Time and Energy Benefits of Using Automatic Optimization Compilers for NPDP Tasks
Abstract
:1. Introduction
2. NPDP Code Optimization
- The Nussinov algorithm [15] (RNA folding)
- The Zuker algorithm [16] (RNA folding)
- The Smith–Waterman algorithm [17] (aligning sequences)
- The Needleman–Wunsch algorithm [5] (aligning sequences)
- The Smith–Waterman algorithm for three sequences [17] (aligning sequences)
- The counting algorithm [18] (RNA folding)
- The McCaskill’s kernel [19] (RNA folding)
- MEA [20] (RNA folding)
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| NPDP | Non-serial polyadic dynamic programming |
| SW | Smith–Waterman |
| NW | Needleman–Wunsch |
| ATF | Affine Transformation Framework |
| MEA | Prediction of maximum expected accuracy |
| RNA | Ribonucleic acid |
| DAPT | Dependence Approximation Program Transformation |
| ECC | Error correction code |
References
- Mullapudi, R.T.; Bondhugula, U. Tiling for Dynamic Scheduling. In Proceedings of the 4th International Workshop on Polyhedral Compilation Techniques, Vienna, Austria, 20–22 January 2014. [Google Scholar]
- Wonnacott, D.; Jin, T.; Lake, A. Automatic tiling of “mostly-tileable” loop nests. In Proceedings of the 5th International Workshop on Polyhedral Compilation Techniques, Amsterdam, The Netherlands, 19–21 January 2015. [Google Scholar]
- Chowdhury, R.; Ganapathi, P.; Tschudi, S.; Tithi, J.J.; Bachmeier, C.; Leiserson, C.E.; Solar-Lezama, A.; Kuszmaul, B.C.; Tang, Y. Autogen: Automatic Discovery of Efficient Recursive Divide-8-Conquer Algorithms for Solving Dynamic Programming Problems. ACM Trans. Parallel Comput. 2017, 4, 4. [Google Scholar] [CrossRef]
- Bielecki, W.; Blaszynski, P.; Poliwoda, M. 3D parallel tiled code implementing a modified Knuth’s optimal binary search tree algorithm. J. Comput. Sci. 2021, 48, 101246. [Google Scholar] [CrossRef]
- Needleman, S.B.; Wunsch, C.D. A General Method Applicable to the Search for Similarities in the Amino Acid Sequence of Two Proteins. In Molecular Biology; Elsevier: Amsterdam, The Netherlands, 1989; pp. 453–463. [Google Scholar] [CrossRef]
- Xue, J. Loop Tiling for Parallelism; Kluwer Academic Publishers: Norwell, MA, USA, 2000. [Google Scholar]
- Palkowski, M.; Bielecki, W. Parallel tiled Nussinov RNA folding loop nest generated using both dependence graph transitive closure and loop skewing. BMC Bioinform. 2017, 18, 290. [Google Scholar] [CrossRef] [PubMed]
- Bondhugula, U.; Hartono, A.; Ramanujam, J.; Sadayappan, P. A practical automatic polyhedral parallelizer and locality optimizer. SIGPLAN Not. 2008, 43, 101–113. [Google Scholar] [CrossRef]
- Bielecki, W.; Palkowski, M. A Parallelizing and Optimizing Compiler-TRACO. 2013. Available online: http://traco.sourceforge.net (accessed on 1 August 2023).
- Bielecki, W.; Poliwoda, M. Automatic Parallel Tiled Code Generation Based on Dependence Approximation. In Parallel Computing Technologies, Proceedings of the 16th International Conference, PaCT 2021, Kaliningrad, Russia, 13–18 September 2021; Malyshkin, V., Ed.; Springer International Publishing: Cham, Switzerland, 2021; pp. 260–275. [Google Scholar]
- Palkowski, M.; Bielecki, W. NPDP benchmark suite for the evaluation of the effectiveness of automatic optimizing compilers. Parallel Comput. 2023, 116, 103016. [Google Scholar] [CrossRef]
- Schone, R.; Ilsche, T.; Bielert, M.; Velten, M.; Schmidl, M.; Hackenberg, D. Energy Efficiency Aspects of the AMD Zen 2 Architecture. In Proceedings of the 2021 IEEE International Conference on Cluster Computing (CLUSTER), Portland, OR, USA, 7–10 September 2021. [Google Scholar] [CrossRef]
- Kelly, W.; Maslov, V.; Pugh, W.; Rosser, E.; Shpeisman, T.; Wonnacott, D. New User Interface for Petit and Other Extensions. User Guide 1996, 1, 996. [Google Scholar]
- Verdoolaege, S. Integer Set Library—Manual. Technical Report. 2011. Available online: https://compsys-tools.ens-lyon.fr/iscc/isl.pdf (accessed on 1 August 2023).
- Nussinov, R.; Pieczenik, G.; Griggs, J.R.; Kleitman, D.J. Algorithms for loop matchings. Siam J. Appl. Math. 1978, 35, 68–82. [Google Scholar] [CrossRef]
- Zuker, M.; Stiegler, P. Optimal computer folding of large RNA sequences using thermodynamics and auxiliary information. Nucleic Acids Res. 1981, 9, 133–148. [Google Scholar] [CrossRef] [PubMed]
- Palkowski, M.; Bielecki, W. Parallel Tiled Codes Implementing the Smith-Waterman Alignment Algorithm for Two and Three Sequences. J. Comput. Biol. 2018, 25, 1106–1119. [Google Scholar] [CrossRef] [PubMed]
- Freiburg Bioinformatics Group. Freiburg RNA Tools, Teaching RNA Algorithms. 2022. Available online: https://rna.informatik.uni-freiburg.de/teaching (accessed on 1 August 2023).
- McCaskill, J.S. The equilibrium partition function and base pair binding probabilities for RNA secondary structure. Biopolymers 1990, 29, 1105–1119. [Google Scholar] [CrossRef] [PubMed]
- Lu, Z.J.; Gloor, J.W.; Mathews, D.H. Improved RNA secondary structure prediction by maximizing expected pair accuracy. RNA 2009, 15, 1805–1813. [Google Scholar] [CrossRef] [PubMed]
- Knuth, D.E. Optimum binary search trees. Acta Inform. 1971, 1, 14–25. [Google Scholar] [CrossRef]
- Palkowski, M.; Bielecki, W. Accelerating Minimum Cost Polygon Triangulation Code with the TRACO Compiler. In Proceedings of the Communication Papers of the 2018 Federated Conference on Computer Science and Information Systems, FedCSIS 2018, Poznań, Poland, 9–12 September 2018; pp. 111–114. [Google Scholar] [CrossRef]
- The Polyhedral Benchmark Suite. 2022. Available online: http://www.cse.ohio-state.edu/pouchet/software/polybench/ (accessed on 1 August 2023).
- McMahon, F.H. The Livermore Fortran Kernels: A Computer Test of the Numerical Performance Range; Technical Report UCRL-53745; Lawrence Livermore National Laboratory: Livermore, CA, USA, 1986. [Google Scholar]
- NAS Benchmarks Suite. 2013. Available online: http://www.nas.nasa.gov (accessed on 1 August 2023).
- Standard Performance Evaluation Corporation (SPEC). SPEChpc 2021 Benchmark Suites. 2021. Available online: https://www.spec.org/hpc2021/ (accessed on 1 August 2023).
- Chen, Z.; Gong, Z.; Szaday, J.J.; Wong, D.C.; Padua, D.; Nicolau, A.; Veidenbaum, A.V.; Watkinson, N.; Sura, Z.; Maleki, S.; et al. Lore: A loop repository for the evaluation of compilers. In Proceedings of the 2017 IEEE International Symposium on Workload Characterization (IISWC), Seattle, WA, USA, 1–3 October 2017; pp. 219–228. [Google Scholar]
- UTDSP Benchmark Suite. 2012. Available online: http://www.eecg.toronto.edu/corinna/DSP/infrastructure/UTDSP.html (accessed on 1 August 2023).
- Pozo, R.; Miller, B. SciMark 4.0. National Institute of Standards and Technology (NIST). 2018. Available online: https://math.nist.gov/scimark2/ (accessed on 1 August 2023).
- Bondhugula, U. Compiling affine loop nests for distributed-memory parallel architectures. In Proceedings of the SC13: International Conference for High Performance Computing, Networking, Storage and Analysis, Denver, CO, USA, 17–22 November 2013; ACM: New York, NY, USA, 2013. SC ’13. pp. 33:1–33:12. [Google Scholar] [CrossRef]
- Zhao, C.; Sahni, S. Cache and energy efficient algorithms for Nussinov’s RNA Folding. BMC Bioinform. 2017, 18, 518. [Google Scholar] [CrossRef]
- Li, J.; Ranka, S.; Sahni, S. Multicore and GPU algorithms for Nussinov RNA folding. BMC Bioinform. 2014, 15, S1. [Google Scholar] [CrossRef]
- Frid, Y.; Gusfield, D. An improved Four-Russians method and sparsified Four-Russians algorithm for RNA folding. Algorithms Mol. Biol. 2016, 11, 22. [Google Scholar] [CrossRef] [PubMed]
- Tchendji, V.K.; Youmbi, F.I.K.; Djamegni, C.T.; Zeutouo, J.L. A Parallel Tiled and Sparsified Four-Russians Algorithm for Nussinov’s RNA Folding. IEEE/ACM Trans. Comput. Biol. Bioinform. 2022, 20, 1795–1806. [Google Scholar] [CrossRef] [PubMed]
- OpenMP Architecture Review Board. OpenMP Application Program Interface, Version 4.0. 2012. Available online: https://www.openmp.org/wp-content/uploads/OpenMP4.0.0.pdf (accessed on 1 August 2023).
- Caamaño, J.M.M.; Selva, M.; Clauss, P.; Baloian, A.; Wolff, W. Full runtime polyhedral optimizing loop transformations with the generation, instantiation, and scheduling of code-bones. Concurr. Comput. Pract. Exp. 2017, 29, e4192. [Google Scholar] [CrossRef]
- Baskaran, M.M.; Hartono, A.; Tavarageri, S.; Henretty, T.; Ramanujam, J.; Sadayappan, P. Parameterized tiling revisited. In Proceedings of the 8th annual IEEE/ACM International Symposium on Code Generation and Optimization, Toronto, ON, Canada, 24–28 April 2010; ACM: New York, NY, USA, 2010. CGO ’10. pp. 200–209. [Google Scholar]
- Bielecki, W.; Palkowski, M.; Poliwoda, M. Automatic code optimization for computing the McCaskill partition functions. In Proceedings of the Annals of Computer Science and Information Systems, Sofia, Bulgaria, 4–7 September 2022. [Google Scholar] [CrossRef]
- Mahjoub, S.; Golsorkhtabaramiri, M.; Amiri, S.S.S.; Hosseinzadeh, M.; Mosavi, A. A New Combination Method for Improving Parallelism in Two and Three Level Perfect Nested Loops. IEEE Access 2022, 10, 74542–74554. [Google Scholar] [CrossRef]
- Chatradhi, N.K. Kernel Driver Amd_Energy. 2023. Available online: https://github.com/amd/amd_energy (accessed on 1 August 2023).
- Palkowski, M. Finding Free Schedules for RNA Secondary Structure Prediction. In Artificial Intelligence and Soft Computing, Proceedings of the 15th International Conference, ICAISC 2016, Zakopane, Poland, 12–16 June 2016; Rutkowski, L., Korytkowski, M., Scherer, R., Tadeusiewicz, R., Zadeh, L.A., Zurada, J.M., Eds.; Springer International Publishing: Berlin/Heidelberg, Germany, 2016; Part II; pp. 179–188. [Google Scholar]
- Grabein, A.; Bhaskaran, S. Latest Top500 List Highlights World’s Fastest and Most Energy Efficient Supercomputers Are Powered by AMD. 2023. Available online: https://ir.amd.com/news-events/press-releases/detail/1131/latest-top500-list-highlights-worlds-fastest-and-most (accessed on 11 August 2023).

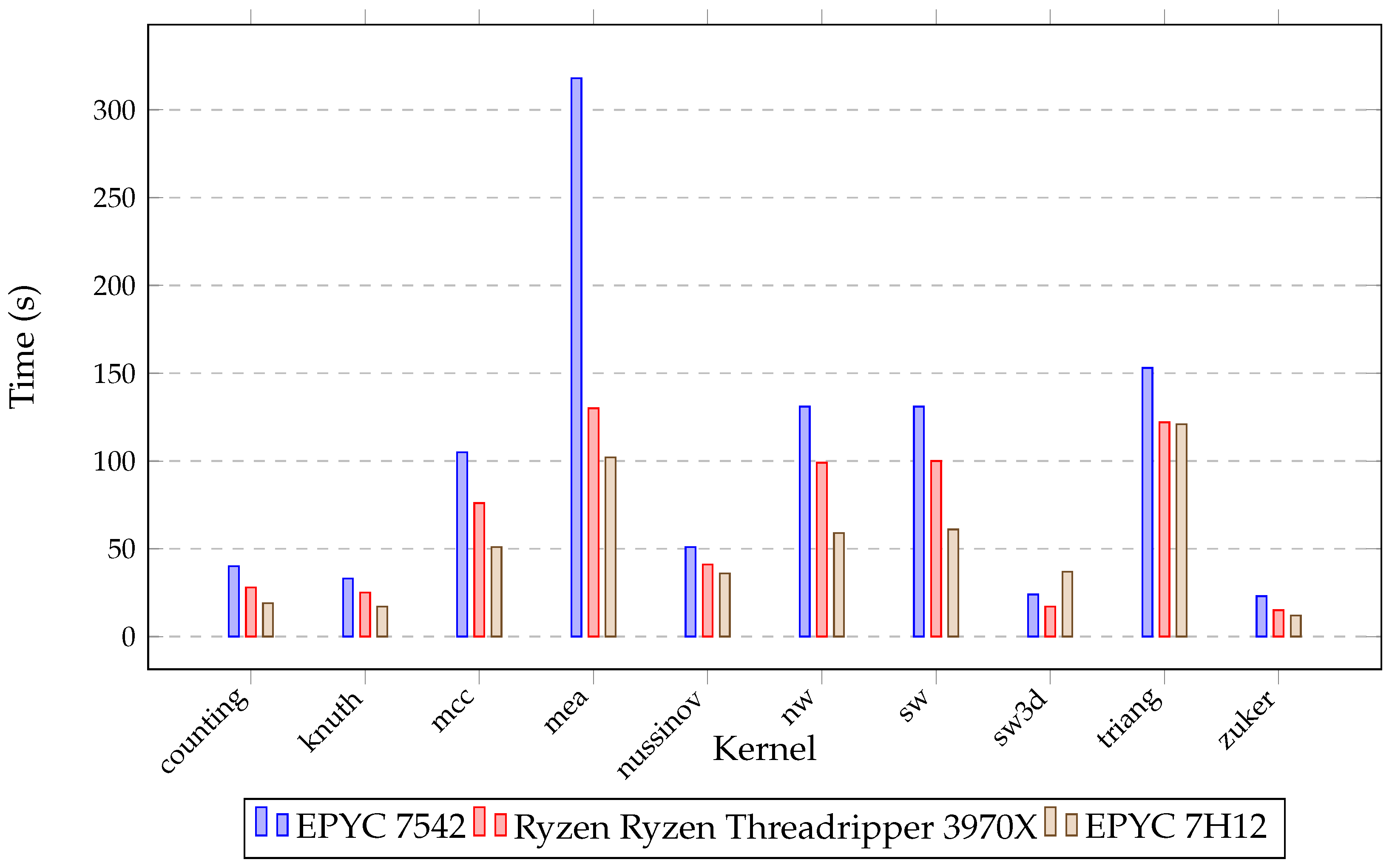
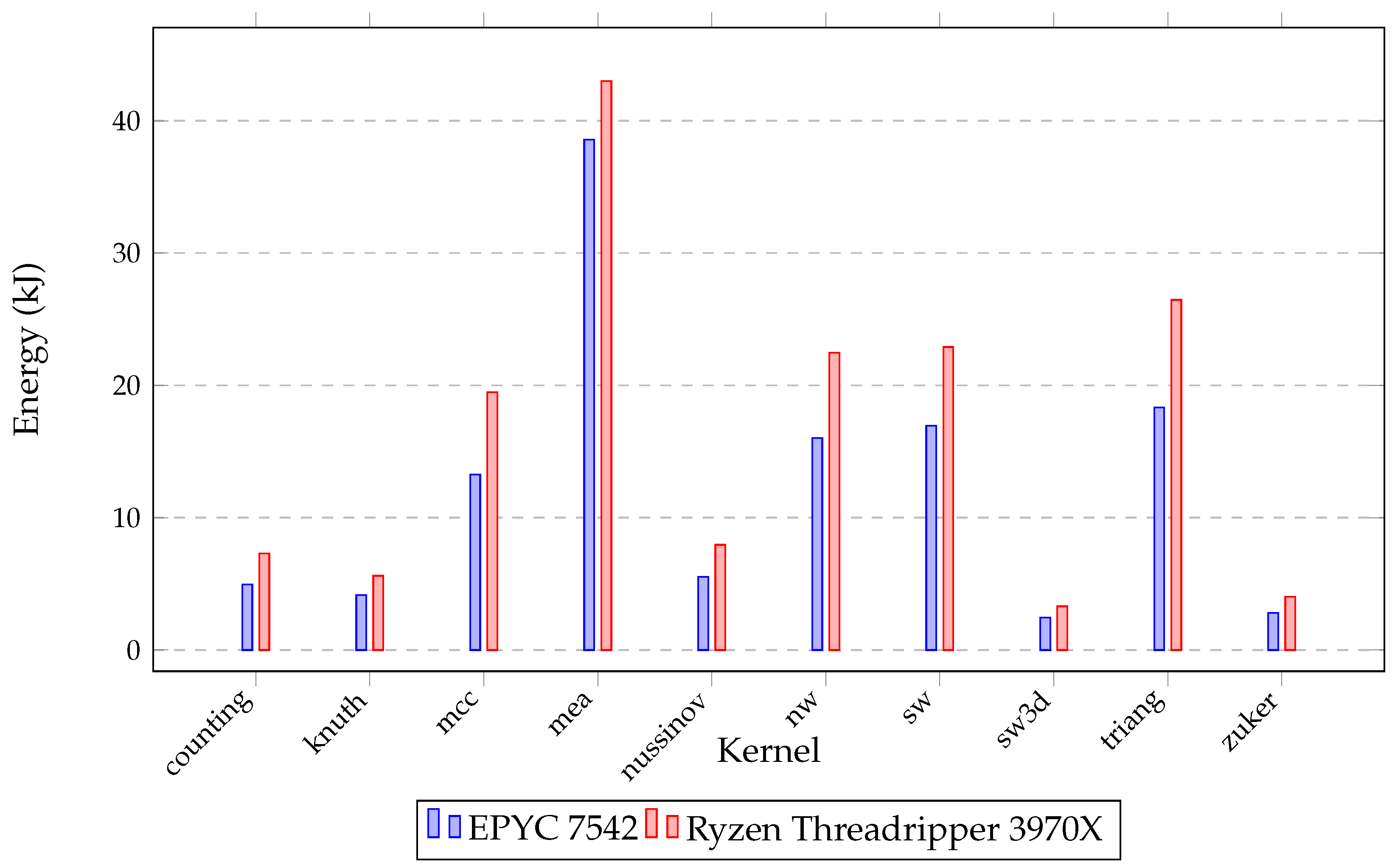
| Processor | Base Clock (GHz) | Turbo Clock (GHz) | Number of Cores | Number of Threads | Cache (MB) | RAM (GB) |
|---|---|---|---|---|---|---|
| EPYC 7542 | 2.9 | 3.4 | 32 | 64 | 128 | 256 |
| EPYC 7H12 | 2.6 | 3.3 | 64 | 128 | 256 | 64 |
| Ryzen Threadripper 3970X | 3.7 | 4.5 | 32 | 64 | 144 | 128 |
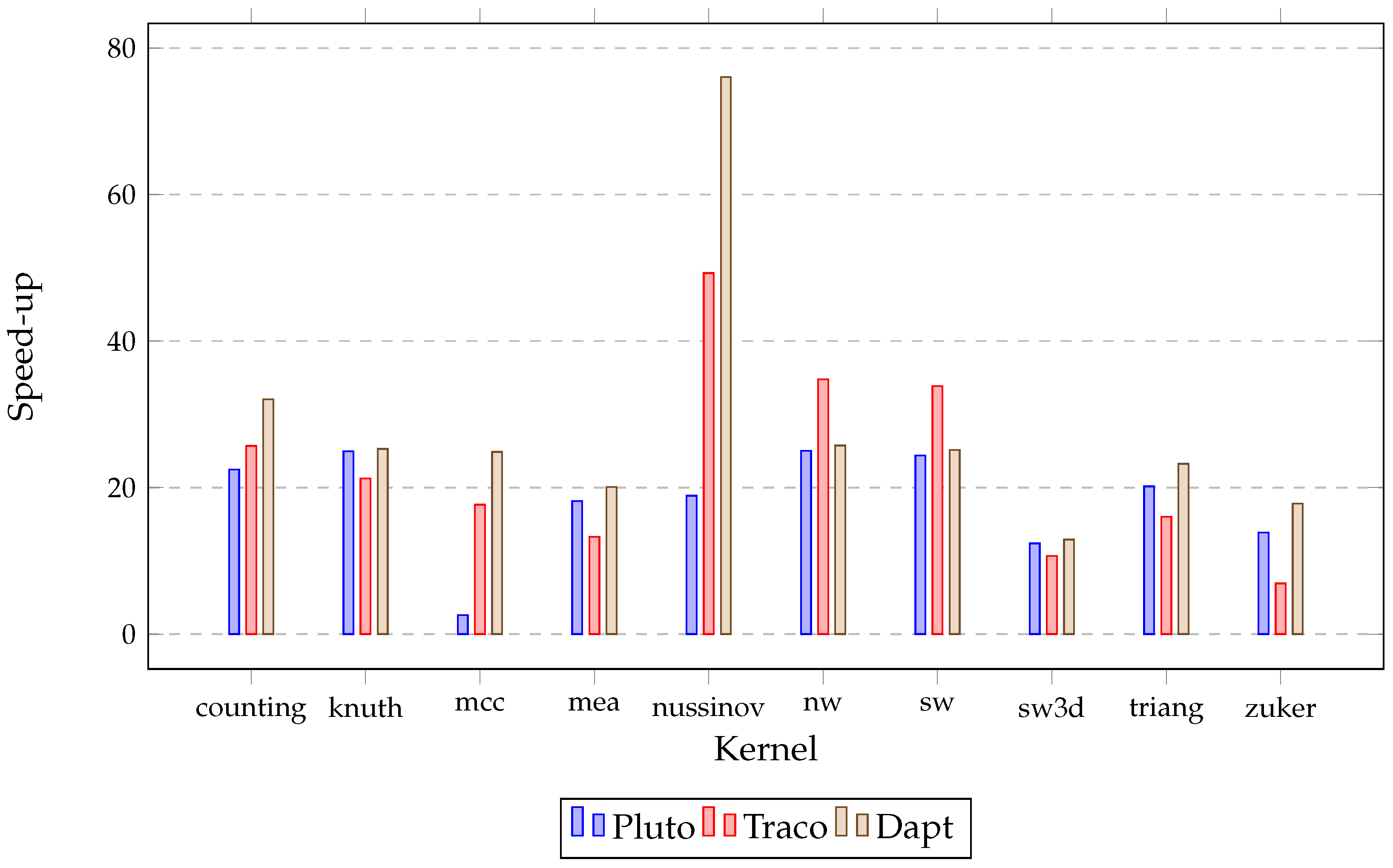
| Benchmark | Size | Original | Pluto | Traco | Dapt |
|---|---|---|---|---|---|
| counting | 10,000 | 1282.01 | 57.1 | 49.9 | 40.01 |
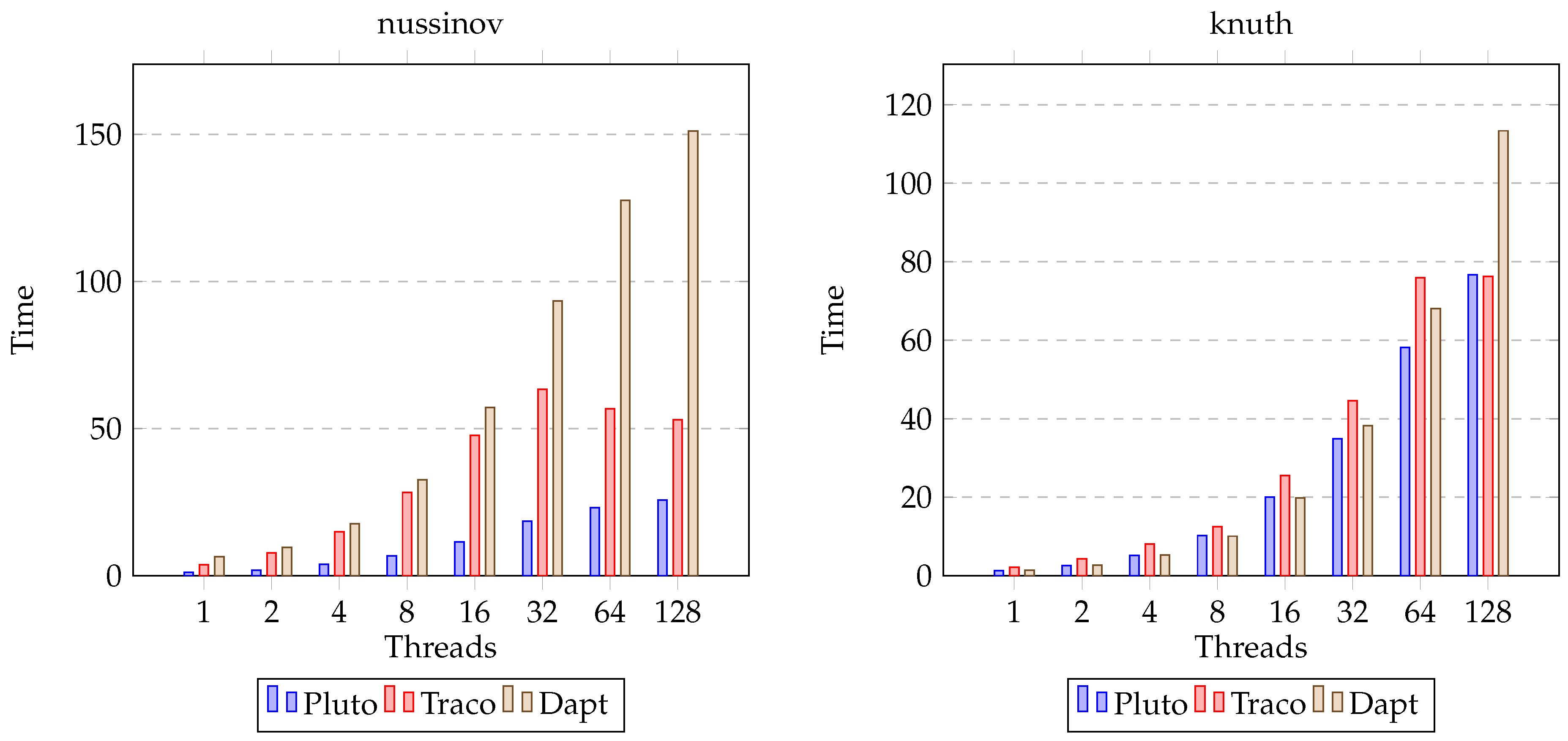
| knuth | 10,000 | 855.42 | 34.29 | 40.3 | 33.84 |
| mcc | 10,000 | 2632.3 | 1021.43 | 149.02 | 105.85 |
| mea | 2500 | 6397.83 | 352.42 | 481.98 | 318.77 |
| nussinov | 10,000 | 3880.43 | 205.33 | 78.74 | 51.03 |
| nw | 10,000 | 4567.33 | 182.56 | 131.33 | 177.32 |
| sw | 10,000 | 4483.13 | 183.96 | 132.46 | 178.33 |
| sw3d | 500 | 309.87 | 25.01 | 29.07 | 24.02 |
| triang | 10,000 | 3574.98 | 177.32 | 223.32 | 153.76 |
| zuker | 2000 | 415.55 | 29.98 | 60.02 | 23.3 |
| Benchmark | Size | Original | Pluto | Traco | Dapt |
|---|---|---|---|---|---|
| counting | 10,000 | 63,781 | 6212 | 5942 | 4946 |
| knuth | 10,000 | 41,317 | 4238 | 4873 | 4153 |
| mcc | 10,000 | 127,733 | 49,207 | 19,089 | 13,265 |
| mea | 2500 | 343,617 | 42,311 | 57,750 | 38,564 |
| nussinov | 10,000 | 184,569 | 24,189 | 8854 | 5519 |
| nw | 10,000 | 220,543 | 20,236 | 16,023 | 19,995 |
| sw | 10,000 | 215,963 | 20,541 | 16,960 | 20,349 |
| sw3d | 500 | 15,253 | 2478 | 2577 | 2450 |
| triang | 10,000 | 175,934 | 20,889 | 27,236 | 18,321 |
| zuker | 2000 | 20,124 | 3244 | 5394 | 2796 |
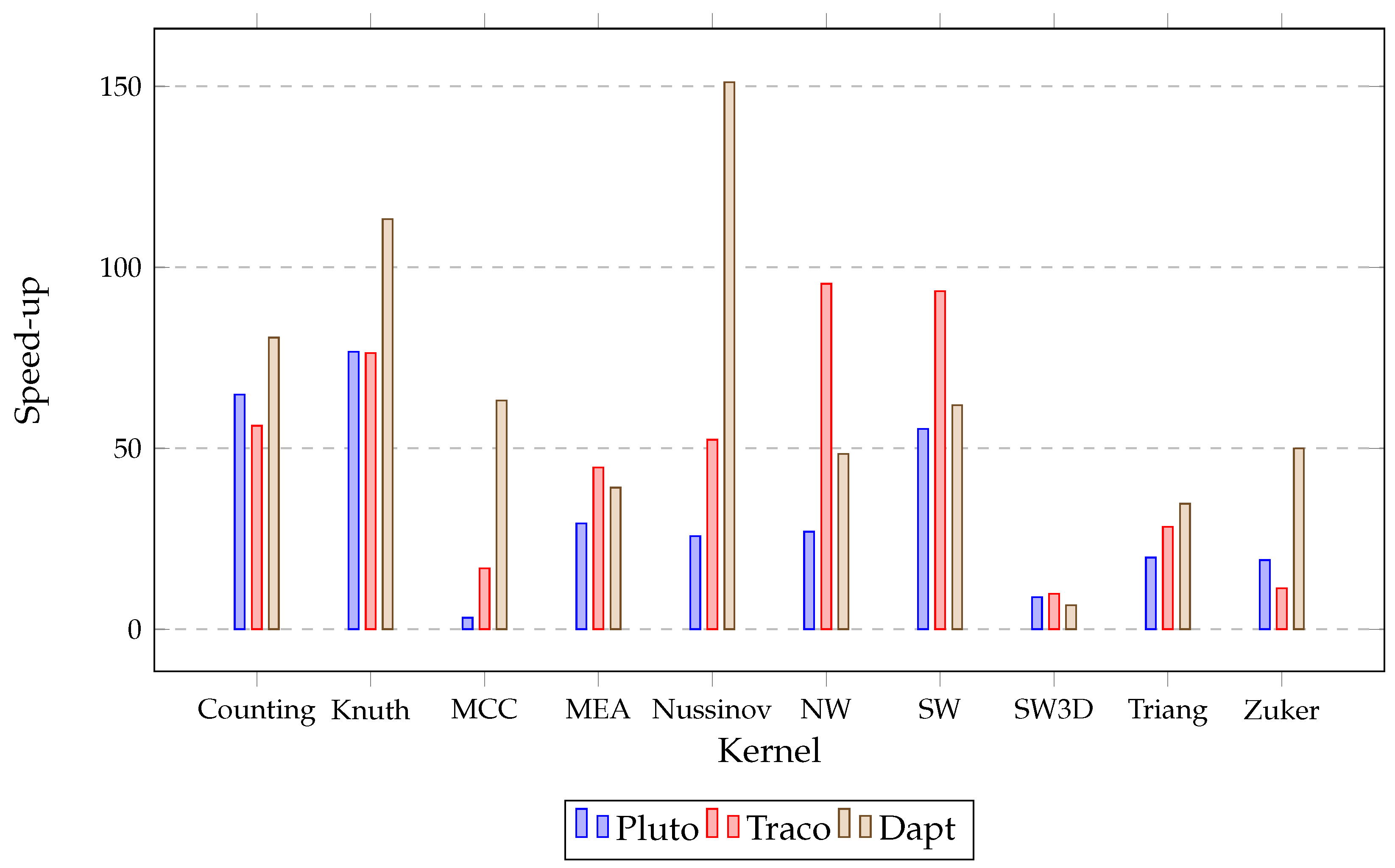
| Benchmark | Size | Original | Pluto | Traco | Dapt |
|---|---|---|---|---|---|
| counting | 10,000 | 1531.82 | 23.62 | 27.26 | 19.01 |
| knuth | 10,000 | 1939.06 | 25.3 | 25.42 | 17.11 |
| mcc | 10,000 | 3274.18 | 1033.98 | 194.6 | 51.84 |
| mea | 2500 | 4578.58 | 156.56 | 102.52 | 117.02 |
| nussinov | 10,000 | 5583.45 | 217.04 | 106.7 | 36.94 |
| nw | 10,000 | 5643.59 | 209.38 | 59.13 | 116.58 |
| sw | 10,000 | 5726.92 | 103.55 | 61.3 | 92.54 |
| sw3d | 500 | 363.64 | 41.05 | 37.18 | 54.92 |
| triang | 10,000 | 4207.33 | 212.6 | 148.7 | 121.37 |
| zuker | 2000 | 628.89 | 32.93 | 55.59 | 12.59 |
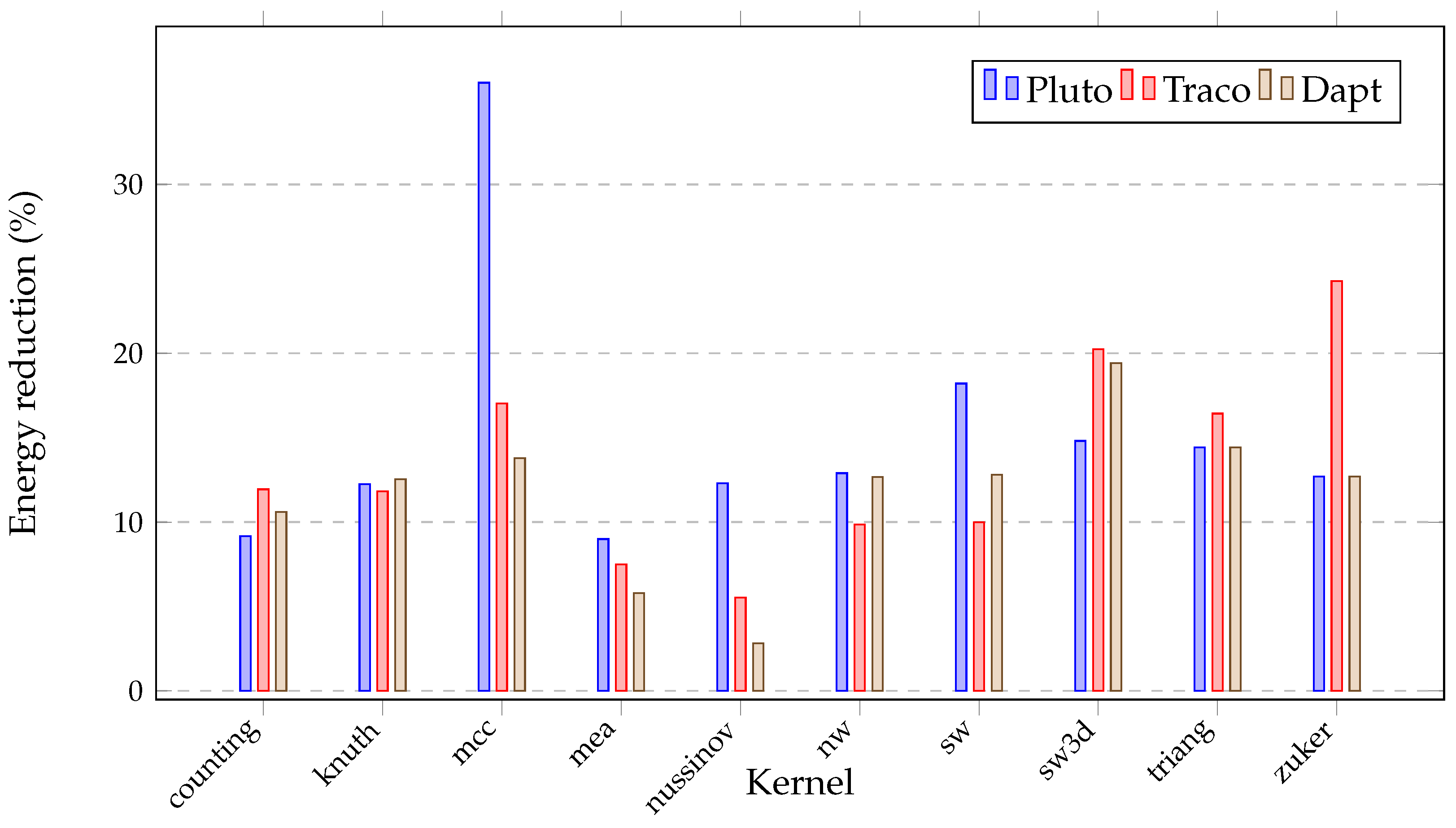
| Benchmark | Compiler | Serial Time (s) | Time (s) | Speed-Up | Energy (kJ) | Instr. per Cycle | Cache Misses (%) |
|---|---|---|---|---|---|---|---|
| Pluto | 30.68 | 32.71 | 6.31 | 0.76 | 35.94 | ||
| counting | Traco | 1003.51 | 34.93 | 28.73 | 8.22 | 0.72 | 22.51 |
| Dapt | 28.77 | 34.88 | 7.30 | 0.77 | 22.37 | ||
| Pluto | 25.94 | 25.26 | 5.54 | 0.29 | 25.68 | ||
| knuth | Traco | 655.36 | 25.07 | 26.14 | 5.35 | 0.26 | 21.81 |
| Dapt | 25.48 | 25.72 | 5.68 | 0.29 | 25.15 | ||
| Pluto | 755.70 | 2.66 | 50.93 | 1.86 | 49.68 | ||
| mcc | Traco | 2007.32 | 110.66 | 18.14 | 24.08 | 0.27 | 47.63 |
| Dapt | 76.89 | 26.11 | 19.49 | 0.35 | 34.12 | ||
| Pluto | 151.87 | 22.56 | 36.83 | 3.17 | 17.86 | ||
| mea | Traco | 3426.01 | 148.96 | 23.00 | 65.66 | 3.11 | 15.23 |
| Dapt | 130.53 | 26.25 | 52.99 | 2.98 | 14.02 | ||
| Pluto | 215.39 | 20.05 | 47.58 | 0.05 | 54.43 | ||
| nussinov | Traco | 4319.22 | 81.38 | 53.07 | 15.61 | 0.19 | 19.98 |
| Dapt | 41.51 | 104.05 | 7.94 | 0.57 | 7.22 | ||
| Pluto | 142.39 | 24.22 | 28.03 | 0.44 | 33.94 | ||
| nw | Traco | 3448.38 | 99.77 | 34.56 | 22.47 | 0.39 | 30.85 |
| Dapt | 144.45 | 23.87 | 28.91 | 0.37 | 33.64 | ||
| Pluto | 152.33 | 22.72 | 29.55 | 0.37 | 31.73 | ||
| sw | Traco | 3461.01 | 100.52 | 34.43 | 22.90 | 0.39 | 31.54 |
| Dapt | 146.33 | 23.65 | 29.34 | 0.37 | 34.57 | ||
| Pluto | 17.66 | 13.93 | 3.09 | 1.91 | 4.54 | ||
| sw3d | Traco | 245.98 | 22.22 | 11.07 | 3.43 | 1.57 | 5.42 |
| Dapt | 17.51 | 14.05 | 3.29 | 1.71 | 4.41 | ||
| Pluto | 132.28 | 20.40 | 27.37 | 0.96 | 22.58 | ||
| triang | Traco | 2698.32 | 140.29 | 19.23 | 30.35 | 0.68 | 38.45 |
| Dapt | 122.93 | 21.95 | 26.64 | 0.91 | 22.55 | ||
| Pluto | 22.69 | 20.07 | 4.56 | 1.91 | 8.47 | ||
| zuker | Traco | 455.37 | 49.08 | 9.28 | 7.66 | 1.71 | 7.17 |
| Dapt | 15.91 | 28.62 | 4.01 | 1.71 | 8.21 |
| Features/Machine | EPYC | Ryzen Threadripper |
|---|---|---|
| Target: | Server/datacenter platform | Efficient workstation platform |
| Characteristics: | Dual CPU configuration supports - up to 2 TB RAM in 8 channels - doubling number of cores (up to 128) and threads (up to 256) | Single socket supports - up to 256 GB RAM in 4 channels - 64 total processing cores (and 128 threads) |
| Frequency: | Boost/Turbo around 3.2 GHz, | Boost speed of 4.3–4.5 GH |
| Software: | Machine learning; scientific simulations like CFD | VFX, video, and rendering; CAD; media and entertainment |
| Strong points: | Higher core count, scalability, better efficiency/performance per watt, easier cooling | Availability, more compatible motherboards, Windows 11 support with drivers, lower price |
| EEC support: | Yes | Only PRO version |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Palkowski, M.; Gruzewski, M. Time and Energy Benefits of Using Automatic Optimization Compilers for NPDP Tasks. Electronics 2023, 12, 3579. https://doi.org/10.3390/electronics12173579
Palkowski M, Gruzewski M. Time and Energy Benefits of Using Automatic Optimization Compilers for NPDP Tasks. Electronics. 2023; 12(17):3579. https://doi.org/10.3390/electronics12173579
Chicago/Turabian StylePalkowski, Marek, and Mateusz Gruzewski. 2023. "Time and Energy Benefits of Using Automatic Optimization Compilers for NPDP Tasks" Electronics 12, no. 17: 3579. https://doi.org/10.3390/electronics12173579
APA StylePalkowski, M., & Gruzewski, M. (2023). Time and Energy Benefits of Using Automatic Optimization Compilers for NPDP Tasks. Electronics, 12(17), 3579. https://doi.org/10.3390/electronics12173579






