Abstract
The detection of skin lesions involves a resource-intensive and time-consuming process, necessitating specialized equipment and the expertise of dermatologists within medical facilities. Lesion segmentation, as a critical aspect of skin disorder assessment, has garnered substantial attention in recent research pursuits. In response, we developed a portable automatic dermatology detector and proposed a dual-CAM weakly supervised bootstrapping model for skin lesion detection. The hardware system in our device utilizes a modular and miniaturized design, including an embedded board, dermatoscope, and display, making it highly portable and easy to use in various settings. Our software solution uses a convolutional neural network (CNN) with a dual-class activation map (CAM) weakly supervised bootstrapping model for skin lesion detection. The model boasts two key characteristics: the integration of segmentation and classification networks, and the utilization of a dual CAM structure for precise lesion localization. We conducted an evaluation of our method using the ISIC2016 and ISIC2017 datasets, which yielded findings that demonstrate an AUC of 86.3% for skin lesion classification for ISIC2016 and an average AUC of 92.9% for ISIC2017. Furthermore, our system achieved diagnostic results of significant reference value, with an average AUC of 92% when tested on real-life skin. The experimental results underscore the portable device’s capacity to provide reliable diagnostic information for potential skin lesions, thereby demonstrating its practical applicability.
1. Introduction
Skin cancer is a rapidly growing type of cancer worldwide. In 2021, the American Cancer Association reported 108,480 new cases of skin cancer and 11,990 deaths in the United States [1]. Skin cancer has high morbidity and mortality rates, with a survival rate of over 95% for early diagnosis and less than 20% for late detection [2]. Early detection and treatment of melanoma is crucial, as it can be visually examined by dermatologists on the skin surface. Nonetheless, the process of visual examination using a dermatoscope is characterized by time-intensive efforts, necessitating elevated skills and focused attention. The accuracy of outcomes is intricately tied to the level of expertise possessed by the medical practitioner. As a result, there has been growing interest in developing computer-aided diagnosis (CAD) systems that can assist dermatologists in improving the accuracy, efficiency, and objectivity of diagnosis while addressing these challenges.
In CAD systems, skin lesion segmentation and classification are the two most important tasks in skin cancer detection. The segmentation task is used to detect the locations and boundaries of lesions, and the classification task is used to diagnose types of skin lesions. i.e., seborrheic keratosis (SK), melanoma (MELA), and nevus (NE). In the current study, the above skin lesion image segmentation and classification tasks face three challenges: (1) training segmentation and classification networks require a large number of labeled datasets; (2) complex backgrounds such as hair, blood vessels, air bubbles, and other interferents affect the quality of lesion segmentation, as shown in Figure 1; and (3) segmentation and classification tasks are relatively independent, and their intrinsic connections cannot be tapped to enhance each other.
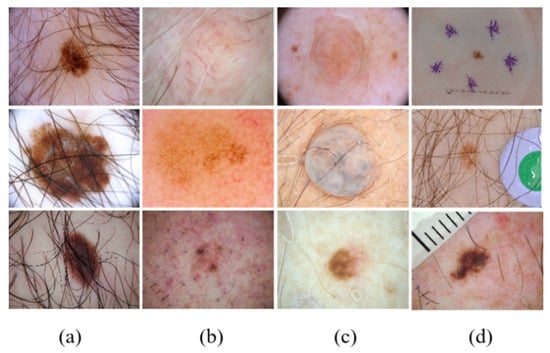
Figure 1.
Examples of typical skin lesion images with a complex background. (a) Hair-affected cases, (b) lesions similar to blood vessels, (c) cases with bubble interference, and (d) cases with other interferents.
Numerous automatic skin lesion segmentation and classification methods have been proposed in the literature [3,4,5,6,7,8]. Among these, deep convolutional neural network (DCNN)-based approaches have shown great success in medical image segmentation and classification [4,5,6,7,8]. In skin lesion segmentation (SLS), the proposed methods are typically based on fully convolutional networks (FCNs) [3] and U-Net [4]. For instance, Kaymak et al. [5] proposed a new FCN architecture for skin lesion image segmentation by modifying the convolutional neural network (CNN) architecture. Lei et al. [6] achieved superior segmentation performance by integrating a skip connection and dense convolution U-Net-based segmentation module with a dual discrimination module. In skin lesion classification (SLC), CNN-based approaches have demonstrated remarkable results and surpassed other traditional methods that require manual feature extraction. Recently, Zhang et al. [7] proposed an attention residual learning convolutional neural network (ARL-CNN) model for skin lesion classification, while Tang et al. [8] introduced a novel two-stage multi-modal learning algorithm (FusionM4Net) for multi-label skin disease classification. However, it is worth noting that the above studies rely on a large number of labeled datasets. Unfortunately, obtaining labeled samples is challenging, and most easily obtainable images are unlabeled.
Weakly supervised semantic segmentation (WSSS) was proven to be a cost-effective approach for semantic segmentation by utilizing weak labels instead of pixel-level masks. Several WSSS methods were proposed, including using image-level labels [9], bounding box annotations [10], and scribbles [11]. Liang et al. proposed a reiterative learning framework for weakly annotated biomedical images [12], and Liu et al. introduced cross-image region mining with a region prototypical network for weakly supervised segmentation [13]. Despite the success of WSSS in image semantic segmentation, there is still a lack of research on utilizing image-level labels to cascade multiple class activation maps (CAMs) for improved target localization.
It is widely acknowledged that segmentation plays a crucial role in computer vision applications, as it provides regions of interest for classification that allow for the extraction of discriminative features. On the other hand, classification has the advantage of providing local fine-grained information for images of the same class with similar features. However, despite their complementary nature, segmentation and classification are often treated as separate tasks, and their potential synergies, such as utilizing inter-class similarities and intra-class differences to facilitate each other, are rarely explored in current research.
In this paper, we propose a dual-CAM weakly supervised bootstrapping model for skin lesion detection. The model consists of three modules: a primary segmentation network module, a dual-CAM-guided classification network module, and a secondary segmentation network module. First, the primary segmentation network generates a rough lesion mask. Then, in the dual-CAM module, the rough mask and category labels are cascaded as pixel-level and image-level labels, respectively, to obtain CAM as a pseudo mask that accurately localizes and classifies skin lesions. Third, the localization information learned using dual-CAM is transferred to the rough mask generated with primary segmentation and fed into the secondary segmentation network, which accurately performs lesion segmentation. In addition, we use softmax cross-entropy loss (SCE) to activate the converged CAM1 with binary cross-entropy loss (BCE) for a second time to reduce mask ambiguity. Furthermore, inspired by the widespread use of Dice loss in biomedical image segmentation to address class imbalance, we compose a cascade loss using BCE and SCE for CAM1 and CAM2, respectively, and jointly use it with Dice loss. We extensively evaluate the proposed method using both the ISIC2016 and ISIC2017 datasets. Compared with other skin lesion segmentation and classification methods, our approach achieves high-quality masks and superior segmentation performance metrics.
The contributions of this paper can be summarized as follows:
- A novel approach for skin lesion segmentation is proposed by integrating segmentation and classification networks to extract common features of images.
- A weakly supervised semantic segmentation method for obtaining lesion pseudo masks is presented, achieved using a dual CAM cascade of image-level and pixel-level labels, which are fed into the segmentation network to transfer localization information and improve lesion segmentation accuracy.
- A portable skin lesion segmentation system was developed and successfully applied to real human skin testing, making the results of this study clinically relevant.
2. Related Work
2.1. Skin Lesion Segmentation and Classification
DCNN-based methods are widely used for skin lesion segmentation, with many approaches based on the Unet architecture and its variations. For instance, Li et al. [14] used a U-Net model trained on a manually created hair mask dataset to segment hair from skin lesion images. Tang et al. [15] proposed a separable-Unet with stochastic weight averaging, which enhanced the pixel-level discriminative representation capability of FCNs by capturing context feature channel correlation and higher semantic feature information. Recently, attention mechanisms have become popular in skin segmentation, such as attention DeepLabv3+ [16] and the active learning ensemble with a multi-model fusion method (ALEM) [17], adaptive dual attention module (ADAM) [18], attention synergy network (AS-Net) [19], and comprehensive attention convolutional neural network (CA-Net) [20]. These methods aim to leverage more global clues for more accurate segmentation. Lightweight networks are also a hot topic of research. For instance, Khouloud et al. [21] proposed an inception residual network for skin lesion segmentation and classification. Xie et al. [22] adapted the DeepLabv3+ network to a skin lesion segmentation task using mutual bootstrapping deep convolutional neural networks. Sarker et al. [23] proposed a lightweight and fully automatic skin lesion segmentation model that achieved precise segmentation with minimum resources.
Despite the success of these approaches, classification and segmentation are typically studied as separate tasks, and information between them is rarely exploited to facilitate each other. Nonetheless, some models have achieved state-of-the-art effects on dermatological segmentation and classification tasks, such as the feature adaptive transformers network (FAT-Net) [24] and the fully transformer network (FTN) [25].
2.2. SLS Based on the Less Labeled Sample Strategy
In the medical domain, obtaining labeled data for image segmentation tasks that require pixel-level annotation, such as skin lesion segmentation, is challenging due to its limited availability. To tackle this issue, several strategies were proposed, such as semi-supervised learning [26], active learning [17], self-supervised learning [27], and weakly supervised learning [28]. For instance, Li et al. [26] proposed a semi-supervised method for skin lesion segmentation, while Punn et al. [27] introduced a self-supervised learning framework that used unsupervised redundancy reduction to acquire data representation. Meanwhile, Chu et al. [29] applied multi-task learning to weakly supervised lesion segmentation. However, these methods rely on a small number of labeled datasets, which may lead to suboptimal performance due to the deviation in the training process. In contrast, active learning provides an alternative strategy to query samples in an interactive manner for iteration.
Recently, weakly supervised learning has gained popularity, and our proposed segmentation algorithm also uses this approach [30]. Weakly supervised learning typically consists of three steps: (1) obtaining a localization map of the target using a classification model, (2) generating pixel-level labels for the image-level labeled images based on the localization map, and (3) the segmentation model trained with the pixel-level labels. Specifically, we obtain lesion pseudo-masks using a dual CAM cascade of image-level and pixel-level labels with a weakly supervised semantic segmentation (WSSS), which enables us to transfer the localization information for accurate lesion segmentation, even in the presence of complex backgrounds.
2.3. Loss Function for Semantic Segmentation
In the realm of image processing, image segmentation and classification share a fundamental connection as image segmentation inherently involves pixel-level classification. The selection of appropriate loss functions for deep learning models engaged in image segmentation holds paramount importance as they significantly influence the algorithm’s learning process. Machine learning loss functions can be classified into four types: distribution-based, including BCE and SCE; region-based, like Dice loss; boundary-based, exemplified by Hausdorff distance loss; and composite [31]. Cross-entropy [32] is a prevalent metric used to quantify the disparity between two probability distributions of a random variable or a set of events. Cross-entropy losses, such as BCE and SCE, find extensive application in classification tasks, demonstrating remarkable efficacy in pixel-level segmentation.
The Dice coefficient serves as a measure of similarity between two images, and Dice loss [33] has emerged as a widely used loss function in biomedical image segmentation, primarily addressing the challenge of class imbalance. Building upon these foundations, Liang et al. [12] combined BCE and Dice loss to craft a specialized loss function for biomedical image segmentation, which facilitated accelerated model convergence and prevented local minima entrapment. Similarly, Xie et al. [22] devised a rank loss function and synergistically integrated it with Dice loss to contend with class imbalance and the disparities between hard and easy pixels within segmentation networks. Inspired by these advancements, we introduce a cascade loss mechanism that integrates BCE and SCE for CAM1 and CAM2, respectively, harmoniously combined with Dice loss. Our empirical investigations underscore the effectiveness of this amalgamated loss function in accelerating model convergence and effectively addressing challenges related to class imbalance.
3. Method
The portable system designed in this study comprises both hardware and software components, primarily addressing the segmentation and classification of skin lesion images. The operational framework of the system can be summarized as follows: Initially, skin lesion region images are captured using a dermatoscope (digital microscope). Subsequently, these images are fed into the segmentation network model, which delineates the contours of the lesions. Finally, the classification network provides suggestions regarding the lesion type. The subsequent sections provide a detailed explanation of both the hardware and software systems.
3.1. Hardware System
As shown in Figure 2, the proposed portable skin disease detection system consists of an image acquisition system, a display, and an embedded card. The image acquisition system is connected to the embedded card with a data cable and collects images using a skin microscope installed on a stand, which can magnify up to 1000 times. We designed the display and processor to be integrated into a compact box for convenient portability. The embedded card is placed at the bottom of the box with a ventilation port on the cover for hardware protection and heat dissipation. The display is located at the top and supported by the cover at a certain angle for easy viewing. The models of the skin microscope, display, and embedded card are 3-in-1 1000×, 5-inch HDMI LCD display, and Jetson Xavier NX, respectively. The main technical parameters are listed in Table 1.
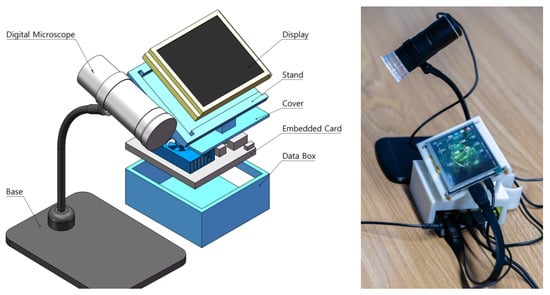
Figure 2.
Simulated zoomed-view schematic diagram showing the portable skin lesion detector and its corresponding physical representation.

Table 1.
Main Technical Parameters of the Measurement System.
3.2. Software Solution
The proposed method, illustrated in Figure 3, is composed of three main modules: a primary segmentation network module (PSN), a dual-CAM-guided classification network module (DCCN), and a secondary segmentation network module (SSN). The PSN and SSN collectively aim to obtain segmented images of skin lesions. Structurally akin, they both use the architecture of DeepLabv3+. However, the SSN distinguishes itself by integrating the lesion localization information, CAM2, obtained from the DCCN into its feature ensemble. In contrast, the DCCN serves as a classification network utilizing pixel and image labels to garner accurate lesion localization information. First, the PSN generates a rough lesion mask. Then, in the dual-CAM module, the rough mask and category labels are combined as pixel-level and image-level labels, respectively, to obtain class activation maps as pseudo-masks. These masks help accurately localize and classify skin lesions, as shown in Figure 4. Third, the localization information learned with dual-CAM is transferred to the rough mask generated using the PSN, and it is fed to the SSN for accurate lesion segmentation. Finally, we input the acquired skin lesion images into the trained model and obtain the diagnostic reference conclusion after performing the segmentation and classification steps.
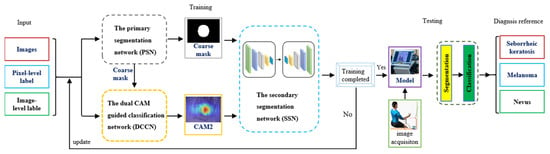
Figure 3.
Overview of the dual-CAM weakly supervised bootstrapping model for skin lesion detection.
3.2.1. PSN
The primary segmentation network (PSN) plays a crucial role in providing the dual-CAM-guided classification network (DCCN) with prior knowledge of the lesion location, which improves its localization and identification capabilities. We train the PSN on a training set , consisting of n images, to obtain a coarse lesion mask. Here, represents the image pixel and represents its pixel label, where denotes the lesion and denotes the background. To enhance the segmentation performance of the network and overcome the effect of complex backgrounds on lesions, we use a transfer learning approach. Specifically, we pre-train the segmentation network using the MS-COCO and PASCAL VOC 2012 datasets using classical DeepLabv3+ as the segmentation network and aligned Xception as the backbone network. Additionally, we modify the decoder by adding a 1 × 1 convolutional layer before the final upsampling and adjust the channels to 1. The parameters of this convolutional layer are randomly initialized and activated using the ReLU function.
3.2.2. DCCN
In the dual-CAM-guided classification network module (DCCN), the rough mask and category labels are cascaded as pixel-level and image-level labels, respectively, to obtain the final-CAM as a pseudo-mask to help it accurately localize and classify skin lesions. To train this network, we use the dataset containing m images, where denotes the image pixel and denotes the category label of the image. For the skin lesion classification task in this paper, has three categories: seborrheic keratosis (SK), melanoma (MELA), and nevus (NE), denoted as . We use the popular classification network Xception as the backbone to extract features. The algorithm in this section is shown in Figure 4 and described in detail as follows.
The PSN generates both images and masks, which are then fed into Xception. Meanwhile, the features are also inputted into the fully connected (FC) layer for prediction after undergoing global average pooling (GAP). As a result, the prediction logits can be represented as:
where F and G represent abbreviations for FC and GPA, respectively. Then, the BCE loss is calculated using P and the pixel-level label as stated in Equation (2):
where denotes the prediction logit of the d-th class, is the sigmoid function, and M is the total number of foreground object classes. Based on the features and the weights of the corresponding FC layers, the CAM is obtained for each image extraction, as stated in Equation (3):
Next, we use CAM-1 as a pseudo-mask applied to to extract the corresponding class of features . We calculate the multiplication of elements between CAM-1 and each channel of using Equation (4):
where and denote the single channels before and after multiplication (using Equation (3)), respectively, and C is the number of feature category maps (i.e., channels). With this step, we finish training the pixel-level label classifier. Next, we use FC2 to further train the image-level label classifier. As such, we define new prediction logits for x:
Using this method, we transform the multi-label image model, which utilizes binary cross-entropy (BCE), into a single-label feature model that utilizes a softmax cross-entropy (SCE) loss function. The SCE loss is expressed as follows:
where and denote the m-th element of y and , respectively. We use to update the model.
Following the retraining process, we extract CAM2 from image x for each category m using the following procedure:
where is the classification weight corresponding to the m-th class.
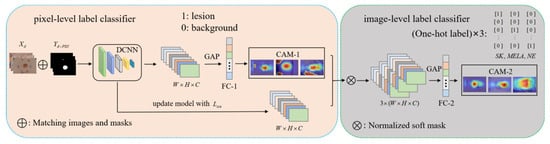
Figure 4.
The DCCN framework consists of two main components: an upper pixel-level label classification module and a lower image-level label classification module. In the left module, images and coarse masks generated with the PSN are fed to the backbone network, which is routinely trained using the multi-label classifier of the BCE. CAM is extracted for each category and then applied to feature maps generated after the update to obtain category-specific features. In the right module, category-specific features and their single label are used to train a multi-class classifier with SCE loss. The loss gradient is back-propagated through the entire network.
3.2.3. SSN
The secondary segmentation network (SSN) stands as a pivotal module harnessing the lesion localization insight, CAM2, obtained from the original input image and the dual CAM-guided classification network (DCCN). This combination is instrumental in achieving precision in lesion segmentation images. Using a structure reminiscent of DeepLabv3+, the SSN unfolds in a series of deliberate steps. Primarily, the original input image is encoded with the Xception backbone network, supported using pre-trained weights. In this encoding phase, the integration of dilated convolutions facilitates a wider local field of view. Subsequently, a feature pyramid undertakes the extraction of multi-scale features. Consecutively, the pinpointed lesion localization information, CAM2, sourced from the DCCN, is seamlessly integrated into the ensemble of features. Ultimately, the decoding process leads the SSN to accurately delineate lesion segmentation outcomes. The architecture of SSN is depicted in Figure 5, which shares the same network structure as a PSN but incorporates location information into the segmentation process. Specifically, the encoder-generated feature maps are concatenated with CAM2 and then passed through 1 × 1 convolution. The resulting feature map generated using CAM2 is then fed to the decoder for fine-grained segmentation.
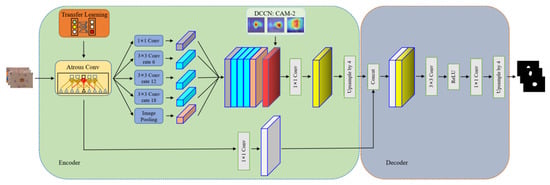
Figure 5.
In the encoder, images are fed into a pre-trained deep convolutional network. Feature extraction is carried out using different rates of atrous convolution, and the resulting feature maps are merged with precise lesion location feature maps outputted from the dual CAM-guided classification network (DCCN). Next, feature compression is performed using 1 × 1 convolution. The decoder module is similar to DeepLabv3+, but we made modifications to the backbone network Xception to better fit our network. We achieved this by removing the final convolution layer and replacing it with a 1 × 1 convolution layer that is activated using the ReLU function. Finally, we upsample the output to obtain the lesion segmentation results.
3.2.4. Hybrid Loss Function
Given that our proposed model involves separate classification and segmentation tasks during training, it is crucial to utilize specific loss functions to ensure fast and stable convergence. To address the intra- and inter-class imbalance problem in training data, we use Dice loss for segmenting both the PSN and SSN networks. In addition, we use cascading losses with the combination of binary cross-entropy (BCE) and focal symmetric cross-entropy (SCE) to construct the DCCN, which extracts precise location information. To combine these losses into a single cohesive metric, we define a hybrid loss function as:
where is the Dice loss, defined in Equation (9), and is the classification cascade loss, defined in Equation (10).
where y represents the ground truth and p signifies the network’s prediction.
4. Experiments and Results
4.1. Datasets
Two datasets provided by the International Skin Imaging Collaboration (ISIC), namely, the ISIC 2016 and ISIC 2017 benchmark datasets, are utilized to assess the proposed segmentation approach. The ISIC 2016 skin lesion challenge dataset [34] comprises 900 images (727 non-melanoma and 173 melanoma) for training and 379 images (304 nonmelanoma and 75 melanoma) for testing, with pixel sizes ranging from 566 × 679 to 2848 × 4228. The ISIC 2017 skin lesion challenge dataset [35] includes 2000 images (1626 non-melanoma and 374 melanoma) for training and 600 images (483 nonmelanoma and 117 melanoma) for testing. The pixel sizes of the images range from 453 × 679 to 4499 × 6748. As part of our data preprocessing, we uniformly resized the image pixels to 224 × 224 and expanded our training data using resources from the ISIC website.
4.2. Performance Evaluation
We evaluate the performance of our proposed segmentation approach using the evaluation metrics suggested in ISIC 2016 and ISIC 2017, including pixel-wise accuracy (ACC), sensitivity (SEN), specificity (SPE), the Dice coefficient (DIC), the Jaccard index (JAI), and the area under the curve (AUC). These metrics are defined in Equations (11)–(16).
The pixel-wise accuracy (ACC) calculation formula is as follows:
The sensitivity (SEN) calculation formula is as follows:
The specificity (SPE) calculation formula is as follows:
The Dice coefficient (DIC) calculation formula is as follows:
The Jaccard index (JAI) calculation formula is defined as follows:
The area under curve (AUC) calculation formula is defined as follows:
where is the true positive rate and is the false positive rate. and () indicate the confidence scores for negative and positive cases, respectively. The definitions of TP, TN, FP, and FN are listed in Table 2.

Table 2.
Definitions of TP, TN, FP, and FN.
4.3. Implementation Details
To mitigate the influence of complex backgrounds in images and to augment the training dataset, we utilized various data augmentation techniques. These techniques included adding random noise to the image, randomly cropping the center of the training image from 50% to 100% of the original image, randomly rotating from 0 to 10 degrees, shearing from 0 to 0.1 radians, shifting from 0 to 20 pixels, scaling 120% of the width and height, and flipping both horizontally and vertically. The resulting enhanced images were then resized to 224 × 224 for training purposes. We set the batchsize of the PSN and SSN to 16 and the batchsize of the DCCN to 32, using Adam as the optimizer with a learning rate of 0.001. All experiments were conducted using a platform featuring an Intel(R) Core (TM) i7-7820X CPU @ 3.60 GHz and four 12 GB NVIDIA GeForce 2080 Ti GPUs.
4.4. Classification Experiments and Results
We conducted a comparison between the DCCN network used for classification in this paper and several representative methods for skin lesion classification, and the results are listed in Table 3 and Table 4. We also provide a visualization of the segmentation and classification results for various modules in our proposed model, including the PSN, CAM1, CAM2, SSN, and segmentation ground truth, and classification of diagnostic results, as shown in Figure 6. For the ISIC2016 dataset, the methods for comparison include the top five methods of the ISIC 2016 Lesion Classification Challenge, Team-CUMED, Team-GTDL, Team-BF-TB Thiemo, Team-ThrunLab, and Team-Jordan Yap, as well as synergic deep learning (SDL, 2019) [36], global-part CNN model with data-transformed ensemble learning (GP-CNN-DTEL, 2020) [37], deep attention branch networks (DABN-ELW, 2021) [38] and deep metric attention learning CNN (DeMAL-CNN, 2022) [39]. For the ISIC2017 dataset, the methods for comparison include SDL, GP-CNN-DTEL, DABN-ELW, and DeMAL-CNN.

Table 3.
Classification performance (%) comparative result with different methods on ISIC 2016 dataset.

Table 4.
Classification performance (%) comparative result with different methods using ISIC 2017 for melanoma and seborrheic keratosis images.
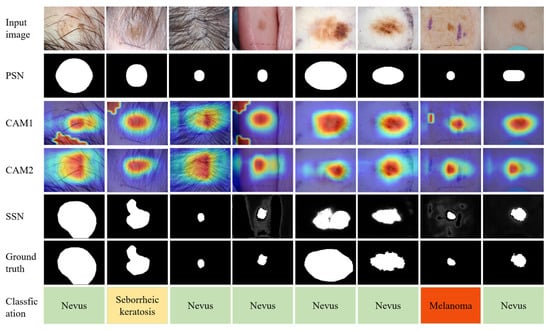
Figure 6.
Results of some different modules in the model proposed in this paper. First row: Input Images. Second row: The mask generated with the primary segmentation network (PSN). Third row: Localization map generated with CAM1. Fourth row: Localization map generated with CAM2. Fifth row: The mask generated with the secondary segmentation network module (SSN). Sixth row: Segmentation ground truth. Last row: Classification of diagnostic results.
As illustrated in Table 3 and Table 4, following precise lesion localization using the SSN, the images achieve optimal segmentation results. Both datasets exhibit superior performance on the DCCN classification network. On the ISIC 2016 dataset, in comparison with state-of-the-art methods, our model demonstrates an almost 1% enhancement in SEN. Moreover, ACC and AUN slightly outperform other methods, while SPE slightly lags behind Team-Jordan Yap and GP-CNN-DTEL. For the ISIC2017 dataset, we obtained the best performance in SEN and AUC in the Melanoma category and AUC in the Seborrheic Keratosis category, while not achieving significant improvement in other metrics. Improving the network performance in these metrics will be one of our future research directions.
4.5. Hybrid Loss and the Effect on Experimental Results
In this study, we introduce a novel hybrid loss that merges binary cross-entropy loss (BCE) and softmax cross-entropy loss (SCE), individually assigned to CAM1 and CAM2. This hybrid loss is used in conjunction with the Dice loss. To assess the efficacy of this hybrid loss and its impact on experimental outcomes, we conducted comprehensive experiments using the ISIC2017 dataset. We conducted separate experiments using BCE, SCE, and a combination of both as classification loss, measuring and recording the corresponding AUC values. The results, as depicted in Figure 7, highlight a significant observation. It becomes evident that using either BCE or SCE alone as the loss function for the DCCN model is less effective compared with the synergistic application of both losses. Furthermore, in the process of selecting the appropriate loss functions for CAM1 and CAM2, a noteworthy finding emerged. Using BCE for CAM1 and SCE for CAM2 yielded the most favorable classification AUC values, as evidenced by the illustrated red curve.
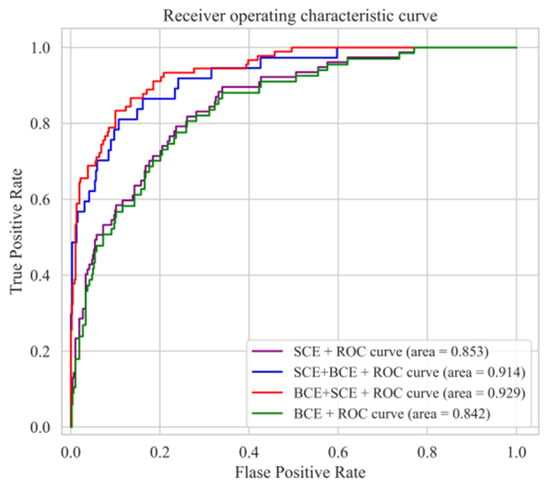
Figure 7.
ROC curves obtained using different loss function combinations.
4.6. Effectiveness of the Dual CAM
To verify the effectiveness of the dual CAM, we conducted a series of experiments using the DCCN network using only CAM1 with pixel-level labels, only CAM2 with image-level labels, and a combination of both. As illustrated in Figure 8, the combination of dual CAMs resulted in the highest AUC values, confirming its efficacy. This highlights how the DCCN network cascades the coarse mask and category labels as pixel-level and image-level labels, respectively, to obtain the best CAM as a pseudo-mask that aids the SSN network in precisely localizing and segmenting skin lesions. Furthermore, our proposed model achieved an AUC value exceeding 0.8 during the training phase when the number of samples reached 50%, equivalent to the performance of the other two cases that required 75% of the number of samples. This indicates that our method requires fewer labeled samples than other approaches while delivering superior classification performance, a promising strategy for reducing the cost of training deep learning networks that typically require large amounts of labeled data. This is particularly relevant in practice.
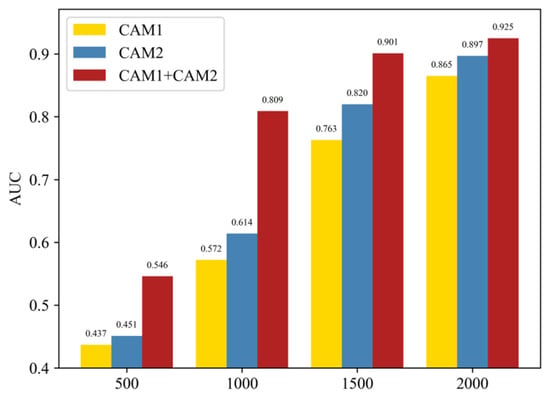
Figure 8.
Comparison of AUC values between the dual CAM model and the separate CAM model using the ISIC2017 validation set with varying numbers of labeled training images (500, 1000, 1500, and 2000).
4.7. System Testing
We tested the efficacy of the portable detector described in this paper on the skin of four volunteers, two males and two females, and obtained promising results, as illustrated in Figure 9. Our device produced clear and detailed images of the skin lesion area, and when coupled with the segmentation and classification algorithm presented in this paper, it achieved precise segmentation of the lesion area. The performance metrics of our method, as compared with the Unet and ResNet_Unet algorithms are shown in Table 5. The performance results demonstrate that our approach significantly outperforms these classical algorithms. It should be noted that since we lack professional medical knowledge to make clear diagnostic results, the test results of this experiment are based on reference conclusions obtained from the healthy physical conditions of four volunteers. In addition, the masks we labeled were not very accurate, which resulted in lower segmentation metrics.
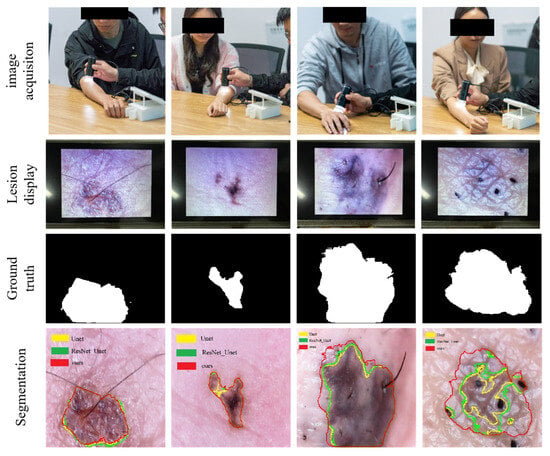
Figure 9.
This figure presents the segmentation results of a portable system tested on volunteers. The segmentation contour lines are illustrated with yellow for Unet, green for ResNet_Unet, and red for the method proposed in this paper.

Table 5.
Segmentation performance (%) comparative result with classical algorithms on human skin images.
5. Conclusions
In this study, we introduced a novel portable automated dermatology detector and put forward a dual-CAM weakly supervised bootstrapping model designed for the segmentation of skin lesions. The model comprises three key components: a PSN responsible for generating preliminary lesion masks, a DCCN used for accurate lesion localization with the utilization of coarse masks and lesion categories as pixel-level and image-level labels, respectively, and an SSN tasked with transferring the localization data to the preliminary lesion masks to achieve precise segmentation. Our method’s efficacy was evaluated extensively in terms of skin lesion segmentation and classification performance across the ISIC2016 and ISIC2017 datasets. Moreover, the successful application of the developed portable dermatology detector underscores its clinical significance. The performance of the portable system in human testing yielded satisfactory results. Looking ahead, we intend to explore the implementation of a lightweight network to decrease the hardware demands and consequently reduce the device’s overall cost. Additionally, we aim to establish stronger collaboration with dermatologists to establish a shared skin lesion database, with the ultimate goal of enhancing diagnostic accuracy and providing timely medical intervention for patients.
Author Contributions
Data curation, methodology, and writing—original draft, H.Q.; investigation, L.S., Y.Y. and L.Z.; data curation, J.L.; writing—review and editing, H.Q., Z.D., L.S., Y.Y. and H.Z. and Q.L. Funding acquisition, Q.L. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported in part by the National Natural Science Foundation of China under Grants 62073129, U21A20490, and 62293510 and the Hunan Provincial Natural Science Foundation of China (2022JJ10020).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Informed consent was obtained from all subjects involved in this study.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer statistics, 2022. CA Cancer J. Clin. 2022, 72, 7–33. [Google Scholar] [CrossRef] [PubMed]
- Balch, C.M.; Gershenwald, J.E.; Soong, S.-J.; Thompson, J.F.; Atkins, M.B.; Byrd, D.R.; Buzaid, A.C.; Cochran, A.J.; Coit, D.G.; Ding, S.; et al. Final version of 2009 AJCC melanoma staging and classification. J. Clin. Oncol. 2009, 27, 6199–6206. [Google Scholar] [CrossRef] [PubMed]
- Long, J.; Shelhamer, E.; Darrell, T. Fully convolutional networks for semantic segmentation. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Boston, MA, USA, 7–12 June 2015; pp. 3431–3440. [Google Scholar]
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional networks for biomedical image segmentation. In Proceedings of the Medical Image Computing and Computer-Assisted Intervention–MICCAI 2015: 18th International Conference, Munich, Germany, 5–9 October 2015; Proceedings, Part III 18. Springer International Publishing: Berlin/Heidelberg, Germany, 2015; pp. 234–241. [Google Scholar]
- Kaymak, R.; Kaymak, C.; Ucar, A. Skin lesion segmentation using fully convolutional networks: A comparative experimental study. Expert Syst. Appl. 2020, 161, 113742. [Google Scholar] [CrossRef]
- Lei, B.; Xia, Z.; Jiang, F.; Jiang, X.; Ge, Z.; Xu, Y.; Qin, J.; Chen, S.; Wang, T.; Wang, S. Skin lesion segmentation via generative adversarial networks with dual discriminators. Med. Image Anal. 2020, 64, 101716. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Xie, Y.; Xia, Y.; Shen, C. Attention residual learning for skin lesion classification. IEEE Trans. Med. Imaging 2019, 38, 2092–2103. [Google Scholar] [CrossRef]
- Tang, P.; Yan, X.; Nan, Y.; Xiang, S.; Krammer, S.; Lasser, T. FusionM4Net: A multi-stage multi-modal learning algorithm for multi-label skin lesion classification. Med. Image Anal. 2022, 76, 102307. [Google Scholar] [CrossRef]
- Liu, Y.; Wu, Y.H.; Wen, P.; Shi, Y.; Qiu, Y.; Cheng, M.M. Leveraging instance-, image-and dataset-level information for weakly supervised instance segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 2020, 44, 1415–1428. [Google Scholar] [CrossRef]
- Wang, J.; Xia, B. Bounding box tightness prior for weakly supervised image segmentation. In Proceedings of the Medical Image Computing and Computer Assisted Intervention–MICCAI 2021: 24th International Conference, Strasbourg, France, 27 September–1 October 2021; Proceedings, Part II. Springer International Publishing: Cham, Switzerland, 2021; pp. 526–536. [Google Scholar]
- Lin, D.; Dai, J.; Jia, J.; He, K.; Sun, J. Scribblesup: Scribble-supervised convolutional networks for semantic segmentation. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 3159–3167. [Google Scholar]
- Liang, Q.; Nan, Y.; Coppola, G.; Zou, K.; Sun, W.; Zhang, D.; Wang, Y.; Yu, G. Weakly supervised biomedical image segmentation by reiterative learning. IEEE J. Biomed. Health Inform. 2018, 23, 1205–1214. [Google Scholar] [CrossRef]
- Liu, W.; Kong, X.; Hung, T.-Y.; Lin, G. Cross-image region mining with region prototypical network for weakly supervised segmentation. IEEE Trans. Multimed. 2021, 25, 1148–1160. [Google Scholar] [CrossRef]
- Li, W.; Raj, A.N.J.; Tjahjadi, T.; Zhuang, Z. Digital hair removal by deep learning for skin lesion segmentation. Pattern Recognit. 2021, 117, 107994. [Google Scholar] [CrossRef]
- Tang, P.; Liang, Q.; Yan, X.; Xiang, S.; Sun, W.; Zhang, D.; Coppola, G. Efficient skin lesion segmentation using separable-Unet with stochastic weight averaging. Comput. Methods Programs Biomed. 2019, 178, 289–301. [Google Scholar] [CrossRef] [PubMed]
- Azad, R.; Asadi-Aghbolaghi, M.; Fathy, M.; Escalera, S. Attention deeplabv3+: Multi-level context attention mechanism for skin lesion segmentation. In Proceedings of the Computer Vision–ECCV 2020 Workshops, Glasgow, UK, 23–28 August 2020; Proceedings, Part I 16. Springer International Publishing: Berlin/Heidelberg, Germany, 2020; pp. 251–266. [Google Scholar]
- Liang, Q.; Qin, H.; Zeng, H.; Long, J.; Sun, W.; Zhang, D.; Wang, Y. Active Learning Integrated Portable Skin Lesion Detection System Based on Multi-model Fusion. IEEE Sens. J. 2023, 23, 9898–9908. [Google Scholar] [CrossRef]
- Wu, H.; Pan, J.; Li, Z.; Wen, Z.; Qin, J. Automated skin lesion segmentation via an adaptive dual attention module. IEEE Trans. Med. Imaging 2020, 40, 357–370. [Google Scholar] [CrossRef] [PubMed]
- Hu, K.; Lu, J.; Lee, D.; Xiong, D.; Chen, Z. AS-Net: Attention Synergy Network for skin lesion segmentation. Expert Syst. Appl. 2022, 201, 117112. [Google Scholar] [CrossRef]
- Gu, R.; Wang, G.; Song, T.; Huang, R.; Aertsen, M.; Deprest, J.; Ourselin, S.; Vercauteren, T.; Zhang, S. CA-Net: Comprehensive attention convolutional neural networks for explainable medical image segmentation. IEEE Trans. Med. Imaging 2020, 40, 699–711. [Google Scholar] [CrossRef]
- Khouloud, S.; Ahlem, M.; Fadel, T.; Amel, S. W-net and inception residual network for skin lesion segmentation and classification. Appl. Intell. 2022, 52, 3976–3994. [Google Scholar] [CrossRef]
- Xie, Y.; Zhang, J.; Xia, Y.; Shen, C. A mutual bootstrapping model for automated skin lesion segmentation and classification. IEEE Trans. Med. Imaging 2020, 39, 2482–2493. [Google Scholar] [CrossRef]
- Sarker, M.K.; Rashwan, H.A.; Akram, F.; Singh, V.K.; Banu, S.F.; Chowdhury, F.U.; Choudhury, K.A.; Chambon, S.; Radeva, P.; Puig, D.; et al. SLSNet: Skin lesion segmentation using a lightweight generative adversarial network. Expert Syst. Appl. 2021, 183, 115433. [Google Scholar] [CrossRef]
- Wu, H.; Chen, S.; Chen, G.; Wang, W.; Lei, B.; Wen, Z. FAT-Net: Feature adaptive transformers for automated skin lesion segmentation. Med. Image Anal. 2022, 76, 102327. [Google Scholar] [CrossRef]
- He, X.; Tan, E.-L.; Bi, H.; Zhang, X.; Zhao, S.; Lei, B. Fully transformer network for skin lesion analysis. Med. Image Anal. 2022, 77, 102357. [Google Scholar] [CrossRef]
- Li, X.; Wu, Y.; Dai, S. Semi-supervised medical imaging segmentation with soft pseudo-label fusion. Appl. Intell. 2023, 1–13. [Google Scholar] [CrossRef]
- Punn, N.S.; Agarwal, S. BT-Unet: A self-supervised learning framework for biomedical image segmentation using barlow twins with U-net models. Mach. Learn. 2022, 111, 4585–4600. [Google Scholar] [CrossRef]
- Bonechi, S. ISIC_WSM: Generating Weak Segmentation Maps for the ISIC archive. Neurocomputing 2023, 523, 69–80. [Google Scholar] [CrossRef]
- Chu, T.; Li, X.; Vo, H.V.; Summers, R.M.; Sizikova, E. Improving weakly supervised lesion segmentation using multi-task learning. In Proceedings of the Fourth Conference on Medical Imaging with Deep Learning, Lübeck, Germany, 7–9 July 2021; pp. 60–73. [Google Scholar]
- Wei, Y.; Xiao, H.; Shi, H.; Jie, Z.; Feng, J.; Huang, T.S. Revisiting dilated convolution: A simple approach for weakly- and semi- supervised semantic segmentation. In Proceedings of the 2018 IEEE/CVF Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–22 June 2018; pp. 7268–7277. [Google Scholar]
- Jadon, S. A survey of loss functions for semantic segmentation. In Proceedings of the 2020 IEEE Conference on Computational Intelligence in Bioinformatics and Computational Biology (CIBCB), Via del Mar, Chile, 27–29 October 2020; pp. 1–7. [Google Scholar]
- Yi-de, M.; Qing, L.; Zhi-Bai, Q. Automated image segmentation using improved PCNN model based on cross-entropy. In Proceedings of the 2004 International Symposium on Intelligent Multimedia, Video and Speech Processing, Hong Kong, China, 20–22 October 2004; pp. 743–746. [Google Scholar]
- Sudre, C.H.; Li, W.; Vercauteren, T.; Ourselin, S.; Jorge Cardoso, M. Generalised dice overlap as a deep learning loss function for highly unbalanced segmentations. In Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support; Springer International Publishing: Cham, Switzerland, 2017; pp. 240–248. [Google Scholar]
- Gutman, D.; Codella, N.C.; Celebi, E.; Helba, B.; Marchetti, M.; Mishra, N.; Halpern, A. Skin lesion analysis toward melanoma detection: A challenge at the international symposium on biomedical imaging (ISBI) 2016, hosted by the international skin imaging collaboration (ISIC). arXiv 2016, arXiv:1605.01397. [Google Scholar]
- Al-Masni, M.A.; Al-Antari, M.A.; Choi, M.-T.; Han, S.-M.; Kim, T.-S. Skin lesion segmentation in dermoscopy images via deep full resolution convolutional networks. Comput. Methods Programs Biomed. 2018, 162, 221–231. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Xie, Y.; Wu, Q.; Xia, Y. Medical image classification using synergic deep learning. Med. Image Anal. 2019, 54, 10–19. [Google Scholar] [CrossRef]
- Tang, P.; Liang, Q.; Yan, X.; Xiang, S.; Zhang, D. GP-CNN-DTEL: Global-part CNN model with data-transformed ensemble learning for skin lesion classification. IEEE J. Biomed. Health Inform. 2020, 24, 2870–2882. [Google Scholar] [CrossRef] [PubMed]
- Ding, S.; Wu, Z.; Zheng, Y.; Liu, Z.; Yang, X.; Yang, X.; Yuan, G.; Xie, J. Deep attention branch networks for skin lesion classification. Comput. Methods Programs Biomed. 2021, 212, 106447. [Google Scholar] [CrossRef]
- He, X.; Wang, Y.; Zhao, S.; Yao, C. Deep metric attention learning for skin lesion classification in dermoscopy images. Complex Intell. Syst. 2022, 8, 1487–1504. [Google Scholar] [CrossRef]
- Matsunaga, K.; Hamada, A.; Minagawa, A.; Koga, H. Image classification of melanoma, nevus and seborrheic keratosis by deep neural network ensemble. arXiv 2017, arXiv:1703.03108. [Google Scholar]
- Díaz, I.G. Incorporating the knowledge of dermatologists to convolutional neural networks for the diagnosis of skin lesions. arXiv 2017, arXiv:1703.01976. [Google Scholar]
- Menegola, A.; Tavares, J.; Fornaciali, M.; Li, L.T.; Avila, S.; Valle, E. RECOD titans at ISIC challenge 2017. arXiv 2017, arXiv:1703.04819. [Google Scholar]
- Bi, L.; Kim, J.; Ahn, E.; Feng, D. Automatic skin lesion analysis using large-scale dermoscopy images and deep residual networks. arXiv 2017, arXiv:1703.04197. [Google Scholar]
- Yang, X.; Zeng, Z.; Yeo, S.Y.; Tan, C.; Tey, H.L.; Su, Y. A novel multi-task deep learning model for skin lesion segmentation and classification. arXiv 2017, arXiv:1703.01025. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).