EEG Emotion Recognition Based on Federated Learning Framework
Abstract
1. Introduction
- 1.
- This paper extends the FL method to EEG signal-based emotion recognition field and evaluates its accuracy in the DEAP and SEED datasets. Our validation shows that the FL method can lead to higher model accuracy;
- 2.
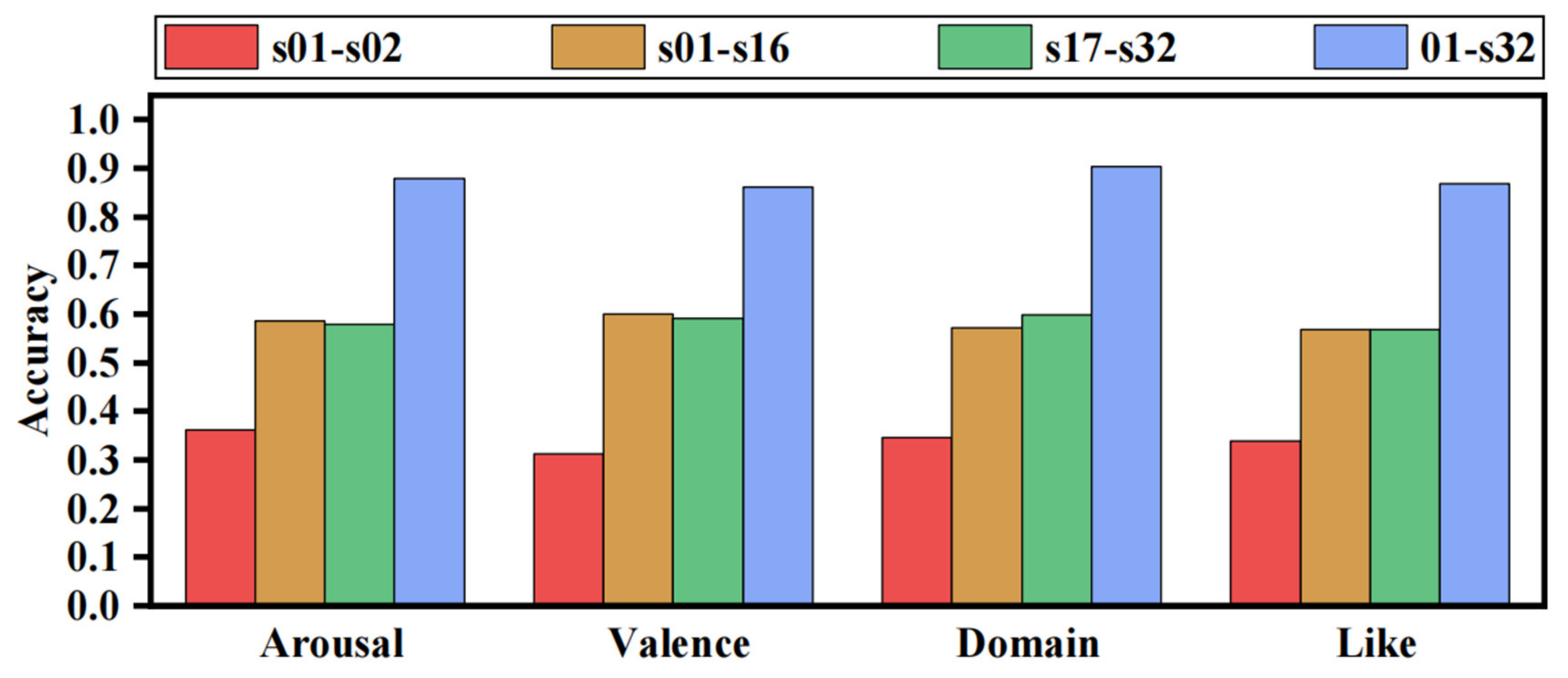
- We constructed different DEAP datasets for evaluating the effect of the diversity of training data on emotion recognition models. It was verified that the accuracy of the emotion recognition model using EEG signals is highly dependent on subjects and that increasing the diversity of subjects can substantially improve the model’s generalization performance, demonstrating the need for the FL method to be applied in this domain;
- 3.
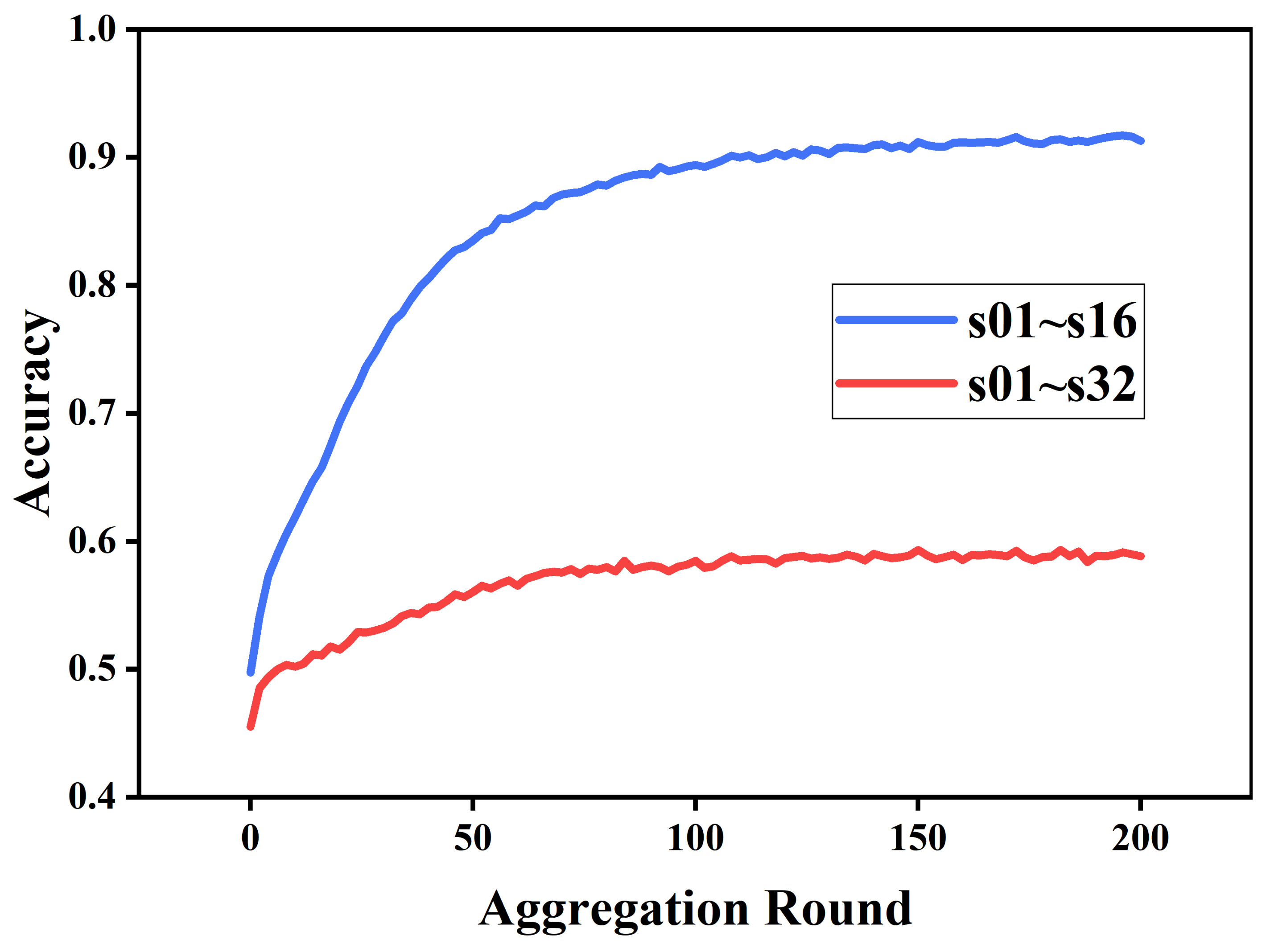
- The impact of the FL method on the accuracy and convergence speed of the emotion recognition model when trained on EEG data with N-IID distribution was evaluated by simulating the N-IID distribution of the inter-client DEAP dataset. Compared with the IID distribution, there is a substantial decrease in the accuracy of the FL-trained emotion recognition model under the N-IID distribution.
2. Materials and Methods
2.1. Electroencephalography—Emotion Recognition Dataset
2.2. Signal Pre-Processing
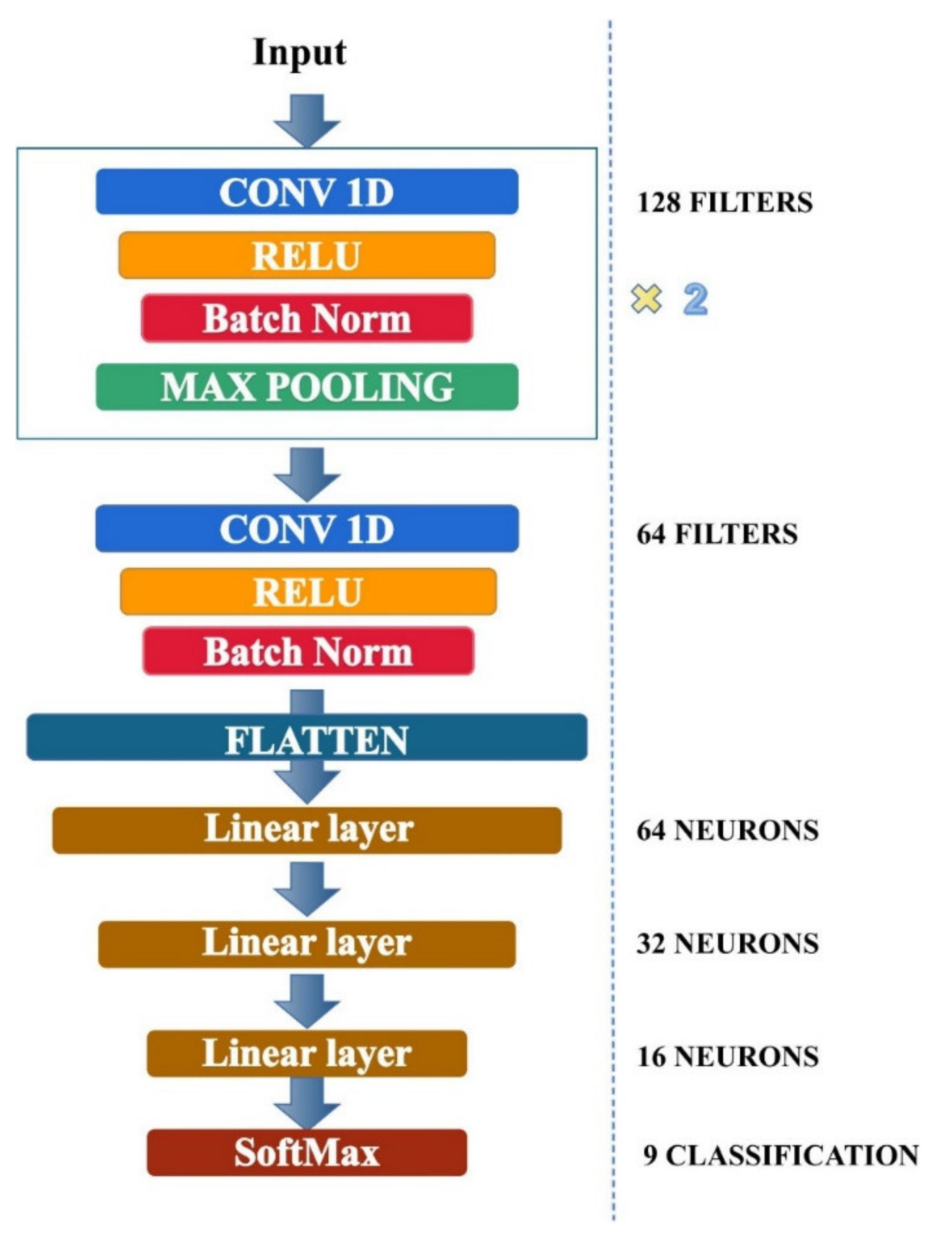
2.3. Emotion Recognition Model
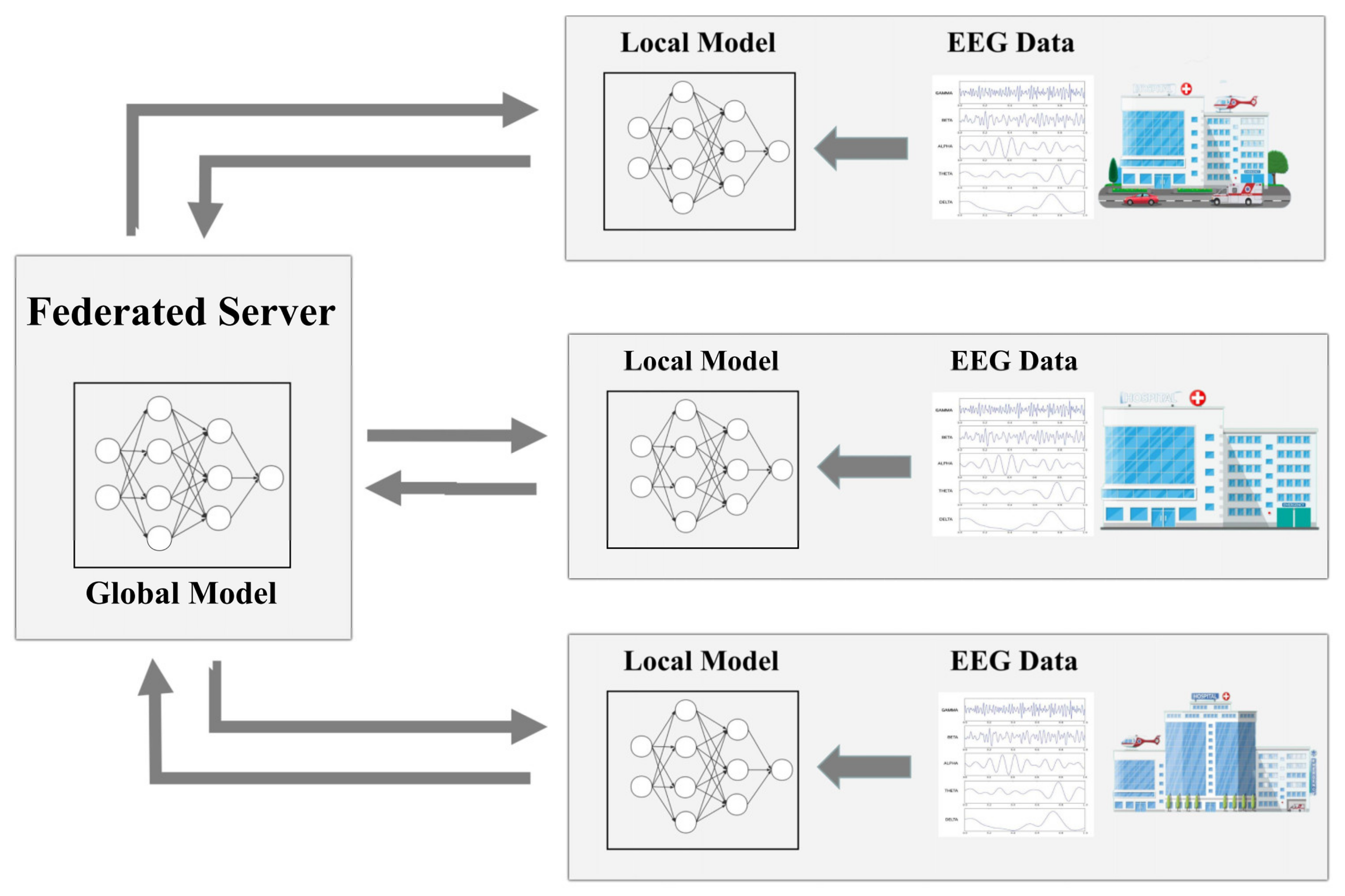
2.4. Federated Learning Algorithm
3. Results
3.1. Experimental Setup
3.2. Experimental Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Fragopanagos, N.; Taylor, J. Emotion recognition in human–computer interaction. Neural Netw. 2005, 18, 389–405. [Google Scholar] [CrossRef] [PubMed]
- Yin, Y.; Zheng, X.; Hu, B.; Zhang, Y.; Cui, X. EEG emotion recognition using fusion model of graph convolutional neural networks and LSTM. Appl. Soft Comput. 2020, 100, 106954. [Google Scholar] [CrossRef]
- Li, Y. A Survey of EEG Analysis based on Graph Neural Network. In Proceedings of the 2021 2nd International Conference on Electronics, Communications and Information Technology (CECIT), Sanya, China, 29 December 2021; pp. 151–155. [Google Scholar] [CrossRef]
- Huang, X.; Zhao, G.; Hong, X.; Zheng, W.; Pietikäinen, M. Spontaneous facial micro-expression analysis using Spatiotemporal Completed Local Quantized Patterns. Neurocomputing 2016, 175, 564–578. [Google Scholar] [CrossRef]
- Huang, X.; Wang, S.-J.; Liu, X.; Zhao, G.; Feng, X.; Pietikainen, M. Discriminative Spatiotemporal Local Binary Pattern with Revisited Integral Projection for Spontaneous Facial Micro-Expression Recognition. IEEE Trans. Affect. Comput. 2017, 10, 32–47. [Google Scholar] [CrossRef]
- Yin, Z.; Zhao, M.; Wang, Y.; Yang, J.; Zhang, J. Recognition of emotions using multimodal physiological signals and an ensemble deep learning model. Comput. Methods Programs Biomed. 2017, 140, 93–110. [Google Scholar] [CrossRef]
- Abadi, M.K.; Kia, M.; Subramanian, R.; Avesani, P.; Sebe, N. Decoding affect in videos employing the MEG brain signal. In Proceedings of the 2013 10th IEEE International Conference and Workshops on Automatic Face and Gesture Recognition (FG), Shanghai, China, 22–26 April 2013; pp. 1–6. [Google Scholar] [CrossRef]
- Jirayucharoensak, S.; Pan-Ngum, S.; Israsena, P. EEG-Based Emotion Recognition Using Deep Learning Network with Principal Component Based Covariate Shift Adaptation. Sci. World J. 2014, 2014, 627892. [Google Scholar] [CrossRef]
- Yang, B.; Han, X.; Tang, J. Three class emotions recognition based on deep learning using staked autoencoder. In Proceedings of the 2017 10th International Congress on Image and Signal Processing, BioMedical Engineering and Informatics (CISP-BMEI), Shanghai, China, 14–16 October 2017; pp. 1–5. [Google Scholar] [CrossRef]
- Wang, X.-H.; Zhang, T.; Xu, X.-M.; Chen, L.; Xing, X.-F.; Chen, C.L.P. EEG Emotion Recognition Using Dynamical Graph Convolutional Neural Networks and Broad Learning System. In Proceedings of the 2018 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), Madrid, Spain, 3–6 December 2018; pp. 1240–1244. [Google Scholar] [CrossRef]
- Cheng, J.; Chen, M.; Li, C.; Liu, Y.; Song, R.; Liu, A.; Chen, X. Emotion recognition from multi-channel eeg via deep forest. IEEE J. Biomed. Health. 2021, 25, 453–464. [Google Scholar] [CrossRef]
- George, F.P.; Shaikat, I.M.; Hossain, P.S.F.; Parvez, M.Z.; Uddin, J. Recognition of emotional states using EEG signals based on time-frequency analysis and SVM classifier. Int. J. Electr. Comput. Eng. (IJECE) 2019, 9, 1012–1020. [Google Scholar] [CrossRef]
- Fdez, J.; Guttenberg, N.; Witkowski, O.; Pasquali, A. Cross-Subject EEG-Based Emotion Recognition Through Neural Networks with Stratified Normalization. Front. Neurosci. 2021, 15, 626277. [Google Scholar] [CrossRef]
- Succetti, F.; Rosato, A.; Di Luzio, F.; Ceschini, A.; Panella, A.M. A FAST DEEP LEARNING TECHNIQUE FOR WI-FI-BASED HUMAN ACTIVITY RECOGNITION. Prog. Electromagn. Res. 2022, 174, 127–141. [Google Scholar] [CrossRef]
- Gong, D.; Ma, T.; Evans, J.; He, A.S. Deep Neural Networks for Image Super-Resolution in Optical Microscopy by Using Modified Hybrid Task Cascade U-Net. Prog. Electromagn. Res. 2021, 171, 185–199. [Google Scholar] [CrossRef]
- Islam, R.; Islam, M.; Rahman, M.; Mondal, C.; Singha, S.K.; Ahmad, M.; Awal, A.; Islam, S.; Moni, M.A. EEG Channel Correlation Based Model for Emotion Recognition. Comput. Biol. Med. 2021, 136, 104757. [Google Scholar] [CrossRef]
- Huang, D.; Chen, S.; Liu, C.; Zheng, L.; Tian, Z.; Jiang, D. Differences first in asymmetric brain: A bi-hemisphere discrepancy convolutional neural network for EEG emotion recognition. Neurocomputing 2021, 448, 140–151. [Google Scholar] [CrossRef]
- Acharya, D.; Jain, R.; Panigrahi, S.S.; Sahni, R.; Deshmukh, S.P.; Bhardwaj, A. Multi-class Emotion Classification Using EEG Signals. In International Advanced Computing Conference; Springer: Singapore, 2021; pp. 474–491. [Google Scholar] [CrossRef]
- Rudakov, E.; Laurent, L.; Cousin, V.; Roshdi, A.; Fournier, R.; Nait-Ali, A.; Beyrouthy, T.; Al Kork, S. Multi-Task CNN model for emotion recognition from EEG Brain maps. In Proceedings of the 2021 4th International Conference on Bio-Engineering for Smart Technologies (BioSMART), Online, 8–10 December 2021; pp. 1–4. [Google Scholar] [CrossRef]
- Solangi, Z.A.; Solangi, Y.A.; Chandio, S.; Aziz, M.B.S.A.; bin Hamzah, M.S.; Shah, A. The future of data privacy and security concerns in Internet of Things. In Proceedings of the 2018 IEEE International Conference on Innovative Research and Development (ICIRD), Bangkok, Thailand, 11–12 May 2018; pp. 1–4. [Google Scholar] [CrossRef]
- General Data Protection Regulation (GDPR). Available online: https://www.epsu.org/sites/default/files/article/files/GDPR_FINAL_EPSU.pdf (accessed on 10 October 2022).
- Valentin, O.; Ducharme, M.; Crétot-Richert, G.; Monsarrat-Chanon, H.; Viallet, G.; Delnavaz, A.; Voix, J. Validation and benchmarking of a wearable EEG acquisition platform for real-world applications. IEEE T. Biomed. Circ. S. 2019, 13, 103–111. [Google Scholar] [CrossRef]
- McMahan, B.; Moore, E.; Ramage, D.; Hampson, S.; Arcas, B.A. Communication-Efficient Learning of Deep Networks from Decentralized Data H. Artif. Intell. Stat. 2017, 54, 10. [Google Scholar]
- Theodora, S.B.; Chen, R.; Theofanie, M.; Olshevsky, A.; Paschalidis, I.C.; Shi, W. Federated learning of predictive models from federated electronic health records. Int. J. Med Inform. 2018, 112, 59–67. [Google Scholar] [CrossRef]
- Agbley, B.L.Y.; Li, J.; Haq, A.U.; Bankas, E.K.; Ahmad, S.; Agyemang, I.O.; Kulevome, D.; Ndiaye, W.D.; Cobbinah, B.; Latipova, S. Multimodal Melanoma Detection with Federated Learning. In Proceedings of the 2021 18th International Computer Conference on Wavelet Active Media Technology and Information Processing (ICCWAMTIP), Chengdu, China, 17–19 December 2021; pp. 238–244. [Google Scholar] [CrossRef]
- Malekzadeh, M.; Hasircioglu, B.; Mital, N.; Katarya, K.; Ozfatura, M.E.; Gündüz, D. Dopamine: Differentially Private Federated Learning on Medical Data. arXiv 2021, arXiv:2101.11693. [Google Scholar]
- Feki, I.; Ammar, S.; Kessentini, Y.; Muhammad, K. Federated learning for COVID-19 screening from Chest X-ray images. Appl. Soft Comput. 2021, 106, 107330. [Google Scholar] [CrossRef]
- Zhang, W.; Zhou, T.; Lu, Q.; Wang, X.; Zhu, C.; Sun, H.; Wang, Z.; Lo, S.K.; Wang, F.-Y. Dynamic-Fusion-Based Federated Learning for COVID-19 Detection. IEEE Internet Things J. 2021, 8, 15884–15891. [Google Scholar] [CrossRef]
- Li, X.; Huang, K.; Yang, W.; Wang, S.; Zhang, Z. On the Convergence of FedAvg on Non-IID Data. arXiv 2019, arXiv:1907.02189. [Google Scholar]
- Koelstra, S.; Muhl, C.; Soleymani, M.; Lee, J.-S.; Yazdani, A.; Ebrahimi, T.; Pun, T.; Nijholt, A.; Patras, I. DEAP: A Database for Emotion Analysis; Using Physiological Signals. IEEE Trans. Affect. Comput. 2011, 3, 18–31. [Google Scholar] [CrossRef]
- Zheng, W.-L.; Lu, B.-L. Investigating Critical Frequency Bands and Channels for EEG-Based Emotion Recognition with Deep Neural Networks. IEEE Trans. Auton. Ment. Dev. 2015, 7, 162–175. [Google Scholar] [CrossRef]
- Cimtay, Y.; Ekmekcioglu, E. Investigating the Use of Pretrained Convolutional Neural Network on Cross-Subject and Cross-Dataset EEG Emotion Recognition. Sensors 2020, 20, 2034. [Google Scholar] [CrossRef] [PubMed]
- Al-Fahoum, A.S.; Al-Fraihat, A.A. Methods of EEG Signal Features Extraction Using Linear Analysis in Frequency and Time-Frequency Domains. ISRN Neurosci. 2014, 2014, 730218. [Google Scholar] [CrossRef]
- Al-Qazzaz, N.K.; Sabir, M.K.; Ali, S.; Ahmad, S.A.; Grammer, K. Effective EEG Channels for Emotion Identification over the Brain Regions using Differential Evolution Algorithm. In Proceedings of the 2019 41st Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Berlin, Germany, 23–27 July 2019; Volume 2019, pp. 4703–4706. [Google Scholar] [CrossRef]
- Wang, J.; Charles, Z.; Xu, Z.; Joshi, G.; McMahan, H.B.; Al-Shedivat, M.; Andrew, G.; Avestimehr, S.; Daly, K.; Data, D.; et al. A Field Guide to Federated Optimization. arXiv 2021, arXiv:2107.06917. [Google Scholar]
- Li, Q.; Diao, Y.; Chen, Q.; He, B. Federated Learning on Non-IID Data Silos: An Experimental Study. In Proceedings of the 2022 IEEE 38th International Conference on Data Engineering (ICDE), Online, 9–12 May 2022; pp. 965–978. [Google Scholar] [CrossRef]
- Luo, Y.; Fu, Q.; Xie, J.; Qin, Y.; Wu, G.; Liu, J.; Jiang, F.; Cao, Y.; Ding, X. EEG-Based Emotion Classification Using Spiking Neural Networks. IEEE Access 2020, 8, 46007–46016. [Google Scholar] [CrossRef]
- Nawaz, R.; Cheah, K.H.; Nisar, H.; Yap, V.V. Comparison of different feature extraction methods for EEG-based emotion recognition. Biocybern. Biomed. Eng. 2020, 40, 910–926. [Google Scholar] [CrossRef]
- Topic, A.; Russo, M. Emotion recognition based on EEG feature maps through deep learning network. Eng. Sci. Technol. Int. J. 2021, 24, 1442–1454. [Google Scholar] [CrossRef]
- Galvão, F.; Alarcão, S.; Fonseca, M. Predicting Exact Valence and Arousal Values from EEG. Sensors 2021, 21, 3414. [Google Scholar] [CrossRef]
- Li, X.; Zhang, Y.; Tiwari, P.; Song, D.; Hu, B.; Yang, M.; Zhao, Z.; Kumar, N.; Marttinen, P. EEG based Emotion Recognition: A Tutorial and Review. ACM Comput. Surv. 2022. [Google Scholar] [CrossRef]

| Channel | DEAP Index | Seed Index | Channel | DEAP Index | Seed Index |
|---|---|---|---|---|---|
| AF3 | 1 | 3 | AF4 | 17 | 4 |
| F3 | 2 | 7 | F4 | 19 | 11 |
| F7 | 3 | 5 | F8 | 20 | 13 |
| FC5 | 4 | 15 | FC6 | 21 | 21 |
| T4 | 7 | 23 | T8 | 25 | 31 |
| P7 | 11 | 41 | P8 | 29 | 49 |
| O0 | 13 | 58 | O2 | 31 | 60 |
| Method | DEAP-Valence | DEAP-Arousal | DEAP-Dominance | DEAP-Liking | SEED |
|---|---|---|---|---|---|
| 1 client | 0.8569 | 0.8638 | 0.8793 | 0.8582 | 0.8225 |
| FL-5 clients | 0.8843 | 0.8974 | 0.9074 | 0.8719 | 0.8572 |
| FL-10 clients | 0.8794 | 0.8870 | 0.9039 | 0.8687 | 0.8463 |
| Class | Class 1 | Class 2 | Class 3 | Class 4 | Class 5 | Class 6 | Class 7 | Class 8 | Class 9 | Distribution | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Client | |||||||||||
| Client 1 | 1 | 1028 | 921 | 34 | 58 | 384 | 755 | 20 | 8249 |  | |
| Client 2 | 34 | 748 | 9 | 85 | 2708 | 358 | 15 | 576 | 50 |  | |
| Client 3 | 154 | 3036 | 4 | 1358 | 515 | 211 | 150 | 4665 | 0 |  | |
| Client 4 | 1008 | 54 | 243 | 1072 | 1120 | 278 | 142 | 784 | 83 |  | |
| Client 5 | 0 | 329 | 142 | 3979 | 3 | 639 | 654 | 816 | 1587 |  | |
| Client 6 | 2101 | 466 | 882 | 1415 | 12 | 185 | 3687 | 277 | 0 | 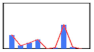 | |
| Client 7 | 2428 | 3069 | 6329 | 0 | 0 | 0 | 0 | 0 | 0 |  | |
| Client 8 | 397 | 471 | 633 | 121 | 3 | 4531 | 1934 | 607 | 31 | 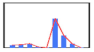 | |
| Client 9 | 3386 | 785 | 355 | 496 | 726 | 3153 | 57 | 2255 | 0 |  | |
| Client 10 | 491 | 14 | 482 | 1440 | 4855 | 261 | 2606 | 0 | 0 |  | |
| All Data | 10,000 | 10,000 | 10,000 | 10,000 | 10,000 | 10,000 | 10,000 | 10,000 | 10,000 |  | |
| Method. | Valence | Arousal | Dominance | Liking |
|---|---|---|---|---|
| IID | 0.9200 | 0.9102 | 0.9222 | 0.9286 |
| N-IID | 0.7633 | 0.7590 | 0.7999 | 0.7633 |
| Method | Valence | Arousal | Dominance | Liking |
|---|---|---|---|---|
| Luo et al. [37] | 0.7400 | 0.7800 | 0.8000 | 0.8627 |
| Acharya et al. [18] | 0.8507 | 0.8383 | 0.8143 | 0.8574 |
| Nawaz et al. [38] | 0.7896 | 0.7762 | 0.7760 | / |
| Topic et al. [39] | 0.7772 | 0.7661 | / | / |
| Galvao et al. [40] | 0.8984 | 0.8983 | / | / |
| FL-10 Client | 0.9200 | 0.9102 | 0.9222 | 0.9286 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, C.; Liu, H.; Qi, W. EEG Emotion Recognition Based on Federated Learning Framework. Electronics 2022, 11, 3316. https://doi.org/10.3390/electronics11203316
Xu C, Liu H, Qi W. EEG Emotion Recognition Based on Federated Learning Framework. Electronics. 2022; 11(20):3316. https://doi.org/10.3390/electronics11203316
Chicago/Turabian StyleXu, Chang, Hong Liu, and Wei Qi. 2022. "EEG Emotion Recognition Based on Federated Learning Framework" Electronics 11, no. 20: 3316. https://doi.org/10.3390/electronics11203316
APA StyleXu, C., Liu, H., & Qi, W. (2022). EEG Emotion Recognition Based on Federated Learning Framework. Electronics, 11(20), 3316. https://doi.org/10.3390/electronics11203316







