A Multi-Task Dense Network with Self-Supervised Learning for Retinal Vessel Segmentation
Abstract
1. Introduction
2. Related works
3. Method
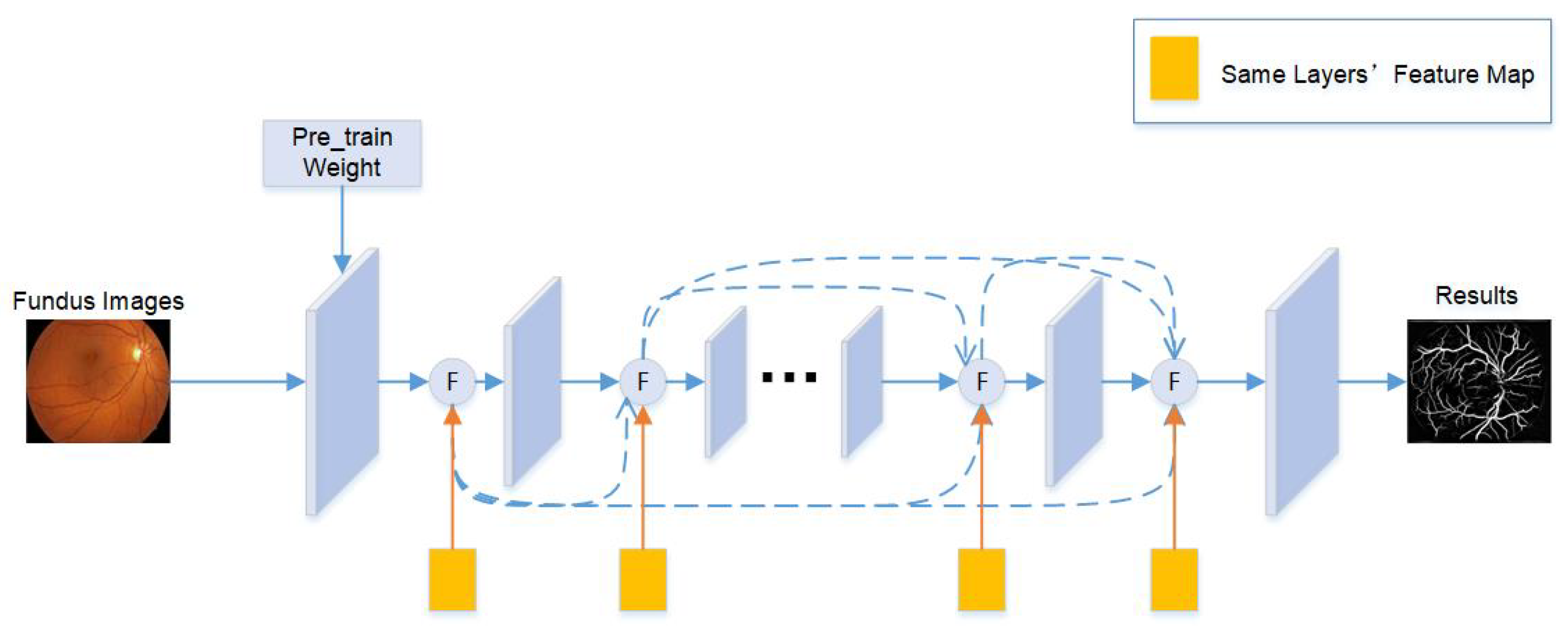
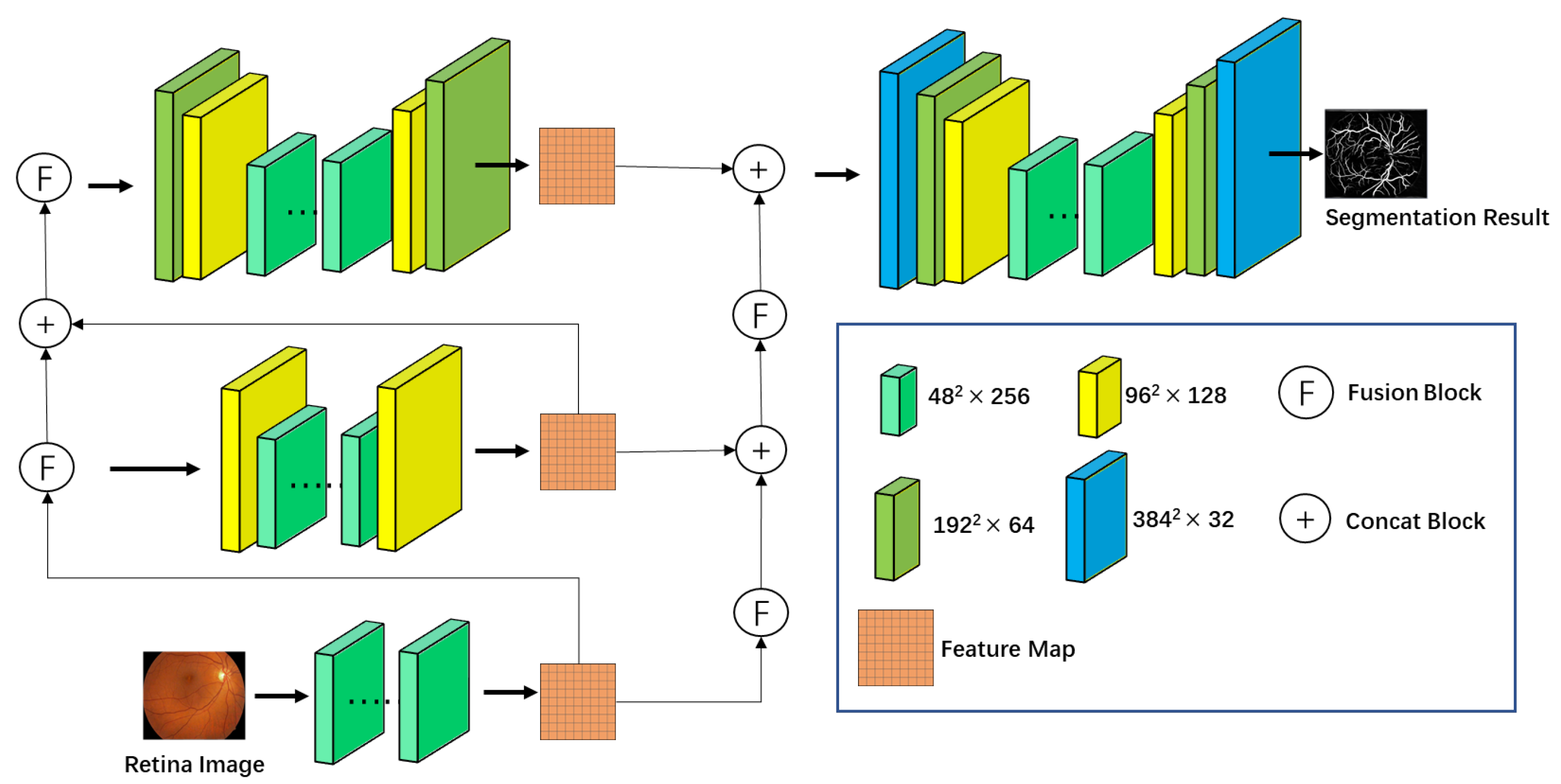
3.1. Network Architecture
3.2. Pre-Training Task Aggregation
3.3. Target Segmentation Tasks
4. Experiments and Results
4.1. Implements Details
4.2. Datasets
4.3. Evaluation Metrics
- Score 5, the segmentation results are excellent (the clarity of the retinal vascular structure is between 80% and 100%, and the vessel segmentation results are not inferior to human eye observation);
- Score 4, the segmentation results are favorable (the clarity of retinal vascular structure is between 60% and 80%, and the vessel segmentation result has few differences with human eye observation, and a few capillaries can not be distinguished);
- Score 3, the segmentation results are borderline (the clarity of retinal vascular structure is between 40–60%, there is a gap between the vascular segmentation result and human eye observation, and a few capillaries are missing);
- Score 2, the segmentation results are weakly poor (retinal vascular structure clarity between 20–40%, the disparity between vascular segmentation results and human eye observation, and a large number of capillaries are missing);
- Score 1, the segmentation results are strongly poor (the clarity of retinal vascular structure is between 0–20%, there is a big gap between the vascular segmentation result and human eye observation, and the main vessels are missing).
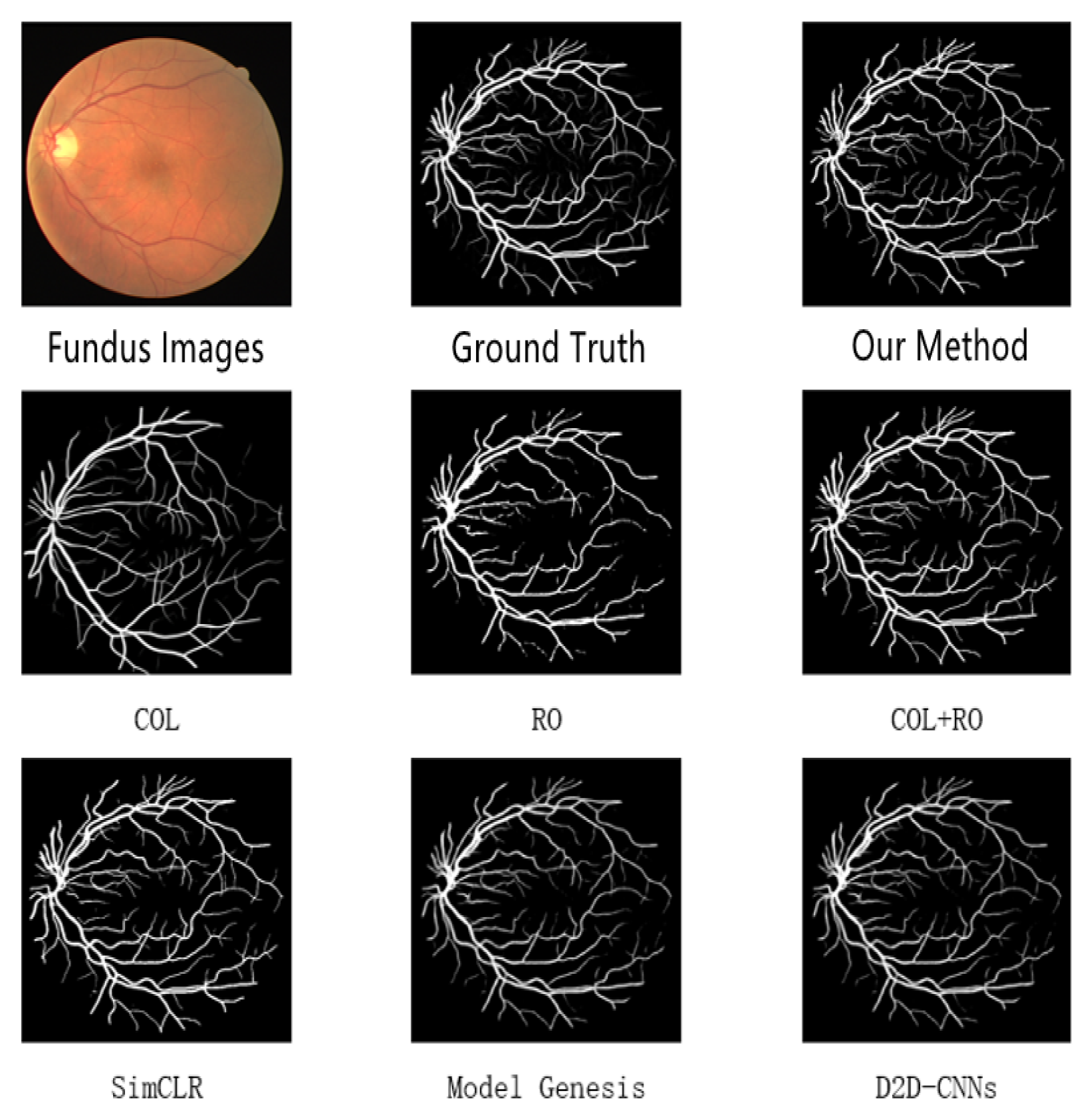
4.4. Results and Analysis
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Balyen, L.; Peto, T. Promising artificial intelligence-machine learning-deep learning algorithms in ophthalmology. Asia-Pac. J. Ophthalmol. 2019, 8, 264–272. [Google Scholar]
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional networks for biomedical image segmentation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Munich, Germany, 5–9 October 2015; Springer: Berlin/Heidelberg, Germany, 2015; pp. 234–241. [Google Scholar]
- He, Q.; Zou, B.; Zhu, C.; Liu, X.; Fu, H.; Wang, L. Multi-Label Classification Scheme Based on Local Regression for Retinal Vessel Segmentation. In Proceedings of the 2018 25th IEEE International Conference on Image Processing (ICIP), Athens, Greece, 7–10 October 2018. [Google Scholar]
- Fraz, M.M.; Remagnino, P.; Hoppe, A.; Uyyanonvara, B.; Rudnicka, A.R.; Owen, C.G.; Barman, S.A. Blood vessel segmentation methodologies in retinal images—A survey. Comput. Methods Programs Biomed. 2012, 108, 407–433. [Google Scholar] [CrossRef] [PubMed]
- Roy, K.; Chaudhuri, S.S.; Roy, P.; Chatterjee, S.; Banerjee, S. Transfer Learning Coupled Convolution Neural Networks in Detecting Retinal Diseases Using OCT Images. In Intelligent Computing: Image Processing Based Applications; Springer: Berlin/Heidelberg, Germany, 2020; pp. 153–173. [Google Scholar]
- Yue, K.; Zou, B.; Chen, Z.; Liu, Q. Retinal vessel segmentation using dense U-net with multiscale inputs. J. Med. Imaging 2019, 6, 034004. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Hu, G.; Shang, L.; Geng, J. Adaptive tracking extraction of vessel centerlines in coronary arteriograms using Hessian matrix. J.-Tsinghua Univ. 2007, 47, 889. [Google Scholar]
- Long, J.; Shelhamer, E.; Darrell, T. Fully convolutional networks for semantic segmentation. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Boston, MA, USA, 7–12 June 2015; pp. 3431–3440. [Google Scholar]
- Wang, D.; Haytham, A.; Pottenburgh, J.; Saeedi, O.; Tao, Y. Hard attention net for automatic retinal vessel segmentation. IEEE J. Biomed. Health Inform. 2020, 24, 3384–3396. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Qiu, S.; He, H. Dual encoding u-net for retinal vessel segmentation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Shenzhen, China, 13–17 October 2019; Springer: Berlin/Heidelberg, Germany, 2019; pp. 84–92. [Google Scholar]
- Ma, Y.; Hao, H.; Xie, J.; Fu, H.; Zhang, J.; Yang, J.; Wang, Z.; Liu, J.; Zheng, Y.; Zhao, Y. ROSE: A retinal OCT-angiography vessel segmentation dataset and new model. IEEE Trans. Med. Imaging 2020, 40, 928–939. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Li, Y.; Hu, Y.; Ma, K.; Zhou, S.K.; Zheng, Y. Rubik’s cube+: A self-supervised feature learning framework for 3d medical image analysis. Med. Image Anal. 2020, 64, 101746. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Sodha, V.; Pang, J.; Gotway, M.B.; Liang, J. Models genesis. Med. Image Anal. 2021, 67, 101840. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Yao, L.; Zhou, T.; Dong, J.; Zhang, Y. Momentum contrastive learning for few-shot COVID-19 diagnosis from chest CT images. Pattern Recognit. 2021, 113, 107826. [Google Scholar] [CrossRef] [PubMed]
- Doersch, C.; Zisserman, A. Multi-task self-supervised visual learning. In Proceedings of the IEEE International Conference on Computer Vision, Venice, Italy, 22–29 October 2017; pp. 2051–2060. [Google Scholar]
- Zhu, J.; Li, Y.; Zhou, S.K. Aggregative Self-Supervised Feature Learning from a Limited Sample. arXiv 2020, arXiv:2012.07477. [Google Scholar]
- He, Y.; Yang, G.; Yang, J.; Chen, Y.; Kong, Y.; Wu, J.; Tang, L.; Zhu, X.; Dillenseger, J.L.; Shao, P.; et al. Dense biased networks with deep priori anatomy and hard region adaptation: Semi-supervised learning for fine renal artery segmentation. Med. Image Anal. 2020, 63, 101722. [Google Scholar] [CrossRef] [PubMed]
- Staal, J.; Abràmoff, M.D.; Niemeijer, M.; Viergever, M.A.; Van Ginneken, B. Ridge-based vessel segmentation in color images of the retina. IEEE Trans. Med. Imaging 2004, 23, 501–509. [Google Scholar] [CrossRef] [PubMed]
- Ballerini, L.; Fetit, A.E.; Wunderlich, S.; Lovreglio, R.; McGrory, S.; Valdes-Hernandez, M.; MacGillivray, T.; Doubal, F.; Deary, I.J.; Wardlaw, J.; et al. Retinal Biomarkers Discovery for Cerebral Small Vessel Disease in an Older Population. In Communications in Computer and Information Science; Springer International Publishing: Cham, Switzerland, 2020; pp. 400–409. [Google Scholar] [CrossRef]
- Hoover, A.; Kouznetsova, V.; Goldbaum, M. Locating blood vessels in retinal images by piecewise threshold probing of a matched filter response. IEEE Trans. Med. Imaging 2000, 19, 203–210. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Isola, P.; Efros, A.A. Colorful image colorization. In European Conference on Computer Vision; Springer: Berlin/Heidelberg, Germany, 2016; pp. 649–666. [Google Scholar]
- Gidaris, S.; Singh, P.; Komodakis, N. Unsupervised representation learning by predicting image rotations. arXiv 2018, arXiv:1803.07728. [Google Scholar]
- Chen, T.; Kornblith, S.; Norouzi, M.; Hinton, G. A simple framework for contrastive learning of visual representations. In International Conference on Machine Learning; PMLR: Toronto, ON, Canada, 2020; pp. 1597–1607. [Google Scholar]
- Zhou, Z.; Sodha, V.; Siddiquee, M.M.R.; Feng, R.; Tajbakhsh, N.; Gotway, M.B.; Liang, J. Models genesis: Generic autodidactic models for 3d medical image analysis. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Shenzhen, China, 3–17 October 2019; Springer: Cham, Switzerland, 2019; pp. 384–393. [Google Scholar]



| Methods | Subjective Metrics | Objective Metrics | ||
|---|---|---|---|---|
| MOS | DS | AUC | AC | |
| COL [21] | 3.7 | 0.734 | 0.7234 | 0.7907 |
| RO [22] | 2.8 | 0.735 | 0.7217 | 0.8783 |
| COL + RO | 3.5 | 0.947 | 0.8061 | 0.8158 |
| SimCLR [23] | 3.3 | 0.721 | 0.8826 | 0.9397 |
| Our Method | 4 | 0.962 | 0.9494 | 0.9274 |
| Model Genesis [24] | 3.9 | 0.96 | 0.9796 | 0.9246 |
| D2D-CNNs [9] | 3.5 | 0.917 | 0.9602 | 0.9867 |
| Pre-Train Dataset | Target Dataset | ACC |
|---|---|---|
| Drive | Vampire | 0.9067 |
| Drive | Drive | 0.9620 |
| Vampire | Drive | 0.9527 |
| Vampire | Vampire | 0.9130 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tu, Z.; Zhou, Q.; Zou, H.; Zhang, X. A Multi-Task Dense Network with Self-Supervised Learning for Retinal Vessel Segmentation. Electronics 2022, 11, 3538. https://doi.org/10.3390/electronics11213538
Tu Z, Zhou Q, Zou H, Zhang X. A Multi-Task Dense Network with Self-Supervised Learning for Retinal Vessel Segmentation. Electronics. 2022; 11(21):3538. https://doi.org/10.3390/electronics11213538
Chicago/Turabian StyleTu, Zhonghao, Qian Zhou, Hua Zou, and Xuedong Zhang. 2022. "A Multi-Task Dense Network with Self-Supervised Learning for Retinal Vessel Segmentation" Electronics 11, no. 21: 3538. https://doi.org/10.3390/electronics11213538
APA StyleTu, Z., Zhou, Q., Zou, H., & Zhang, X. (2022). A Multi-Task Dense Network with Self-Supervised Learning for Retinal Vessel Segmentation. Electronics, 11(21), 3538. https://doi.org/10.3390/electronics11213538






