1. Introduction
In 2021, the International Diabetes Federation estimated that 537 million adults worldwide have diabetes. Another 541 million are at risk of developing diabetes [
1]. Nerve and blood vessel damage are two complications associated with diabetes. These two complications lead to peripheral neuropathy and vascular disease and are associated with the development of diabetic foot ulcers (DFUs). It is estimated that a quarter of all diabetes patients will develop foot ulcers during the progression of this disease [
2]. Late detection of DFU and poor monitoring lead to lower extremity amputations and increase the likelihood of five-year mortality [
3]. Ulcer detection and monitoring capabilities using deep learning (DL) have recently been widely researched and showed promising results. However, deep learning models require large numbers of annotated data to achieve high accuracy, and collecting and labeling such data can be time-consuming and expensive.
The availability of public data reduces the cost of data collection but does not eliminate it completely. Datasets are formed using pictures from a single country or region [
4,
5,
6], where sample counts are on the order of hundreds and the uniform picture collection protocol limits generalizing capabilities during training. Even if medical institutions collect ulcer images during periodic visits and can grant access to these data, data preparation for a specific machine learning objective (classification, object detection or semantic segmentation) remains challenging and requires medical expertise. Fortunately, previous research in various domains has shown that data augmentation helps enrich the training dataset and contributes to the development of better deep learning models [
7].
In a recent systematic review on monitoring diabetic foot ulcers using deep learning [
8], it was found that some researchers use augmentation excessively and not enough test data are available for validation, while others do not use augmentation or do not report doing so. Chlap et al. have also found that many researchers use traditional data augmentation without investigating its effect on training results [
9]. In their systematic review on medical image augmentation, Garcea et al. found that traditional augmentation techniques are not used to their full potential and are rarely evaluated for their impact on trained model performance [
10]. In his review, Goceri observed that although data augmentation improves model performance, the exact contribution is not investigated [
11]. Additionally, it is not clear which augmentation techniques should be used to achieve the best results with different image types. Shorten and Khoshgoftaar have observed that to this day, there are no guidelines regarding how much augmentation is beneficial for achieving the best-performing model [
12]. Khosla and Saini conducted a comprehensive survey on multi-domain data augmentation techniques to reach a similar conclusion [
13]. As demonstrated with the dog breed example, the effectiveness of training data augmentation is domain- and objective-specific and therefore should be analyzed individually. Related work on augmentation analysis should be reviewed to find methodological approaches that are used in other research and any possible frameworks that could be applied to the subject of diabetic foot ulcers.
In this article, augmentation techniques used in a recent review [
8] on diabetic foot ulcer detection and monitoring are analyzed. The main goal is to better understand which technique improves performance the most and should be considered in future research in the domain of diabetic foot ulcer monitoring. Performed analysis is focused only on classic augmentation techniques and does not cover deep learning augmentation methods that have been found to be effective in other studies in the medical domain [
14,
15]. In this analysis, 11 classic augmentation techniques are used to train individual semantic segmentation models using the U-net architecture [
16], which was modified for melanoma detection [
17] and adapted for FUSeg2021 challenge [
4] dataset evaluation [
18]. The performance of trained models is compared against a benchmark model trained without data augmentation. The results were analyzed and compared to identify the augmentation technique that improves model performance the most, as well as the augmented sample count that is the most feasible. Finally, the three best-performing augmentation techniques were used to create multiple combined datasets. The models trained using them were compared against the benchmark model and best-performing models trained using a single augmentation technique. Additionally, samples from the Medetec dataset [
19] were used to verify the findings.
This article is organized as follows. Similar articles are reviewed in
Section 2. The base dataset, external datasets, training and testing setup, compared augmentation methods and metrics used are presented in
Section 3.
Section 4 presents the augmentation results, analysis of training results by considering different aspects and an overview of testing performance using single and combined augmented training datasets. Known limitations are briefly reviewed in
Section 5. Finally, the discussion in
Section 6 notes the final observations and conclusions.
2. Related Work
The most beneficial augmentation techniques are data-specific, and a one-fits-all approach is not possible. Other research on this subject was reviewed to gain insights into proper test design. Cubuk et al. [
20] used reinforcement learning to find the most beneficial augmentation policy for image classification. Their AutoAugment strategy discretizes search space, where each used augmentation policy is defined by a series of two augmentation operations, probability and magnitude of their application. Although the proposed search strategy is computationally expensive, the authors have demonstrated that learned augmentation strategies are transferable to other datasets. However, the authors do not specify the augmentation operations and the pool of magnitudes that were used. Fast AutoAugment by Lim et al. [
21] optimizes AutoAugment policy search by splitting the dataset into multiple folds and using only part of the fold data for model training. Though the search process is optimized, it remains computationally expensive, and a large dataset is required. Similarly, Ho et al. proposed Population-Based Augmentation [
22], where Population-Based Training [
23] is used to find the augmentation schedule rather than an extensive policy that could be used throughout the training. Inspired by the two previous papers, RandAugment [
24] reduces the augmentation search space (using only two parameters: operation and magnitude) and therefore does not require a separate search phase. It uses fourteen transforms and a randomized grid search. In contrast to RandAugment, TrivialAugment [
25] employs a customized augmentation sampling procedure. The augmentation technique and 1 of 30 discrete magnitudes are randomly sampled for each image. Though the latest automatic augmentation methods have reduced computational complexity, their main application was a classification task, and they used popular natural image datasets (e.g., ImageNet [
26]) with abundant data and a domain-specific search space.
Xu et al. [
27] reused AutoAugment strategies for three-dimensional medical image semantic segmentation. To simplify the search problem, they reduced the search space by using 7 image transformations and 11 discrete values of magnitude for each operation on the left and right bounds. They used stochastic relaxation to reform the search problem and make it solvable using gradient descend. However, the authors do not specify the motivation behind selecting augmentation techniques and left and right bounds for their parameter magnitude.
Liu et al. [
28] have proposed a MedAugment augmentation framework explicitly designed for medical images. The framework uses two transform spaces: six pixel-level (photometric) and eight spatial (geometric) transforms. Sampling complexity is defined by setting the number of consecutive augmentation transforms, which defines the number of possible strategies and the augmentation level, which defines the transform probability threshold and magnitude range. While the authors have shown that the framework outperforms other automatic augmentation methods, there is no search mechanism for optimal performance and additional customization might be required when it is used for DFU images.
Finally, Athalye et al. have demonstrated that the cut–paste strategy [
29] can be used for context-preserving training dataset augmentation, which regularizes the model. Simultaneously, the resulting model achieved a performance on par with that of the model trained using classical augmentation techniques.
None of the reviewed articles directly address the possibility of using diabetic foot ulcer image augmentation to achieve optimal model training. It has been observed that automatic augmentation methods, varying in their levels of customization and streamlining, do not offer a computationally inexpensive approach for selecting a domain-specific augmentation technique. While applying complex methods like AutoAugment or Fast AutoAugment may lead to optimal solution, the search space definition requires prior knowledge. In contrast, this work employs a simple augmentation strategy in which models are trained using data augmented with a single technique and predefined magnitudes. This approach helps us in finding domain-specific augmentation techniques suitable for any foot ulcer dataset.
3. Materials and Methods
In this analysis, image augmentation was tested to assess its impact on performance improvement in semantic segmentation. This section describes the setup and data used for the analysis of augmentation methods. The datasets used are introduced, indicating sources and image characteristics. Augmentation methods and bulk classes are presented to better understand the differences between them. The hardware setup used for this training provides the information that formed the basis for test setup decisions.
3.1. Dataset
In the medical domain, data collection and annotation are time-consuming processes that are subject to ethical and privacy-preserving regulations. Moreover, they are costly due to the required domain-specific medical expertise. To isolate image augmentation analysis from the difficulties introduced during training data preparation, an existing publicly available dataset was used. The augmentation analysis was performed using the publicly available FUSeg2021 foot ulcer semantic segmentation dataset [
4]. The dataset was collected at the Advancing the Zenith of Healthcare (AZH) Wound and Vascular Center, Milwaukee, WI. Images in the dataset were manually annotated by medical students and further refined by nurses and wound specialists to create image masks. For the segmentation challenge, samples were resized and zero-padded to achieve 512-by-512-pixel images; the same image resolution was used for augmentation analysis. The original dataset contains 1210 images, and it was split into test (200 samples), train (810 samples) and validation (200 samples) sets. Only the train and validation subsets, with a total of 1010 samples, have segmentation masks; therefore, this subset was used for training, validation and testing.
Additionally, 38 Medetec [
19] diabetic foot ulcer images augmented using horizontal, vertical and horizontal–vertical flipping were used for trained model performance validation using third-party data. Data annotations were taken from the FUSeg2021 introductory repository [
18]. The resulting 152 224-by-224-pixel images were only used for testing.
3.2. Testing Setup
Augmentation analysis was performed on a single Windows 10 machine equipped with 12 processors running at 3.2 GHz. Python version 3.9.16 was used to program routines for data splitting, augmentation, model training and testing. The original FUSeg2021 dataset train and validation folders were merged into image and mask sets, and then randomly shuffled and split into training, validation and testing sets using ratios of 0.7:0.2:0.1. Augmentation was performed for the training set only, and the resulting images were stored in separate folders named after the augmentation method. Each folder contained both augmented and original images. Since photometric augmentation does not affect image masks, original segmentation masks were reused by copying and renaming files. A more detailed explanation can be found in
Section 3.3.
Each training set created during augmentation was used to train a semantic segmentation network with the U-net [
16] architecture. During each training iteration, validation data were used without shuffling. Models were trained on a Nvidia GeForce RTX 3080 graphical processing unit (GPU) using the Keras library [
30]. Based on GPU capability, a batch size of 8 images was used. Model fitting was performed using Adam optimizer with a learning rate of 0.0001 and a binary cross-entropy loss function. Training iterations were executed for 2000 epochs with early stopping at the 100th epoch after reaching the highest dice score (see
Section 3.4.) value on validation data evaluation.
Keras utility for random seed setting was used to set a random seed to 100 before fitting every model. This ensured that initialization of the model weights remained consistent and did not influence the training results. Augmented data shuffling was also performed using the same random seed number. Validation data were not reshuffled before training, as this had already been performed during dataset splitting.
Microsoft Excel was used for collected data analysis. Model training and testing data were loaded using Power Query and analyzed using standard Excel functionalities.
3.3. Image Augmentation
This section introduces the most common augmentation methods [
8]; only the classical methods [
7] are considered. Classical methods can be grouped into two bulk categories: geometric and photometric. Brightness, contrast, Gaussian blur, saturation and sharpness adjustment operations are considered photometric augmentation techniques and only affect pixel color information (see
Figure 1).
Crop, flip, rotation, scale, shear and shift operations are considered geometric and affect pixel contextual information. In a semantic segmentation task, image and mask augmentations are performed simultaneously (see
Figure 2) to produce a new annotation for the augmented image.
The augmentation process was designed so that, for every augmentation technique, only one parameter (see
Table 1) was modified to produce new training samples.
All the augmentation techniques listed in the above table were implemented using the imgaug library [
31]. Parameter ranges, as provided in the above table, were derived from the metadata of a systematic review [
8] and were considered as a good starting point that was proven in previous research. In past research, the actual parameter values (except for flip augmentation) or rules for their selection have rarely been defined; therefore, we decided to use boundary and middle (original samples) values. This resulted in the first collection of datasets (later denoted as scenario 1). The second collection of datasets (denoted as scenario 2) was created using a finer approach for parameter value selection. This was achieved by using two additional middle parameter values (from the lower and higher value ranges). Additionally, flip augmentation resulted in three training datasets: vertical, horizontal and combined (vertical, horizontal and both). Finally, the whole collection consisted of 24 training datasets.
3.4. Performance Metrics
Individual model performance metrics were continuously monitored during training to ensure correct model training progress. During data analysis, metrics were used for model comparison and research conclusion. The dice coefficient is commonly used for semantic segmentation performance evaluation, though there are also other popular metrics. In this study, we performed comparative analysis, and a single metric provided means for easier comparison. Dice coefficient, precision and recall metrics will be introduced in this section.
Semantic segmentation correctness is evaluated according to correct pixel class allocation. In this research, pixels were classified as belonging or not belonging to a foot ulcer. Based on predicted pixel correctness, four confusion matrix terms were derived. Pixels that were correctly classified as belonging to foot ulcer are denoted as True Positive (
) and those not belonging as True Negative (
). Incorrectly classified foot ulcer pixels are categorized as False Negative (
) and non-ulcer pixels fall into the False Positive (
) category. These four categories were used to derive the performance metrics shown in
Table 2.
4. Results
4.1. Augmentation Results
The augmentation methods described in
Section 3.3 resulted in 24 training datasets, including the original dataset. Although the number of new samples (per image) created during augmentation was similar for most techniques, some methods resulted in a larger dataset. This section aims to illustrate the augmentation results and provide details about the number of samples created during augmentation.
Table 3 specifies the dataset increase for the two augmentation scenarios described in
Section 3.3.
4.2. Training Results
The 24 training datasets mentioned earlier resulted in 24 U-net segmentation models, trained for 140 to 461 epochs.
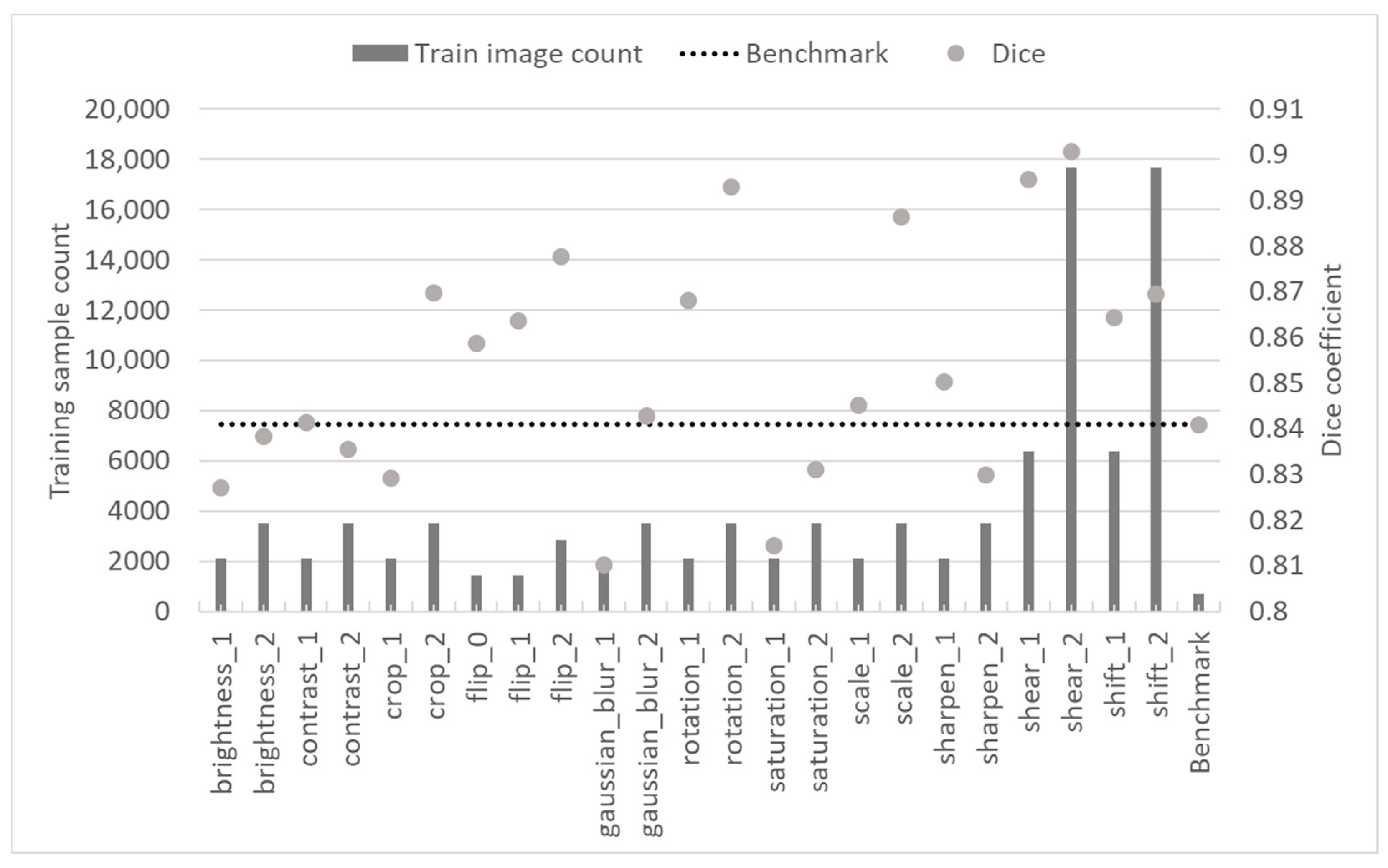
Figure 3 demonstrates the achieved trained model test results.
These data were further analyzed to gain insights into the effect of training epoch count, different augmentation classes, sample count and the methods’ effectiveness on the trained model performance.
4.3. Analysis
To analyze the data, only the best-performing model of each augmentation technique was compared. This provides more clarity and improves comprehension. The underscored indices indicate the augmentation scenario (see
Table 3) that was used to generate the training data. To determine which augmentation technique yielded improvement over the benchmark model trained with the initial training dataset, testing dice scores were compared.
Figure 4 demonstrates that only a subset of augmentation techniques helped improve segmentation results. It can be observed that photometric techniques had at least improved the training dataset and resulted in models that mostly performed similarly to the benchmark one. In contrast, all models trained using data augmented applying geometric transformations improved semantic segmentation test results by at least 2.9%. Rotation, scale and shear were the best-performing augmentation techniques.
Figure 5 demonstrates selected results (as explained above) of model testing with samples from the Medetec dataset. It can be observed that the benchmark results are the worst, and all augmentation techniques improved the model’s performance. Although the difference between the results achieved using photometric and geometric methods was less distinct, the majority of geometric methods outperformed photometric methods. Gaussian blur demonstrated the worst performance, while shear and shift improved benchmark performance by more than 20%.
Models were trained using early stopping with a patience of 100 epochs. This introduced some ambiguity to the training duration and might have affected the model training results. However, this ambiguity was found not to affect segmentation performance after comparing testing results against training duration.
Figure 6 demonstrates that training duration had no direct relationship with the achieved testing results. Though models trained using sheared images took the longest time to train and achieved the best dice coefficient, augmentation specifics are believed to have contributed to prolonged training.
The data sample count generated using different augmentation techniques is another variable that should be considered when designing a training dataset augmentation strategy. The data shown in
Figure 3 were compiled to arrive at the conclusions shown in
Figure 7. It can be observed that all techniques, except two (contrast and sharpness), yielded better results (in absolute value) when more training samples were generated. At the same time, only four techniques (crop, Gaussian blur, rotation and scale) yielded significantly better results when using more training samples. It should be noted that here, there was no obvious relationship between the increase in samples and the augmentation technique class (geometric and photometric). One important observation is that for shear and shift, which represent the two largest training datasets (as shown in
Figure 3), an increase in dataset samples did not contribute much to model performance and should not be considered to avoid unnecessary computational expense.
To conclude the augmentation analysis, datasets generated by the three best-performing techniques were combined to create four additional datasets: three using all possible combinations of two datasets and one combining all three datasets. In
Figure 7, it is shown that augmentation volume increase using the shear technique had a minor effect on model performance; therefore, the smaller (scenario 1) dataset (see
Table 3) was used. For rotation and scale, scenario 2 datasets were used.
Figure 8 compares the testing results of models trained with combined datasets and the top three models reviewed earlier. It can be observed that all new models outperformed the second- and third-best models trained when using individual augmentation techniques, but only two outperformed the model trained using the shear augmentation technique. Rotation and scale alone did not add enough new information compared with the dataset produced using the shear operation. A combination of all three (shear, rotation and scale) datasets was used to train the best-performing model. Finally, it should be noted that augmentation improved the benchmark result by 6.9%.
Figure 9 demonstrates a similar comparison with the Medetec dataset. It can be observed that neither model trained using combined datasets outperformed the model trained solely on the dataset produced using the shear operation. However, it should be noted that, as seen in
Figure 8, the best results were achieved when the combined training dataset contained data augmented using the shear method.
5. Limitations
This analysis was thoughtfully designed and attempted to eliminate any bias or other artifacts that could affect the results. However, a few limitations and directions for future research should be mentioned. Firstly, all augmentation function parameters were subjectively selected and may not necessarily have yielded the best possible performance. Secondly, only classical augmentation techniques were used; the usage of synthetic data might further improve segmentation results for third-party test data.
6. Discussion and Conclusions
Diabetic foot ulcer is a treatable condition when its development is continuously monitored. Semantic segmentation is a tool that can be used to implement this function. The problem arises when there is not enough or not sufficiently diverse data available for segmentation model training. Researchers commonly use various augmentation techniques to artificially increase the dataset size and diversity. However, the effect of augmentation is rarely evaluated or reported. This research aimed to individually evaluate the most popular classical augmentation techniques.
It was demonstrated that with the used dataset, photometric augmentation techniques are not as effective as geometric techniques. The resulting models achieved a performance that varied around the benchmark, without showing significant improvement. In contrast, geometric techniques helped improve the segmentation model results by at least 2.9%, and the best-performing model achieved a 6% increase in dice score. The three best-performing augmentation techniques were shear, rotation and scale. Four different combinations of these three datasets were created to train additional models. It was found that all except one demonstrated improved test results. The best model overall was trained on a dataset created using all three augmentation techniques, achieving a 6.9% performance improvement over the benchmark.
Trained models were additionally tested on diabetic foot image samples from the Medetec database. It was demonstrated that segmentation performance on this third-party dataset was substantially poorer due to different picture resolution, composition and illumination. However, models trained using geometrically augmented data showed a significant result improvement compared with a benchmark model.
Augmented sample count was tested as another variable affecting trained model performance. This analysis used an augmentation procedure that generated at least two training datasets for each augmentation technique. Variable parameters and different increment numbers were used to produce different training sample counts. Results achieved using different volumes of generated samples were compared to evaluate the effect of the increment size on trained model performance. It was found that, with the exception of the contrast and sharpen augmentation technique, the model results improved when more samples were used. However, the shift and shear models (trained with the two largest datasets) improved only by 0.6% when more data were generated. Due to varying results, it is not conclusive whether the augmented sample count has an effect on model performance.
The training duration of each model was optimized using an early stopping condition. Models were trained for an additional 100 epochs after the validation dice score stopped increasing. There was a concern that this approach might have affected the performance of the trained models. However, the results show that there is no direct relationship between model performance and training time.
This study has demonstrated that training dataset augmentation can benefit model test performance. The test results achieved using two different datasets lead to the conclusion that while model performance can be improved even when testing using completely different data, model utilization requires additional fine-tuning using training data from the same dataset. Making concrete recommendations on augmentation setup that could be useful across different datasets is difficult. However, in the context of diabetic foot ulcer semantic segmentation, geometric augmentation techniques yield the best model performance.