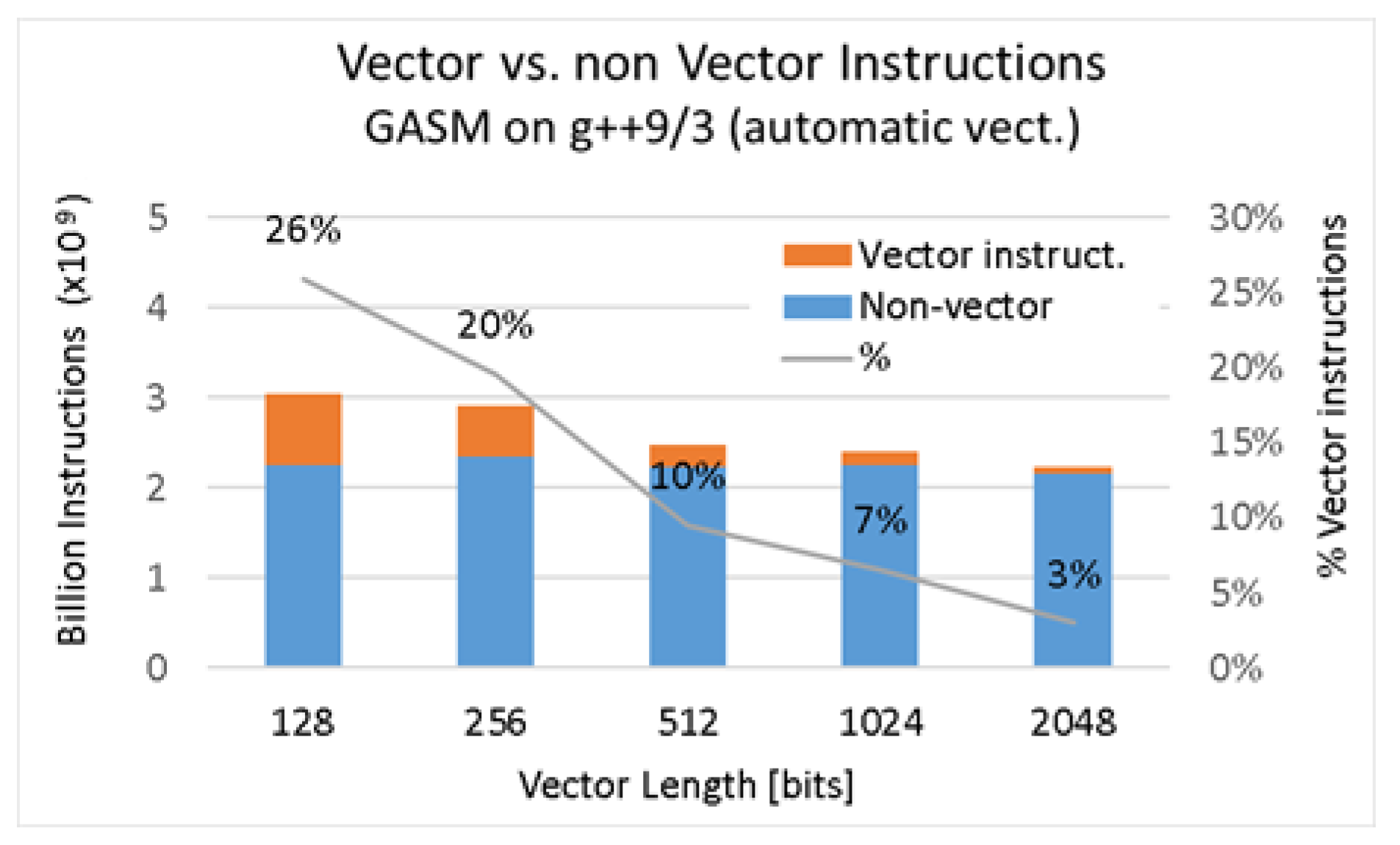
Porting GASM to the SVE architecture was straightforward due to the project’s use of autovectorization, allowing both the armclang++ and g++ compilers (with the -march=armv8-a+sve -O3 flag) to vectorize the entire application automatically. The number and percentage of emulated (vectorized) instructions were measured using the ARM Instruction Emulator (armie). Due to the slower execution time of armie, we performed this test only on the Tiny dataset. The execution time for Racon on the Tiny dataset was approximately s. In comparison, the execution time for the SVE in armie was approximately 350 s, which is approximately 700 times slower. However, since this is just an emulation, comparisons of processing times and speedups are not included in this analysis.
The high percentages of emulated instructions relative to the total number of instructions for both
g++ and
armclang++ indicate that these compilers can vectorize code, even for basic SISD implementations effectively. When comparing the two,
armclang++ shows superior performance in terms of vectorization, resulting in higher percentages. Additionally, in some cases,
armclang++ generates more instructions. The percentages of emulated instructions in the total number of instructions for
armclang++ with autovectorization are presented in
Table 6 and
Figure 5, while the results for
g++ with autovectorization can be found in
Table 7 and
Figure 6.
In addition, we have manually ported parts of SPOA, an implementation of the
Needleman–Wunsch algorithm with linear gap penalties based on SSE/AVX2, to improve its performance. The exact configuration uses Racon with default settings.
Table 8 and
Figure 7 show the percentage of emulated instructions relative to the total number of instructions using manual vectorization on the
armclang++ compiler.
The results show that the manually ported code performs better at lower register sizes, as it was designed for 128–256 registers with SSE&AVX widths. However, for 512 or greater register sizes, the armclang++ compiler is more effective in vectorizing the code. This suggests that a manually implemented approach from scratch may yield better performance than porting concepts from existing code vectorized for a specific architecture.
4. Porting GASM to ARM Using Real Hardware
In
Section 3.1, we studied the vectorization potential of the GASM application using ARM and SVE vector extensions via the
armie emulator. Our results indicate that a significant portion of the application (up to
) can be vectorized using autovectorization. Additionally, manual use of SVE intrinsics can improve performance by greatly reducing the number of instructions. We further tested these assumptions on real hardware that supports SVE. Only the Racon component of the pipeline was considered in these experiments.
The experiments for this study were conducted on the BSC CTE-ARM A64FX cluster, which consists of 192 computer nodes. Each node has the following configuration:
FX100 CPU (ARMv8-A + SVE) – GHz (4 sockets and 12 CPU/socket, total 48 CPUs per node)
32 GB of main memory HBM2
Single Port Infiniband EDR
TofuD network
The A64FX supports three different operation modes—512-bit, 256-bit, and 128-bit wide—and the GASM is designed to support multithreading. Therefore, all experiments were performed using multiple threads.
To evaluate the effectiveness of vectorization techniques, we tested and compared five implementations: a baseline implementation without vectorization, NEON autovectorization, NEON intrinsics using the SIMDe library, SVE autovectorization, and SVE intrinsics. All implementations were built using the armclang compiler. The results of these tests allowed us to determine the most effective vectorization method for our specific application.
The performance results of comparing NEON autovectorization and NEON intrinsics with the baseline implementation are presented in
Table 9. In contrast, the results of comparing SVE autovectorization and SVE intrinsics with the baseline are shown in
Table 10. Our analysis indicates that SVE consistently outperforms NEON in all scenarios. In particular, the SVE autovectorization is faster than the baseline for all input sizes, while the NEON autovectorization is slower than the baseline. Manual vectorization using SVE or NEON also provides significant performance improvements compared to autovectorization.
Our results demonstrate that exporting a few kernels in the application to SVE or NEON intrinsics can improve performance. In the case of NEON, manual vectorization achieved an average speedup of , compared to for autovectorization. For SVE manual vectorization, the average speedup was , the same as that achieved by SVE autovectorization. Furthermore, our findings indicate that the Tiny instance is not as significant as the Medium instance yielded substantial improvement using manual intrinsics. However, further comparisons with larger register widths are needed.
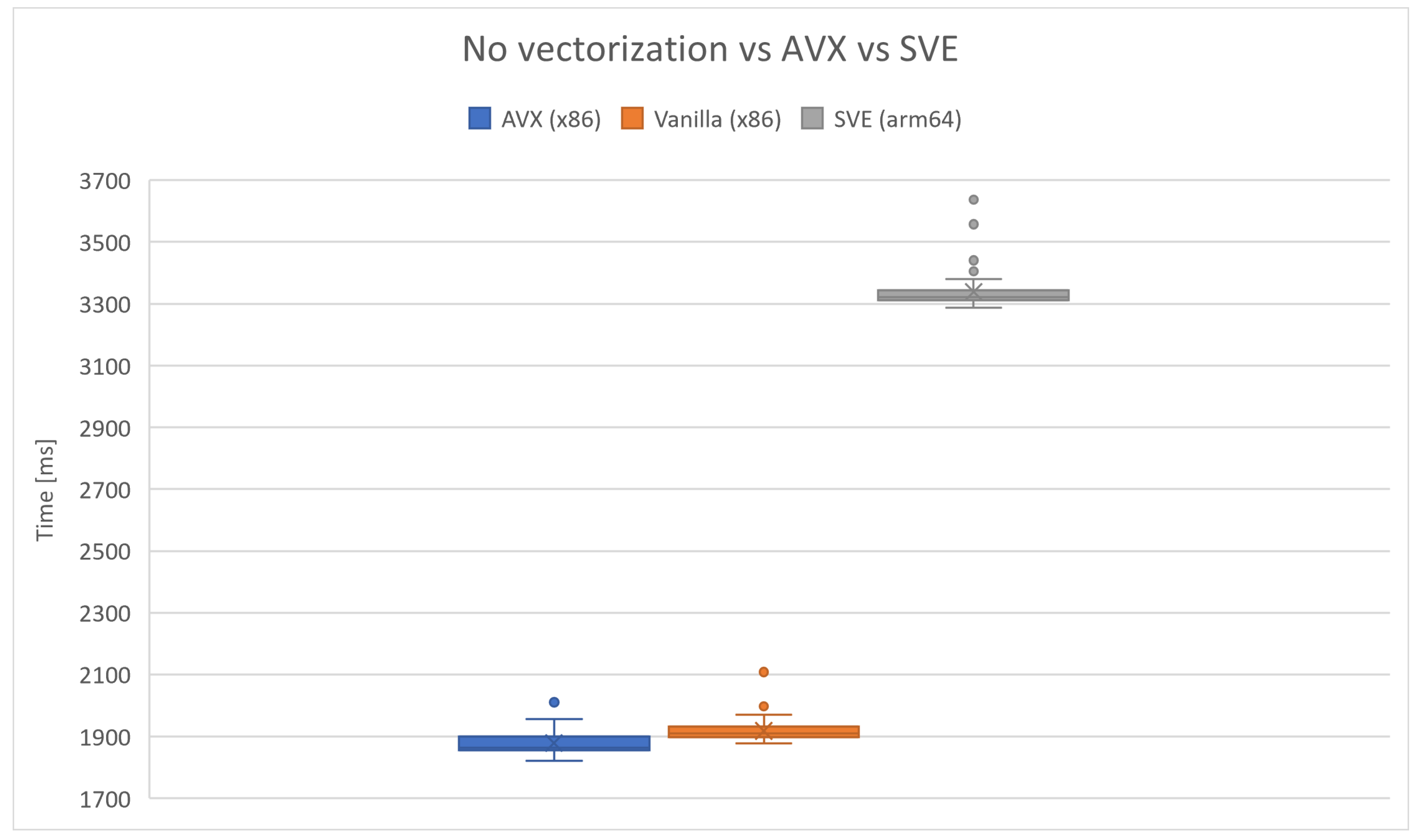
Figure 8 demonstrates the contrast between Racon execution on a
Tiny dataset using nonvectorized code, AVX-enabled code, and SVE-enabled code with
g++ compiler. The x86 evaluation was conducted on an AMD Ryzen 7 5800H Zen3 CPU to highlight the variance on a non-ARM-based CPU, significantly influencing the HPC field. The experiments were iterated
times, revealing that, on average, AVX showcases better performance statistically. The ARM-based machine was AWS EC2
xlarge instance with Neoverse N1 arm64 CPU. The average execution time measured on AVX was
ms, and the execution time on nonvectorized code was
ms. The average execution time on ARM-based machine for SVE was
ms.
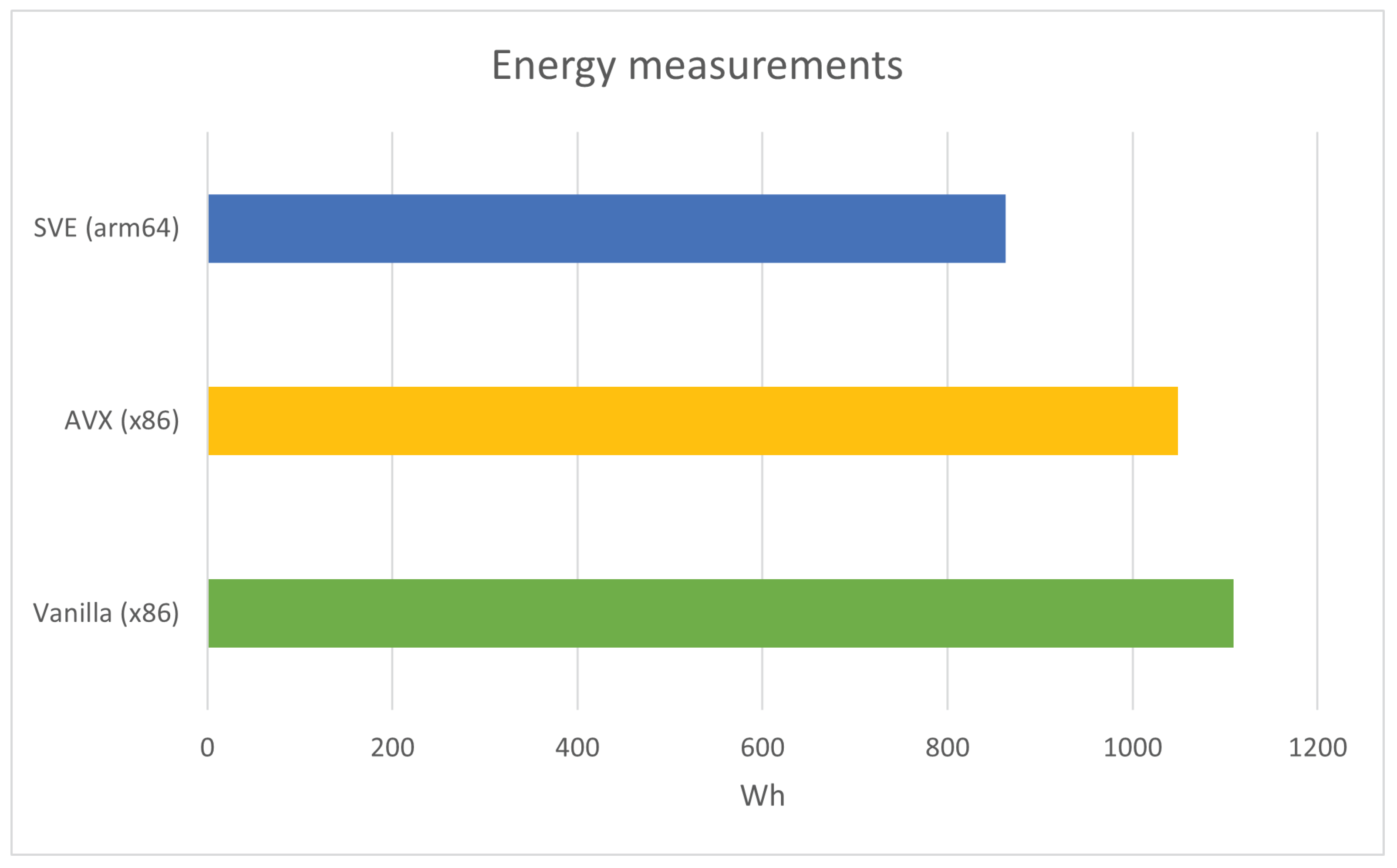
Finally, power measurements were conducted to compare the energy consumption of vectorized (using AVX) and nonvectorized Racon on the x86 architecture. The experiment involved Racon polishing the Tiny dataset for iterations. Power readings were recorded every s.
The results indicate that the energy consumption of the nonvectorized code was
Wh, whereas the energy consumption of the AVX-enabled version was
Wh. Conversely, the energy consumption of the SVE ran on arm64 was
Wh.
Figure 9 compares energy consumption between the nonvectorized, vectorized versions on the x86 CPU and SVE on the ARM-based CPU of the Racon.
5. Dispatcher Architecture
The Racon consensus module includes a dispatcher that automatically selects the most efficient instruction set for the underlying processor infrastructure. Our custom-made dispatcher can select the optimal compiled code segment for each processor architecture at runtime, ensuring efficient performance for various CPU architectures. This allows for faster consensus generation and for improved overall performance of the assembly pipeline.
To improve the performance of our code, we first reimplemented it using the type traits template wrappers technique in C++. Type traits are a powerful tool in C++ that allows for compile-time introspection and manipulation of types, enabling more efficient and optimized code [
37]. In our implementation, we utilized type traits in template metaprogramming to optimize the code for different CPU architectures (e.g., SVE and NEON). Each of these architectures is represented by an enumeration, which allows us to use type traits to determine the properties of these types and generate optimized code accordingly.
Next, we implemented a dispatcher that selects the fastest instruction set based on the CPU features of the system. For this, we used Google’s cpu_features library to retrieve information about the available instruction sets [
38].
After this, we used the
cmake tool to build the binaries for all platforms with optimized flags for improved performance. Finally, according to the current CPU capabilities, the dispatcher starts with the fastest SIMD variant. The overall process is outlined as follows:
Step 1: Utilize type traits template wrapper to adapt the code for optimized performance.
Step 2: Implement a dispatcher that selects the fastest instruction set for a given CPU using the priority list of architectures (e.g., AVX2, SSE4.1, SVE, NEON, and SSE2) and the cpu_features library from Google.
Step 3: Use cmake to build binaries for the specified list of CPU types, utilizing various compiler flags for optimized performance on different platforms.
Step 4: Run the dispatcher to automatically start the fastest SIMD variant according to the current CPU’s capabilities.
The dispatcher code for this study can be found on GitHub [
39].
6. Discussion
Optimizing the GASM pipeline through vectorization primarily focuses on Racon, which accounts for approximately of the total time spent in the pipeline during genome assembling. We have tested the effects of vectorization on performance in various scenarios: automatic vectorization and manual vectorization using a custom CPU dispatcher that automatically selects the fastest available instruction set. Furthermore, we conducted our experiments on different platforms, including the armie simulator using Intel Xeon E5645 GHz, a real ARM-based HPC BSC CTE-ARM A64FX, and a machine equipped with an AMD Ryzen 7 5800H Zen3 CPU.
Following our experiments, we demonstrate that executing on ARM NEON, utilizing intrinsics, results in a speedup of . Moreover, when comparing the speedups achieved through autovectorization versus intrinsics on the NEON architecture, the results indicate greater speedups using autovectorization, specifically a speedup compared to achieved with intrinsics. Conversely, when comparing autovectorization and vectorization using intrinsics on SVE, there is no difference; both exhibit average speedups of , with intrinsics proving better for larger datasets. Those results are due to autovectorization efficiently utilizing intrinsics on NEON.
Finally, we conducted experiments using an x86-based CPU (Zen3) and an arm64 CPU (Neoverse N1), comparing their execution time and energy consumption. Regarding execution time measurements, AVX outperformed the nonvectorized code on the x86 CPU, while SVE on the ARM-based CPU achieved an average speedup of over the nonvectorized code and over SVE. However, concerning energy consumption measurements, the ARM-based SVE consumed less energy compared to AVX on the x86 architecture and less energy compared to the nonvectorized code on the x86 CPU architecture.
7. Conclusions
In this article, we explore the efficiency of the Genome ASseMbly (GASM) pipeline on ARM-based High-Performance Computing (HPC) platforms by focusing on different implementations of the Scalable Vector Extension (SVE) architecture. We compare the performance of the g++9.3 and armclang++20.3 compilers in terms of vectorized versus nonvectorized code using the autovectorization capabilities of each compiler.
Additionally, we compare the autovectorization capabilities of these compilers with hand-tuned code utilizing intrinsics. Despite manually porting GASM to SVE, we found that manually porting complex C++ code to SVE using intrinsics is a significant effort due to the use of incomplete types. Furthermore, the existing vectorized code must be substantially reorganized, making it necessary to vectorize the code from scratch without relying on the existing vectorized version.
On the other hand, recent advances in compiler technology have significantly improved their autovectorization capabilities, making manual porting less necessary when autovectorization is easy to use and efficient. Our experiments found that the nonvectorized version was surprisingly well auto-vectorized by the armclang++ compiler, while the g++ compiler performed somewhat less well.
Future work in this area may include exploring using different ARM processors and memories with GASM vectorization.
Author Contributions
Conceptualization, M.B.; methodology, M.B.; resources, J.K. and M.K.; writing—original draft preparation, K.P.; writing—review and editing M.B. and M.K.; visualization, J.K. and K.P.; funding acquisition, M.K.; project administration, M.K.; validation, J.K., M.B. and M.K. All authors have read and agreed to the published version of the manuscript.
Funding
This project has received funding from the European High-Performance Computing Joint Undertaking (JU) under Framework Partnership Agreement No. 800928 and Specific Grant Agreement No. 101036168 EPI SGA2. The JU receives support from the European Union’s Horizon 2020 research and innovation program and Croatia, France, Germany, Greece, Italy, Netherlands, Portugal, Spain, Sweden and Switzerland. The content of this work is solely the responsibility of the authors.
Data Availability Statement
Data are contained within the article.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
The following abbreviations are used in this manuscript:
| HPC | High-Performance Computing |
| EU | European Union |
| EPI | European Processor Initiative |
| SIMD | Single Instruction Multiple Data |
| CPU | Central Processing Units |
| SIMDe | SIMD Everywhere |
| GASM | Genome ASseMbler |
| POA | Partial Order Alignment |
| GPU | Graphical Processing Units |
| SVE | Scalable Vector Extension |
| SPOA | SIMD POA |
| Racon | Rapid Consensus |
| VLA | Vector Length Agnostic |
| ACLE | ARM C Language Extensions |
| SMRT | Single Molecule Real-Time |
| ONT | Oxford Nanopore Technologies |
| SISD | Single Instruction Single Data |
| GPGPU | General-Purpose GPU |
| AVX | Advanced Vector Extensions |
References
- Lauring, A.S.; Hodcroft, E.B. Genetic variants of SARS-CoV-2—What do they mean? JAMA 2021, 325, 529. [Google Scholar] [CrossRef] [PubMed]
- Ajayan, J.; Nirmal, D.; Tayal, S.; Bhattacharya, S.; Arivazhagan, L.; Fletcher, A.S.A.; Murugapandiyan, P.; Ajitha, D. Nanosheet field effect transistors-a next generation device to keep moore’s law alive: An intensive study. Microelectron. J. 2021, 114, 105141. [Google Scholar] [CrossRef]
- Saxena, S.; Khan, M.Z.; Singh, R. Esacr: An energy saving approach from cloud resources for green cloud environment. In Proceedings of the 2021 10th International Conference on System Modeling & Advancement in Research Trends (SMART), Moradabad, India, 10–11 December 2021; pp. 628–632. [Google Scholar]
- Top 500. Available online: https://www.top500.org/ (accessed on 27 July 2022).
- European High-Performance Computing Joint Undertaking. 3 New r&i Projects to Boost the Digital Sovereignty of Europe. February 2022. Available online: https://eurohpc-ju.europa.eu/3-new-ri-projects-boost-digital-sovereignty-europe-2022-02-03_en (accessed on 27 July 2022).
- EuroHPC. European Processor Initiative. Available online: https://www.european-processor-initiative.eu/ (accessed on 27 July 2022).
- Kovač, M.; Denis, J.-M.; Notton, P.; Walter, E.; Dutoit, D.; Badstuebner, F.; Stilkerich, S.; Feldmann, C.; Dinechin, B.; Stevens, R.; et al. European Processor Initiative: Europe’s Approach to Exascale Computing; CRC Press: Boca Raton, FL, USA, 2022; Chapter 14; p. 18. [Google Scholar]
- Terzo, O.; Martinovič, J. HPC, Big Data, and AI Convergence Towards Exascale: Challenge and Vision, 1st ed.; CRC Press: New York, NY, USA, 2022. [Google Scholar]
- Simd Everywhere. 2022. Available online: https://github.com/simd-everywhere/simde (accessed on 2 October 2023).
- Vaser, R.; Sović, I.; Nagarajan, N.; Šikić, M. Fast and accurate de novo genome assembly from long uncorrected reads. Genome Res. 2017, 27, 737–746. [Google Scholar] [CrossRef] [PubMed]
- Bertil Schmidt and Andreas Hildebrandt. Next-generation sequencing: Big data meets high performance computing. Drug Discov. Today 2017, 22, 712–717. [Google Scholar] [CrossRef] [PubMed]
- Angelova, N.; Danis, T.; Lagnel, J.; Tsigenopoulos, C.S.; Manousaki, T. Snakecube: Containerized and automated pipeline for de novo genome assembly in hpc environments. BMC Res. Notes 2022, 15, 98. [Google Scholar] [CrossRef] [PubMed]
- Zafeiropoulos, H.; Gioti, A.; Ninidakis, S.; Potirakis, A.; Paragkamian, S.; Angelova, N.; Antoniou, A.; Danis, T.; Kaitetzidou, E.; Kasapidis, P.; et al. 0s and 1s in marine molecular research: A regional hpc perspective. GigaScience 2021, 10, giab053. [Google Scholar] [CrossRef] [PubMed]
- Castrignanò, T.; Gioiosa, S.; Flati, T.; Cestari, M.; Picardi, E.; Chiara, M.; Fratelli, M.; Amente, S.; Cirilli, M.; Tangaro, M.A.; et al. Elixir-it hpc@cineca: High performance computing resources for the bioinformatics community. BMC Bioinform. 2020, 21 (Suppl. 10), 352. [Google Scholar] [CrossRef] [PubMed]
- Jacob, A.; Paprzycki, M.; Ganzha, M.; Sanyal, S. Applying SIMD Approach to Whole Genome Comparison on Commodity Hardware; Springer: Berlin/Heidelberg, Germany, 2008; Volume 4967, pp. 1220–1229. [Google Scholar]
- Gao, Y.; Liu, Y.; Ma, Y.; Liu, B.; Wang, Y.; Xing, Y. Erratum to: Abpoa: An simd-based c library for fast partial order alignment using adaptive band. Bioinformatics 2021, 37, 3384. [Google Scholar] [CrossRef] [PubMed]
- McIntosh-Smith, S.; Price, J.; Deakin, T.; Poenaru, A. A performance analysis of the first generation of hpc-optimized arm processors. Concurr. Comput. Pract. Exp. 2019, 31, e5110. [Google Scholar] [CrossRef]
- Calore, E.; Gabbana, A.; Schifano, S.F.; Tripiccione, R. Thunderx2 performance and energy-efficiency for hpc workloads. Computation 2020, 8, 20. [Google Scholar] [CrossRef]
- Sato, M. The supercomputer “fugaku” and arm-sve enabled a64fx processor for energy-efficiency and sustained application performance. In Proceedings of the 2020 19th International Symposium on Parallel and Distributed Computing (ISPDC), Warsaw, Poland, 5–8 July 2020; pp. 1–5. [Google Scholar]
- Stephens, N.; Biles, S.; Boettcher, M.; Eapen, J.; Eyole, M.; Gabrielli, G.; Horsnell, M.; Magklis, G.; Martinez, A.; Premillieu, N.; et al. The arm scalable vector extension. IEEE Micro 2017, 37, 26–39. [Google Scholar] [CrossRef]
- Armejach, A.; Brank, B.; Cortina, J.; Dolique, F.; Hayes, T.; Ho, N.; Lagadec, P.-A.; Lemaire, R.; Lopez-Paradis, G.; Marliac, L.; et al. Mont-blanc 2020: Towards scalable and power efficient european hpc processors. In Proceedings of the 2021 Design, Automation & Test in Europe Conference & Exhibition (DATE), Grenoble, France, 1–5 February 2021; pp. 136–141. [Google Scholar]
- Ogbe, R.; Ochalefu, D.; Olaniru, O. Bioinformatics advances in genomics—A review. Int. J. Curr. Pharm. Rev. Res. 2016, 8. [Google Scholar]
- Pevsner, J. Bioinformatics and Functional Genomics; John Wiley & Sons: Hoboken, NJ, USA, 2015. [Google Scholar]
- Pop, M. Genome assembly reborn: Recent computational challenges. Briefings Bioinform. 2009, 10, 354–366. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Fan, J.; Sun, Z.; Liu, S. Nextpolish: A fast and efficient genome polishing tool for long-read assembly. Bioinformatics 2020, 36, 2253–2255. [Google Scholar] [CrossRef] [PubMed]
- Di Genova, A.; Buena-Atienza, E.; Ossowski, S.; Sagot, M.-F. Efficient hybrid de novo assembly of human genomes with wengan. Nat. Biotechnol. 2021, 39, 422–430. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Li, K.; Li, K.; Xie, X.; Lin, F. Swpepnovo: An efficient de novo peptide sequencing tool for large-scale ms/ms spectra analysis. Int. J. Biol. Sci. 2019, 15, 1787–1801. [Google Scholar] [CrossRef] [PubMed]
- Park, D.-H.; Beaumont, J.; Mudge, T. Accelerating smith-waterman alignment workload with scalable vector computing. In Proceedings of the 2017 IEEE International Conference on Cluster Computing (CLUSTER), Honolulu, HI, USA, 5–8 September 2017; pp. 661–668. [Google Scholar]
- Li, H. Minimap and miniasm: Fast mapping and de novo assembly for noisy long sequences. Bioinformatics 2016, 32, 2103–2110. [Google Scholar] [CrossRef] [PubMed]
- Sović, I.; Križanović, K.; Skala, K.; Šikić, M. Evaluation of hybrid and non-hybrid methods for de novo assembly of nanopore reads. Bioinformatics 2016, 32, 2582–2589. [Google Scholar] [CrossRef] [PubMed]
- Jang, M.; Kim, K.; Kim, K. The performance analysis of arm neon technology for mobile platforms. In Proceedings of the 2011 ACM Symposium on Research in Applied Computation, RACS’11, Miami, FL, USA, 2–5 November 2011; Association for Computing Machinery: New York, NY, USA, 2011; pp. 104–106. [Google Scholar]
- Zhong, D.; Shamis, P.; Cao, Q.; Bosilca, G.; Sumimoto, S.; Miura, K.; Dongarra, J. Using arm scalable vector extension to optimize open mpi. In Proceedings of the 2020 20th IEEE/ACM International Symposium on Cluster, Cloud and Internet Computing (CCGRID), Melbourne, VIC, Australia, 11–14 May 2020; pp. 222–231. [Google Scholar]
- Meyer, N.; Georg, P.; Pleiter, D.; Solbrig, S.; Wettig, T. Sve-enabling lattice qcd codes. In Proceedings of the 2018 IEEE International Conference on Cluster Computing (CLUSTER), Belfast, UK, 10–13 September 2018; pp. 623–628. [Google Scholar]
- Kodama, Y.; Odajima, T.; Matsuda, M.; Tsuji, M.; Lee, J.; Sato, M. Preliminary performance evaluation of application kernels using arm sve with multiple vector lengths. In Proceedings of the 2017 IEEE International Conference on Cluster Computing (CLUSTER), Honolulu, HI, USA, 5–8 September 2017; pp. 677–684. [Google Scholar]
- Li, H. Minimap2: Pairwise alignment for nucleotide sequences. Bioinformatics 2018, 34, 3094–3100. [Google Scholar] [CrossRef] [PubMed]
- Chi, C.C.; Alvarez-Mesa, M.; Bross, B.; Juurlink, B.; Schierl, T. Simd acceleration for hevc decoding. IEEE Trans. Circuits Syst. Video Technol. 2015, 25, 841–855. [Google Scholar] [CrossRef]
- Alexandrescu, A. Modern C++ Design: Generic Programming and Design Patterns Applied; C++ in-depth Series; Addison-Wesley: Boston, MA, USA, 2001. [Google Scholar]
- Google. cpu_features. 2022. Available online: https://github.com/google/cpu_features (accessed on 5 November 2023).
- Mario Brcic. Spoa. 2020. Available online: https://github.com/mbrcic/spoa (accessed on 20 September 2023).
Figure 1.
Genome sequence read using SIMD vectorization used in SPOA [
10].
Figure 1.
Genome sequence read using SIMD vectorization used in SPOA [
10].
Figure 2.
Depiction of SVE vector registers (
to
) and predicate registers (
to
) [
34].
Figure 2.
Depiction of SVE vector registers (
to
) and predicate registers (
to
) [
34].
Figure 3.
Illustration of the GASM pipeline.
Figure 3.
Illustration of the GASM pipeline.
Figure 4.
Average execution time (in milliseconds) and standard deviation of each phase of the GASM pipeline for iterations on a Tiny MB dataset.
Figure 4.
Average execution time (in milliseconds) and standard deviation of each phase of the GASM pipeline for iterations on a Tiny MB dataset.
Figure 5.
Comparison of vector vs. nonvector instructions on armclang++ compiler using automatic vectorization.
Figure 5.
Comparison of vector vs. nonvector instructions on armclang++ compiler using automatic vectorization.
Figure 6.
Comparison of vector vs. nonvector instructions on g++ compiler using automatic vectorization.
Figure 6.
Comparison of vector vs. nonvector instructions on g++ compiler using automatic vectorization.
Figure 7.
Comparison of vector vs. nonvector instructions on armclang++ compiler using manual vectorization.
Figure 7.
Comparison of vector vs. nonvector instructions on armclang++ compiler using manual vectorization.
Figure 8.
Comparison of nonvectorized code and AVX on x86 and SVE on arm64-based machine with g++ compiler run over iterations.
Figure 8.
Comparison of nonvectorized code and AVX on x86 and SVE on arm64-based machine with g++ compiler run over iterations.
Figure 9.
Energy measurement comparison of the nonvectorized (x86), AVX (x86), and SVE (arm64) version of the Racon ran for with g++ compiler.
Figure 9.
Energy measurement comparison of the nonvectorized (x86), AVX (x86), and SVE (arm64) version of the Racon ran for with g++ compiler.
Table 1.
Parameters of the Racon configuration.
Table 1.
Parameters of the Racon configuration.
| Parameter | Value |
|---|
| window_length | 500 |
| quality_threshold | 10 |
| error_threshold | |
| trim | TRUE |
| match | 3 |
| mismatch | |
| gap | |
| type | 0 |
| drop_unpolished_sequences | TRUE |
| num_threads | 24 |
Table 2.
GASM dataset inputs and configuration.
Table 2.
GASM dataset inputs and configuration.
| Dataset | Racon Threads | Size |
|---|
| Tiny | 24 threads | 1.7 MB |
| Medium | 24 threads | 255.2 MB |
Table 3.
Compiler and application-specific flags used to build and run specific implementations.
Table 3.
Compiler and application-specific flags used to build and run specific implementations.
| Implementation | Flags | Notes |
|---|
| Baseline | compiler: -march=armv8-a -O3 -fno-vectorize | -fno-vectorize ensured that autovectorization is disabled |
| NEON autovectorization | compiler: -march=armv8-a+simd -O3 | |
| NEON intrinsics | compiler: -march= armv8-a+simd -O3 | Using SIMDe ever existing x86_64 intrinsics implementation |
| SVE autovectorization | compiler: -march=armv8-a+sve -O3 | |
| SVE intrinsics | compiler: -march=armv8-a+sve | Different codebase was used |
Table 4.
Racon on ARM NEON execution times measurements with g++ and armclang++ compilers and two datasets.
Table 4.
Racon on ARM NEON execution times measurements with g++ and armclang++ compilers and two datasets.
| Racon on ARM NEON |
|---|
| Intrinsics | Compiler | Dataset | Time [s] |
| None | armclang++ | Tiny | 0.50513 |
| Medium | 75.630302 |
| g++ | Tiny | 0.514017 |
| Medium | 94.251879 |
| SIMDe NEON | armclang++ | Tiny | 0.397405 |
| Medium | 68.565859 |
| g++ | Tiny | 0.377458 |
| Medium | 68.236989 |
Table 5.
Performance speedup with g++ and armclang++ compilers and two datasets.
Table 5.
Performance speedup with g++ and armclang++ compilers and two datasets.
| Dataset | Compiler | Speedup |
|---|
| Tiny | g++ | 1.36× |
| armclang++ | × |
| Medium | g++ | 1.38× |
| armclang++ | 1.10× |
Table 6.
Automatic vectorization of GASM on armclang++ compiler.
Table 6.
Automatic vectorization of GASM on armclang++ compiler.
GASM; Compiler: armclang++20.3
(Autovectorization) |
|---|
| Vector Bits | Number of Instructions | Emulated Instructions | Percentage |
| 2048 | 2,029,668,930 | 247,675,608 | 12.20% |
| 1024 | 2,273,011,857 | 419,114,254 | 18.44% |
| 512 | 2,566,715,377 | 695,920,468 | 27.11% |
| 256 | 3,293,028,985 | 1,465,811,709 | 44.51% |
| 128 | 4,669,475,700 | 2,693,015,698 | 57.67% |
Table 7.
Automatic vectorization of GASM on g++ compiler.
Table 7.
Automatic vectorization of GASM on g++ compiler.
GASM; Compiler: g++9.3
(Autovectorization) |
|---|
| Vector Bits | Number of Instructions | Emulated Instructions | Percentage |
| 2048 | 2,226,655,621 | 66,652,365 | 2.99% |
| 1024 | 2,402,486,570 | 157,475,514 | 6.55% |
| 512 | 2,472,852,855 | 234,906,508 | 9.50% |
| 256 | 2,907,590,357 | 569,836,636 | 19.60% |
| 128 | 3,053,568,362 | 792,491,154 | 25.95% |
Table 8.
Manual vectorization of GASM on armclang++ compiler.
Table 8.
Manual vectorization of GASM on armclang++ compiler.
SPOA; Compiler: armclang++20.3
(Manually Ported Code) |
|---|
| Vector Bits | Number of Instructions | Emulated Instructions | Percentage |
| 2048 | 2,368,949,052 | 57,594,277 | 2.4% |
| 1024 | 2,387,255,830 | 194,554,945 | 8.2% |
| 512 | 2,582,357,561 | 436,263,410 | 16.9% |
| 256 | 2,773,083,031 | 637,365,127 | 23.0% |
| 128 | 3,967,910,395 | 1,690,963,198 | 42.6% |
Table 9.
Comparison of NEON autovectorization and NEON intrinsics with baseline implementation.
Table 9.
Comparison of NEON autovectorization and NEON intrinsics with baseline implementation.
| Input | Baseline | NEON | NEON Speedups |
|---|
| | | Autovectorization | Intrinsics | Autovectorization | Intrinsics |
| Tiny | 0.6244 s | 0.6010 s | 0.6318 s | 1.03× | 0.98× |
| Medium | 118.998 s | 83.25 s | 101.53 s | 1.42× | 1.17× |
| AVERAGE | | | | 1.23× | 1.08× |
Table 10.
Comparison of SVE autovectorization and SVE intrinsics with baseline implementation.
Table 10.
Comparison of SVE autovectorization and SVE intrinsics with baseline implementation.
| Input | Baseline | SVE | SVE Speedups |
|---|
| | | Autovectorization | Intrinsics | Autovectorization | Intrinsics |
| Tiny | 0.6244 s | 0.388 s | 0.4473 s | 1.61× | 1.40× |
| Medium | 118.998 s | 78.46 s | 68.84 s | 1.52× | 1.73× |
| AVERAGE | | | | 1.57× | 1.57× |
| Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).