Abstract
Naive Bayes (NB) is one of the essential algorithms in data mining. However, it is rarely used in reality because of the attribute independence assumption. Researchers have proposed many improved NB methods to alleviate this assumption. Among these methods, due to its high efficiency and easy implementation, the filter-attribute-weighted NB methods have received great attentions. However, there still exist several challenges, such as the poor representation ability for a single index and the fusion problem of two indexes. To overcome the above challenges, we propose a general framework of an adaptive two-index fusion attribute-weighted NB (ATFNB). Two types of data description category are used to represent the correlation between classes and attributes, the intercorrelation between attributes and attributes, respectively. ATFNB can select any one index from each category. Then, we introduce a regulatory factor to fuse two indexes, which can adaptively adjust the optimal ratio of any two indexes on various datasets. Furthermore, a range query method is proposed to infer the optimal interval of regulatory factor . Finally, the weight of each attribute is calculated using the optimal value and is integrated into an NB classifier to improve the accuracy. The experimental results on 50 benchmark datasets and a Flavia dataset show that ATFNB outperforms the basic NB and state-of-the-art filter-weighted NB models. In addition, the ATFNB framework can improve the existing two-index NB model by introducing the adaptive regulatory factor . Auxiliary experimental results demonstrate the improved model significantly increases the accuracy compared to the original model without the adaptive regulatory factor .
1. Introduction
The naive Bayes (NB) classifier is a classical classification algorithm. Due to its simplicity and efficiency, it is widely used in many fields such as data mining and pattern recognition.
Assume that a dataset contains m training instances, an instance can be represented by an n-dimensional attribute value vector . The NB classifier uses Equation (1) to predict the class label of the instance .
where C is the set of all possible class labels c, n is the number of attributes, and represents the value of the jth attribute of the ith instance. is the prior probability of class c, and is the conditional probability of the attribute value given the class c, which can be calculated by Equations (2) and (3), respectively.
where represents all the values of the jth attribute in training instances. (·) is a custom function to calculate the number of unique data in C or . denotes the correct class label for the ith instance. (·) is a binary function, which takes the value one if and c are identical and zero otherwise [1].
Duo to the attribute independence assumption, the NB classifier is a simple, stable, easy to implement, and better classification algorithm for various applications. However, real data are complicated and diverse, which make it difficult to satisfy this assumption. Thus, researchers have proposed many methods to reduce the influence of the attribute independence assumption. These methods can be divided into six categories: Structure extension models directed arcs to represent the dependence relationship between attributes [2,3,4,5,6]. Fine-tuning adjusts the probability value to find a good estimation of the desired probability term [7,8]. The purpose of instance selection is to construct NB model on a subset of training set instead of the whole training set [9,10]. Instance weighting consists of assigning instances different weights by different strategies [11,12,13]. Attribute selection is the process of removing redundant attributes [14,15,16,17,18,19]. Distinguished from attribute selection, attribute weighting assigns a weight to each attribute in order to relax the independence assumption and make the NB model more flexible [20,21,22,23,24,25,26,27].
In this paper, we focus our attention on attribute weighting, which is further divided into wrapper methods and filter methods. The wrapper methods optimize the weighted matrix by using gradient descent to improve classification performance. Wu et al. proposed a weighted NB algorithm based on differential evolution, which gradually adjusted the weights of attributes through evolutionary algorithms to improve the prediction results [28]. Zhang et al. proposed two attribute-value-weighting models based on the conditional log-likelihood and mean square error [1]. However, these methods are often less efficient due to the time-consuming optimization process. Another category obtained the weights by analyzing the correlation of attributes [20,21,24,26,27]. Since correlation can be easily and efficiently obtained by various measurement indexes, the computational efficiency of filter methods obviously increases. Related filter methods are detailed introduced in Section 2. Although filter methods have some advantages such as being flexible and computational efficient, there are still two problems. Most of the methods utilize a single index, which expresses the data characteristic, to determine the attribute weight. However, a single index cannot comprehensively discover information about the dataset. In order to fully dig up the information from the dataset, a two-index fusion method was proposed, which could achieve better performance [29]. However, the ratio of the two indexes became the second problem. The method assumed that the contributions of the two indexes were equivalent and ignored the difference in contribution between the two indexes.
To overcome the above problems, we propose an adaptive two-index fusion attribute-weighted naive Bayes (ATFNB) method. ATFNB can select any index from two categories of data description, respectively. The first category describes the correlation between attributes and classes, and the second category describes the intercorrelation between attributes and attributes. Once two indexes have been selected, ATFNB fuses the two indexes by introducing a regulatory factor . Due to the diversity of datasets, the regulatory factor can be adaptively adjusted to get the optimal ratio between the two indexes. Moreover, a range query method is proposed to obtain the optimal value of the regulatory factor . To verify the effectiveness of ATFNB, we conduct extensive experiments on 50 UCI datasets and a Flavia dataset. Experimental results show that ATFNB has a better performance compared to NB and state-of-the-art filter NB models.
To sum up, the main contributions of our work include the following:
- We propose a general framework, ATFNB, which fuses any two indexes from two categories of data description. Our framework can derive all existing filter-attribute-weighted NB models by selecting difference indexes.
- We introduce a regulatory factor , which can adaptively adjust the optimal ratio of two indexes, and shows which index is more important on various datasets.
- A quick range query method is proposed to obtain the optimal value of the regulatory factor . Compared to the traditional method, step-length searching, our method obviously speeds up the optimization.
- The existing two-index NB methods’ performance are significantly improved by introducing the regulatory factor .
The rest of the paper consists of the following parts. Section 2 comprehensively reviews the filter-attribute-weighted methods. Section 3 proposes an adaptive two-index fusion attribute-weighted naive Bayes method. Section 4 presents the experimental datasets, setting and results. Section 5 further discusses the experimental results. Finally, Section 6 summarizes the research and gives some future work.
2. Related Work
Given a dataset D with n attributes and K classes. The naive Bayes weight matrix is shown in Table 1.

Table 1.
The naive Bayes weight matrix.
The naive Bayes method incorporates the attribute weight into the formula as follows:
where is the weight of the jth attribute . The most critical issue of filter-weighted NB methods is how to determine the weight of each attribute, which has attracted more great attention. Many weighted NB methods have been proposed based on various measurements of attribute weighted. Here, we introduce several state-of-the-art filter-weighted NB methods.
Ferreira et al. firstly proposed a weighted naive Bayes method to alleviate the independence assumption, which assigned weights to different attributes [30]. Based on this idea, Zhang et al. presented an attribute-weighted model based on a gain ratio (WNB) [31]. Attributes with a higher gain ratio deserved higher weights in WNB. Therefore, the weight of each attribute can be defined by Equation (5).
where is the gain ratio of attribute [32].
Then, Lee et al. proposed a novel model that used the Kullback–Leibler metric to calculate the weight of each attribute [33]. This model was certain information between each attribute and the corresponding class label c, which was obtained by Kullback–Leibler [34] measuring the difference between the prior distribution and the posterior distribution of the target attributes. The weight value of the jth attribute is shown in Equation (6).
where means the probability of the value , and is a normalization constant. is the average mutual information between the class label c and the attribute value of .
Next, Jiang’s team proposed a series of filter-attribute-weighted methods, which included deep feature weighting (DFW) [26] and correlation-based feature weighting (CFW) [29]. DFW assumed that more independent features should be assigned higher weights. The correlation-based feature selection was used to evaluate the degree of dependence between attributes [35]. According to this selection, the best subset was selected from the attribute space. The weight value assigned to the selected attribute was two, and the weight value assigned to other attributes was one, as shown in Equation (7).
Compared with the above methods with a single index, CFW was the first two-index weighted NB method, which used the attribute-class correlation and the average attribute–attribute intercorrelation to constitute the weight of each attribute. The mutual information was measured from the attribute–class correlation and the attribute–attribute intercorrelation, defined as Equations (8) and (9), respectively.
where and represent the values of attributes and , respectively. is the correlation between the attribute and class C. is the redundancy between two different attributes and . Finally, the weight of the attribute is defined as Equation (10).
where and are the normalized values, which, respectively, represent the maximum correlation and the maximum redundancy.
3. ATFNB
3.1. The General Framework of ATFNB
The filter-weighted NB methods assign a specific weight for each attribute to alleviate the independence assumption. However, there are still some challenges, such as the poor representation ability for a single index and the fusion problem of two indexes. Therefore, we propose an adaptive two-index fusion attribute-weighted NB. The framework of ATFNB is shown in Figure 1. Given a dataset, two indexes are selected from a class–attribute category and an attribute–attribute category, respectively. Then, the regulatory factor is utilized to fuse the two indexes, and adaptively generate the optimal ratio value. Next, the weight of each attribute is calculated via the optimal regulatory factor . Finally, the attribute weights are incorporated into the NB classifier to predict the class labels.
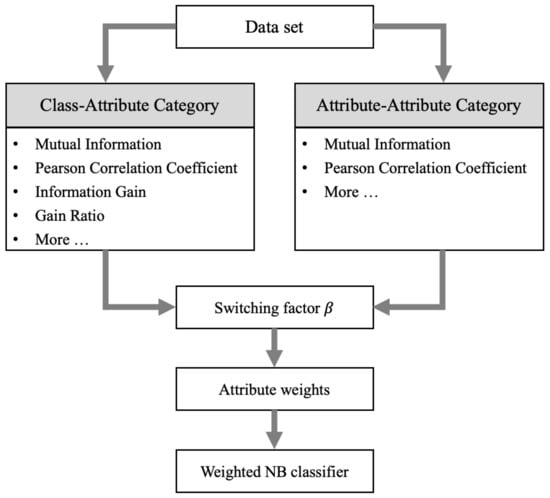
Figure 1.
The general framework adaptive two-index fusion attribute-weighted NB.
3.2. Index Selection
As shown in Figure 1, the ATFNB framework contains two widely used types of the attribute correlation: class–attribute and attribute–attribute. The class–attribute category is to measure the correlation between attributes and classes. The stronger the correlation between an attribute and a class, the more significant the attribute’s contribution to the classification. Thus, the index value is positively correlated with the weight. Common indexes in this category include the mutual information, Pearson correlation coefficient, information gain, gain ratio, etc. The attribute–attribute category is to measure the redundancy between attributes. In order to satisfy the independence assumption of the naive Bayes classifier as much as possible, attributes with high redundancy are assigned small weights. Thus, the weight is inversely correlated to the index value. Common measures of redundancy between attributes include the mutual information and Pearson correlation coefficient, etc. Except for the above six indexes in Figure 1, researchers can add any other index or design new indexes to represent data characteristics.
By selecting different indexes, the ATFNB framework can become any weighted NB model, including the existing weighted NB models. If only the gain ratio is selected from the class–attribute category, ATFNB degenerates into the single-index WNB model. If the class–attribute and attribute–attribute category both choose the mutual information, ATFNB becomes the two-index CFW model. Thus, the index selection is a critical step in the ATFNB framework. Any two indexes selected from the two categories can generate various models, which may achieve different results. In Section 5.3, the classification performance of different index selections is discussed in detail.
3.3. Range Query Method for the Regulatory Factor
Since the significantly discriminative attribute should be highly correlated with the class and has low redundancy with other attributes, its weights should be positively associated with the difference between the class–attribute correlation and attribute–attribute intercorrelation [29]. The mathematics formula of the weight can be defined by Equation (11).
where and represents the values of the selected class–attribute index and attribute–attribute index, respectively. The existing two-index methods, CFW, consider that the contributions of two indexes are equivalent [29]. However, various indexes contain different characteristics, and an equivalent contribution of the two indexes is unreasonable. Thus, we introduce a regulatory factor to adaptively control the ratio of two indexes. After incorporating the regulatory factor , Equation (11) can be rewritten as Equation (12).
where the regulatory factor .
Conventionally, the step-length searching strategy can be applied to search the optimal interval of . However, the accuracy and computational efficiency are affected by the step size. When the step size gets smaller, the optimal interval of is more accurate but the search gets much slower. Thus, we propose a range query method to calculate the optimal interval of the regulatory factor . Firstly, the basic weighted NB model (Equation (4)) is logarithmically transformed, and the detailed transformation process is shown as follows.
where is the probability value that the instance belongs to class c. is the interval when instance is correctly classified.
Based on Equation (13), a probability set can be constructed to store the probability values of instances belonging to different classes. The probability set of is . If the correct label of is , should be greater than the other probability values in . This can be defined as follows:
When the instance is correctly classified, the interval of can be obtained. For m instances, a set contains the m intervals corresponding to each instance. To calculate the optimal interval from G, so that any value in the interval can obtain the same classification accuracy on the training set, the upper and lower bounds of all intervals in G are sorted in ascending order . Any two adjacent values in Q are regarded as the lower and upper bounds of a subinterval. Thus, Q can generate subintervals. The subintervals in satisfying Equation (15) are taken as .
where (·) is a binary function, which takes the value 1 if is a subset of and 0 otherwise, as shown in Equation (16).
According to the above derivation processes, the range query method for the regulatory factor (RQRF) is described in Algorithm 1.
| Algorithm 1: RQRF |
Input: class–attribute (), attribute–attribute (), dataset D For each instance in D: For each class c in C: Calculate and in Equation (13). According and , get . End If instance label is : Find the value that satisfies Equation (14); it is recorded as , otherwise = ∅. End End For each Find the subinterval that conforms to Equation (15). End Output: |
Any value in can achieve a consistent classification accuracy in dataset D, so we choose any value from the optimal interval . After obtaining the value of the regulatory factor, the weight can be calculated by Equation (12).
3.4. The Implementation of ATFNB
The general framework of ATFNB is briefly described in Algorithm 2. According to Algorithm 2, we can see that how to select two indexes and , and how to learn the regulatory factor are two crucial problems. To select and , several indexes are listed in Section 3.2. To learn the value of the regulatory factor , we single out an RQRF algorithm in Section 3.3. Once the value of the regulatory factor is obtained, we can use Equation (12) to calculate the weights of each attribute. Finally, these weights are applied to construct an attribute-weighted NB classifier.
| Algorithm 2: ATFNB Framework |
Input: Training set D, test set X (1) For each attribute in D Calculate (attribute–attribute) index Calculate (class–attribute) index . (2) According to RQRF, the value of the regulatory factor is solved. (3) According to Equation (12), the weight matrix is obtained. (4) According to Equation (4), the class label of each instance in X is predicted. Output: The class label of instances in X |
4. Experiments and Results
4.1. Experimental Data
To verify the effectiveness of ATFNB, a collection of 50 benchmark datasets and 15 groups of the leaf dataset were conducted.
The 50 benchmark classification datasets were chosen from the University of California at Irvine (UCI) repository [36], representing various fields and data characteristics, as listed in Table 2. We used the mean of the corresponding attribute to replace missing data values in each dataset, then applied chi-square-based algorithm to discretize the numerical attribute values [37]. The amount of discretization of each attribute was consistent with the number of types of class labels.

Table 2.
Descriptions of 50 UCI datasets used in the experiments.
The Flavia dataset contains 32 types of leaf and each leaf has 55–77 pieces. Four texture and ten shape features of each leaf were extracted based on grayscale and binary images [38]. We constructed 15 groups for comparative experiments, and in each group we randomly selected 15 kinds of leaves from the whole Flavia dataset. The detailed characteristics of the 15 groups are listed in Table 3. Then, the same preprocessing pipeline as for the UCI dataset was applied to discretize the continuous attributes.

Table 3.
Descriptions of 15 groups from Flavia dataset used in the experiments.
4.2. Experimental Setting
ATFNB is a general framework of attribute-weighted naive Bayes, which can adaptively fuse any two indexes. According to Figure 1, we selected two simple and popular indexes from two categories: the information gain from the class–attribute category and the Pearson correlation coefficient from the attribute–attribute category. Notably, ATFNB refers to a specific NB model fused from the above two indexes in the following experiments, and no longer represents a general framework.
For the class–attribute category, the information gain describes the information content provided by the attribute for the classification. The formula of the information gain is shown in Equation (17).
where is the information entropy. The discrete attribute has V values . indicates that the vth branch node contains all the instances in dataset D, whose value is on the attribute .
For the attribute–attribute category, the Pearson correlation coefficient was used to calculate the correlation between attributes and , and the formula can be written as Equation (18).
where represents the covariance between attributes and , and represent the standard deviation of and , respectively.
After obtaining these two indexes, the weight of each attribute could be calculated as Equation (19).
where is expressed as the normalized value of the attribute’s information gain, and represents the average degree of redundancy between the ith attribute and other attributes. The formula of is shown in Equation (20).
where is expressed as the normalized value between attributes and .
To validate the classification performance, we compared ATFNB to the standard NB classifier and two existing state-of-the-art filter-weighted methods. In addition, the original CFW was a specific model of our framework under the regulatory factor = 0.5. When the regulatory factor of CFW could be adaptively obtained from the dataset, the original CFW evolved into CFW-. Now, we introduce these comparisons and their abbreviations as follows:
- NB: the standard naive Bayes model [39].
- WNB: NB with gain ratio attribute weighting [31].
- CFW: NB with MI class-specific and attribute-specific attribute weighting [29].
- CFW-: CFW with the adaptive regulatory factor .
4.3. The Effectiveness of the Regulatory Factor
The regulatory factor can be adaptively adjusted to obtain the optimal ratio for different datasets. In order to verify the effectiveness and efficiency of the regulatory factor , we compared the RQRF method with the step-length searching (SLS) algorithm. The SLS algorithm generates with 0.01 as the step size. The optimal interval of regulatory factor by the RQRF and SLS algorithms in four datasets are shown in Figure 2.
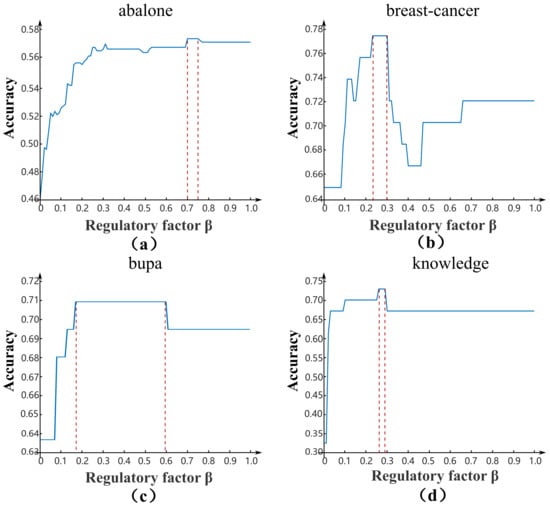
Figure 2.
The optimal regulatory factor of RQRF and SLS algorithms in four datasets. The blue solid line represents the accuracy of each step size by SLS, and the red dotted lines represent the optimal interval obtained by RQRF.
From Figure 2, for both the SLS and RQRF algorithms, the optimal interval of in each dataset was biased. On abalone, the lower bound of the interval of was greater than 0.5. On breast-cancer and knowledge, the upper bound of the interval of was less than 0.5. Only the interval of in bupa contained 0.5. Thus, it could be concluded that the regulatory factor value set as 0.5 was unreasonable for all datasets. In addition, it could be clearly seen that the interval size of regulatory factor was inconsistent. On bupa, the interval size of was the largest. On the contrary, the size was the smallest on knowledge.
The optimal interval of regulatory factor and the runtime calculated by the SLS and RQRF algorithms are shown in Table 4. From Table 4, we can see that two optimal intervals obtained by the SLS and RQRF algorithms had a high coincidence degree. If we reduced the step size of SLS, the coincidence degree between the two algorithms would further improve. Yet, SLS would become very inefficient. For the RQRF method, the runtime was obviously faster than that of the SLS method, and was sped up at least 150 times. Therefore, the RQRF method is not only more accurate than the SLS method, but also more efficient.

Table 4.
The optimal interval of regulatory factor and the runtime from SLS and RQRF methods.
4.4. Experimental Results on UCI Datasets
Table 5 shows the detailed classification accuracy results of five algorithms. All classification accuracies were obtained by averaging the results of 30 independent runs. The five algorithms were used on the same training set and testing set. We conduct a group of experiments on 50 UCI datasets to compare ATFNB with NB, WNB, CFW and CFW- in terms of classification accuracy.

Table 5.
Classification accuracy comparisons for ATFNB versus NB, WNB, CFW, CFW- on UCI datasets.
Compared with WNB, CFW, and NB, the accuracy of ATFNB on 33 datasets was the highest, which far exceeded that of WNB (0 datasets), CFW (8 datasets), and NB (9 datasets). The average accuracy of ATFNB was 83.17%, which was significantly higher than that of the other algorithms, and the improvement on the average accuracy was approximately 3%, 2%, and 2%, respectively.
In addition, the average accuracy of CFW- increased by 1.76% compared with that of CFW. This meant that the adaptive regulatory factor could improve the existing two-index NB model. Compared with ATFNB, the average accuracy of CFW- was higher than that of ATFNB. The reason was that the mutual information (class–attribute) and mutual information (attribute–attribute) were included in CFW-, which had a more powerful representation than the information gain and Pearson correlation coefficient in ATFNB. In Section 5.3, models generated by different combinations of indexes are discussed in detail.
Base on the accuracy result, we used a two-tailed t-test at the level to compare each pair of algorithms beside CFW-. Table 6 summarizes the comparison results on the UCI datasets. From Table 6, ATFNB had significant advantages over the other weighting algorithms. ATFNB was better than WNB (28 wins and 0 loss), CFW (22 wins and 4 losses), and NB (19 wins and 5 losses).

Table 6.
Summary of two-tailed t-test results of classification accuracy with regard to ATFNB on UCI datasets.
Based on the classification accuracy of Table 5, we utilized the Wilcoxon signed-rank test to compare the four algorithms. The Wilcoxon signed-rank test is a nonparametric statistical test, which ranks the performance differences of the two algorithms for each dataset, considering both the sign of the difference and the order of the difference. Table 7 shows the ranks calculated by the Wilcoxon test. In Table 7, the numbers above the diagonal line indicate the sum of ranks for the datasets of the algorithm in the row that is better than the algorithm in the corresponding column (the sum of the ranks for the positive difference, represented by ). Each number below the diagonal is the sum of ranks for the datasets in which the algorithm in the column is worse than the algorithm in the corresponding row (The sum of the ranks for the negative difference, represented by ). According to the critical value table of the Wilcoxon test, for Table 7, when = 0.05 and n = 50, if the smaller of and was equal to or less than 434, we considered that the two classifiers were significantly different, so we rejected the null hypothesis. The significant data in Table 7 (equal to or less than 434) are marked with symbols (•, ∘), as shown in Table 8.

Table 7.
Ranks of the Wilcoxon test with regard to ATFNB on UCI datasets.

Table 8.
Summary of the Wilcoxon test with regard to ATFNB on UCI datasets.
According to the results of the Wilcoxon signed rank-sum test, on the UCI datasets, ATFNB was significantly better than WNB (), CFW () and Standard NB ().
4.5. Experimental Results on Flavia Dataset
In order to further verify the effectiveness of ATFNB, we conducted 15 groups of experiments on the Flavia dataset. We randomly divided the data in each group of experiments 30 times and used a two-tailed t-test for the results of 30 experiments. The detailed results are shown in Table 9.

Table 9.
Classification accuracy comparisons for ATFNB, NB, WNB, CFW, CFW- on Flavia dataset. * indicates that ATFNB was significantly better than its competitors (NB, WNB, CFW) through a two-tailed t-test at the p = 0.05 significance level.
Comparing ATFNB with other existing classifiers (WNB, CFW, NB), the average accuracy of ATFNB was 87.21%, which was significantly higher than that of algorithms, and the improvement on the average accuracy was approximately 3%, 1.5%, and 1%, respectively. In 15 groups of experiments, ATFNB achieved the highest classification accuracy among 10 groups of data, which was far better than NB, WNB, and CFW.
The average accuracy of CFW- was slightly lower than that of ATFNB, but the average accuracy of CFW- was higher than that of CFW. On Flavia, the choice of indexes had little effect on the average accuracy, but adding a regulatory factor to the model could effectively improve the performance of model.
We summarize the results of the two-tailed test in Table 9, as shown in Table 10. In Table 10, ATFNB was better than WNB (nine wins and zero loss), CFW (seven wins and one loss), and NB (nine wins and zero loss).

Table 10.
Summary two-tailed t-test results of classification accuracy with regard to ATFNB on Flavia dataset.
On the basis of Table 9, we used the Wilcoxon signed-rank test to compare the four algorithms. According to the critical value table of the Wilcoxon test, for Table 11, when and , if the smaller of and was equal to or less than 25, we considered that two classifiers were significantly different, so we rejected the null hypothesis.The significant data in Table 11 (equal to or less than 25) are marked with symbols (•,∘), as shown in Table 12.

Table 11.
Ranks of the Wilcoxon test with regard to ATFNB on Flavia dataset.

Table 12.
Summary of the Wilcoxon test with regard to ATFNB on Flavia dataset.
In the Flavia dataset, the ATFNB algorithm had obvious advantages compared with WNB (), CFW (), and standard NB ().
5. Discussion
5.1. The Influence of Instance and Attribute Number
To further analyze the relationship between the performance of ATFNB and the characteristic of a dataset, we observed its performance from the two perspectives of number of instances and number of attributes. In terms of the number of instances, we divided the datasets into two categories: less than 500 instances and greater than or equal to 500 instances. Similarly, according to the number of attributes, we divided attributes into two categories: the number of attributes was less than 15, and the number of attributes was greater than or equal to 15. Then, we combined the above two criteria, which resulted in four divisions. Finally, we calculated the percentage of datasets with the highest classification accuracy of ATFNB and competitors (NB, WNB, CFW) in eight divisions. The detailed results are shown in Table 13.

Table 13.
ATFNB and competitors’ obtained percentage of datasets with the highest classification accuracy in each division. Bold text is to highlight the superiority of the method.
From Table 13, we could clearly find in which circumstances ATFNB performed better than its competitors. Here, we summarize the highlights as follows:
- (1)
- On 78.26% of datasets with less than 500 instances, ATFNB could achieve the highest classification accuracy. On datasets with a number of instances greater than or equal to 500, 56.25% could achieve the maximum classification accuracy. By comparison, ATFNB is more advantageous on datasets with fewer instances.
- (2)
- For datasets with a number of attributes less than 15, the percentage of datasets with the highest classification accuracy for ATFNB (67.74%) was also higher than that with a number of attributes greater than or equal to 15 (63.16%).
- (3)
- When the number of instances was less than 500 and the number of attributes was greater than 15, the percentage of datasets with the highest classification accuracy for ATFNB (87.5%) was significantly higher than that of the other three types of datasets (73.33%, 62.50%, 45.45%).
The performance of ATFNB had obvious advantages on the datasets whose number of instances was smaller than 500, especially when the number of attributes was greater than or equal to 15, such as the dataset “congressional-voting”. By contrast, ATFNB did not perform well on datasets with a large number of instances and attributes. In a word, ATFNB can be perfectly suitable for small data classification and is not limited by dimensions.
5.2. The Distribution of the Regulatory Factor
In Section 4.3, we validated the effectiveness of the regulatory factor in ATFNB. Here, the distributions of the regulatory factor in various datasets are further analyzed. We firstly list the interval of the regulatory factor for the 50 UCI datasets as shown in Table 14. From Table 14, we can summarize that the lower bound of the optimal interval in 11 datasets was greater than 0.5, the upper bound of the optimal interval in 23 datasets was less than 0.5, and the optimal interval of the remaining 16 datasets contained 0.5. In ATFNB, the information gain and Pearson correlation coefficient provided different contributions on the 50 UCI datasets. In addition, these results further demonstrated that the regulatory factor set as a fixed value was unreasonable.

Table 14.
The interval of regulatory factor on 50 UCI datasets.
To further investigate the relationship between the distribution of regulatory factor and the characteristics of the datasets, we applied the same division criteria as in Section 5.1 on the 50 UCI datasets and summarize the detailed results in Table 15. From Table 15, we can observe the preference between the data characteristics and the distribution of the regulatory factor , and summarize the highlights as follows:

Table 15.
The relationship between the regulatory factor in ATFNB and the characteristics of the dataset.
- (1)
- If the number of instances was less than 500, the upper bound value of in 52.17% of the datasets was less than 0.5. If the number of instances was more than 500, the upper bound value of in 40.74% of the datasets was less than 0.5.
- (2)
- From the perspective of the number of attributes, regardless of the number of attributes, the upper bound value of was less than 0.5 in most datasets.
- (3)
- (3) Considering the number of instance and attributes simultaneously, the upper bound value of in 62.50% of the datasets with a number of instances less than 500 and number of attributes greater than 15 was less than 0.5. On the dataset with a number of instances greater than 500 and number of attributes greater than 15, the upper bound value of in 45.46% of the datasets was less than 0.5.
Based on the results in Table 15, the upper bound value of was less than 0.5 in most datasets. We can conclude that ATFNB pays attention to the Pearson correlation coefficient between attributes, especially in small instances and high-dimensional datasets.
5.3. The Impact of Different Index Combinations
The ATFNB framework contains two categories, and each category provides several popular indexes to represent the characteristic of datasets. In order to analyze the impact of different index combinations, we selected any two indexes from two categories, respectively. Excluding the gain ratio from the class–attribute category, six weighted NB models could be constructed as shown in Figure 3. Notably, ATFNB-IP and ATFNB-MM were equal to ATFNB and CFW-, respectively.
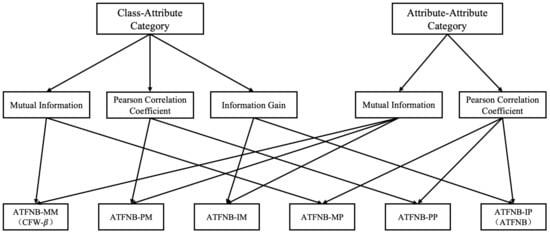
Figure 3.
The index selection of each combination.
Then, we compared the six combinations on the 50 UCI datasets, and the average accuracy of the six combinations are shown in Figure 4. From Figure 4, it can be seen that ATFNB-PP had the lowest average accuracy, but the average accuracy of ATFNB-PP outperformed the basic NB and NB (0.8146), WNB (0.8028), and CFW (0.8169). This further demonstrated the effectiveness of the ATFNB framework with an adaptive regulatory factor. In addition, compared with three indexes the from class–attribute category, the average accuracy of ATFNB-M* (denoted ATFNB-MP and ATFNB-MM) was better than ATFNB-P* and ATFNB-I*. This meant that the mutual information from the class–attribute category was more signification than the Pearson correlation coefficient and information gain.
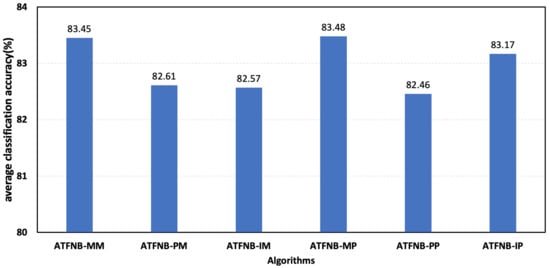
Figure 4.
Average accuracy of six combinations.
5.4. Computation Time
The experimental platform in this paper was an AMD 5900X processor, 3.70 GHz, 32 GB memory, Windows 10 system, and the algorithm was implemented in Matlab2020A.
In order to comprehensively analyze the computation time of our framework, we split ATFNB into three stages, which were the stage of index calculation and conditional probability calculation (Stage 1), the stage of regulatory factor calculation (Stage 2), and the stage of classification (Stage 3). Table 16 shows the computation time of each stage and the whole process for various algorithms on the blood dataset. From Table 16, we find that the main differences were in the Stage 1 and Stage 2. In the Stage 1, the type and number of indexes directly affected the runtime. CFW, CFW-, and ATFNB needed to calculate two indexes, thus these three algorithms took more time than the other two single-index NB methods. In Stage 2, although CFW- and ATFNB introduced a regulator factor, the computation time was very short using the quick range query method. In terms of total time, CFW- and ATFNB only increased time by about 10% compared to CFW, but the accuracy significant improved.

Table 16.
The computation time of each stage for various algorithms on the blood dataset (in seconds).
6. Conclusions and Future Work
In this paper, we proposed an adaptive two-index fusion attribute-weighted NB (ATFNB) to overcome the problems of existing weighted methods, such as the poor representation ability with a single index and the fusion problem of two indexes. ATFNB could select any one index from the attribute–attribute category and the class–attribute category, respectively. Then, a regulatory factor was introduced to fuse two indexes and it was inferred by a range query. Finally, the weight of each attribute was calculated using the optimal value and integrated into an NB classifier to improve the accuracy. The experimental results on 50 benchmark datasets and the Flavia dataset showed that ATFNB outperformed the basic NB classifier and state-of-the-art filter-weighted NB models. In addition, we incorporated the regulatory factor into CFW. The results demonstrated the improved model CFW- had a significantly increased accuracy compared to CFW without the adaptive regulatory factor .
In future work, there are two directions to further improve the NB model. Firstly, ATFNB may consider more than two indexes from different data description categories. Secondly, we hope to design more new indexes to represent the correlation of class–attribute or attribute–attribute.
Author Contributions
Conceptualization, X.Z. and N.Y.; methodology, X.Z.; software, X.Z.; validation, X.Z., Z.Y. and D.W. (Dongyang Wu); formal analysis, N.Y.; investigation, X.Z.; resources, N.Y.; data curation, D.W. (Donghua Wu); writing—original draft preparation, X.Z.; writing—review and editing, X.Z. and L.Z.; visualization, L.Z.; supervision, L.Z.; project administration, L.Z.; funding acquisition, N.Y. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Postgraduate Research & Practice Innovation Program of Jiangsu Province grant number KYCX22_1106.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Zhang, H.; Jiang, L.; Yu, L. Class-specific attribute value weighting for Naive Bayes. Inf. Sci. 2020, 508, 260–274. [Google Scholar] [CrossRef]
- Koivisto, M.; Sood, K. Exact Bayesian structure discovery in Bayesian networks. J. Mach. Learn. Res. 2004, 5, 549–573. [Google Scholar]
- Friedman, N.; Koller, D. Being Bayesian about network structure. A Bayesian approach to structure discovery in Bayesian networks. Mach. Learn. 2003, 50, 95–125. [Google Scholar] [CrossRef]
- Friedman, N.; Geiger, D.; Goldszmidt, M. Bayesian network classifiers. Mach. Learn. 1997, 29, 131–163. [Google Scholar] [CrossRef]
- Jiang, L.; Zhang, H.; Cai, Z. A novel bayes model: Hidden naive bayes. IEEE Trans. Knowl. Data Eng. 2008, 21, 1361–1371. [Google Scholar] [CrossRef]
- Jiang, L.; Wang, S.; Li, C.; Zhang, L. Structure extended multinomial naive Bayes. Inf. Sci. 2016, 329, 346–356. [Google Scholar] [CrossRef]
- Diab, D.M.; El Hindi, K.M. Using differential evolution for fine tuning naïve Bayesian classifiers and its application for text classification. Appl. Soft Comput. 2017, 54, 183–199. [Google Scholar] [CrossRef]
- El Hindi, K. Fine tuning the Naïve Bayesian learning algorithm. AI Commun. 2014, 27, 133–141. [Google Scholar] [CrossRef]
- Ryu, D.; Jang, J.I.; Baik, J. A hybrid instance selection using nearest-neighbor for cross-project defect prediction. J. Comput. Sci. Technol. 2015, 30, 969–980. [Google Scholar] [CrossRef]
- Xie, Z.; Hsu, W.; Liu, Z.; Lee, M.L. Snnb: A selective neighborhood based naive Bayes for lazy learning. In Proceedings of the Pacific-Asia Conference on Knowledge Discovery and Data Mining, Taipei, Taiwan, 6–8 May 2002; Springer: Berlin/Heidelberg, Germany, 2002; pp. 104–114. [Google Scholar]
- Zhang, H.; Jiang, L.; Yu, L. Attribute and instance weighted naive Bayes. Pattern Recognit. 2021, 111, 107674. [Google Scholar] [CrossRef]
- Elkan, C. Boosting and naive bayesian learning. In Proceedings of the International Conference on Knowledge Discovery and Data Mining, Newport Beach, CA, USA, 14–17 August 1997. [Google Scholar]
- Jiang, L.; Cai, Z.; Wang, D. Improving naive Bayes for classification. Int. J. Comput. Appl. 2010, 32, 328–332. [Google Scholar] [CrossRef]
- Chen, J.; Huang, H.; Tian, S.; Qu, Y. Feature selection for text classification with Naïve Bayes. Expert Syst. Appl. 2009, 36, 5432–5435. [Google Scholar] [CrossRef]
- Choubey, D.K.; Paul, S.; Kumar, S.; Kumar, S. Classification of Pima indian diabetes dataset using naive bayes with genetic algorithm as an attribute selection. In Proceedings of the Communication and Computing Systems: Proceedings of the International Conference on Communication and Computing System (ICCCS 2016), Nanjing, China, 19–31 July 2017; pp. 451–455.
- Hall, M.A.; Holmes, G. Benchmarking attribute selection techniques for discrete class data mining. IEEE Trans. Knowl. Data Eng. 2003, 15, 1437–1447. [Google Scholar] [CrossRef]
- Jiang, L.; Cai, Z.; Zhang, H.; Wang, D. Not so greedy: Randomly selected naive Bayes. Expert Syst. Appl. 2012, 39, 11022–11028. [Google Scholar] [CrossRef]
- Lee, C.; Lee, G.G. Information gain and divergence-based feature selection for machine learning-based text categorization. Inf. Process. Manag. 2006, 42, 155–165. [Google Scholar] [CrossRef]
- Deng, X.; Li, Y.; Weng, J.; Zhang, J. Feature selection for text classification: A review. Multimed. Tools Appl. 2019, 78, 3797–3816. [Google Scholar] [CrossRef]
- Lee, C.H. An information-theoretic filter approach for value weighted classification learning in naive Bayes. Data Knowl. Eng. 2018, 113, 116–128. [Google Scholar] [CrossRef]
- Yu, L.; Jiang, L.; Wang, D.; Zhang, L. Toward naive Bayes with attribute value weighting. Neural Comput. Appl. 2019, 31, 5699–5713. [Google Scholar] [CrossRef]
- Jiang, L.; Zhang, L.; Yu, L.; Wang, D. Class-specific attribute weighted naive Bayes. Pattern Recognit. 2019, 88, 321–330. [Google Scholar] [CrossRef]
- Wu, J.; Pan, S.; Zhu, X.; Cai, Z.; Zhang, P.; Zhang, C. Self-adaptive attribute weighting for Naive Bayes classification. Expert Syst. Appl. 2015, 42, 1487–1502. [Google Scholar] [CrossRef]
- Hall, M. A decision tree-based attribute weighting filter for naive Bayes. In Proceedings of the International Conference on Innovative Techniques and Applications of Artificial Intelligence, Cambridge, UK, 11–13 December 2006; Springer: Berlin/Heidelberg, Germany, 2006; pp. 59–70. [Google Scholar]
- Taheri, S.; Yearwood, J.; Mammadov, M.; Seifollahi, S. Attribute weighted Naive Bayes classifier using a local optimization. Neural Comput. Appl. 2014, 24, 995–1002. [Google Scholar] [CrossRef]
- Jiang, L.; Li, C.; Wang, S.; Zhang, L. Deep feature weighting for naive Bayes and its application to text classification. Eng. Appl. Artif. Intell. 2016, 52, 26–39. [Google Scholar] [CrossRef]
- Zhang, L.; Jiang, L.; Li, C.; Kong, G. Two feature weighting approaches for naive Bayes text classifiers. Knowl.-Based Syst. 2016, 100, 137–144. [Google Scholar] [CrossRef]
- Wu, J.; Cai, Z. Attribute weighting via differential evolution algorithm for attribute weighted naive bayes (wnb). J. Comput. Inf. Syst. 2011, 7, 1672–1679. [Google Scholar]
- Jiang, L.; Zhang, L.; Li, C.; Wu, J. A correlation-based feature weighting filter for Naive Bayes. IEEE Trans. Knowl. Data Eng. 2018, 31, 201–213. [Google Scholar] [CrossRef]
- Ferreira, J.; Denison, D.; Hand, D. Weighted Naive Bayes Modelling for Data Mining; Technical Report; Imperial College: London, UK, 2001. [Google Scholar]
- Zhang, H.; Sheng, S. Learning weighted naive Bayes with accurate ranking. In Proceedings of the Fourth IEEE International Conference on Data Mining (ICDM’04), Online. 1 November 2004; pp. 567–570. [Google Scholar]
- Quinlan, J.R. C4.5: Programs for Machine Learning; Elsevier: San Mateo, CA, USA, 2014. [Google Scholar]
- Lee, C.H.; Gutierrez, F.; Dou, D. Calculating feature weights in naive bayes with kullback-leibler measure. In Proceedings of the 2011 IEEE 11th International Conference on Data Mining, Vancouver, BC, Canada, 11–14 December 2011; pp. 1146–1151. [Google Scholar]
- Kullback, S.; Leibler, R.A. On information and sufficiency. Ann. Math. Stat. 1951, 22, 79–86. [Google Scholar] [CrossRef]
- Hall, M.A. Correlation-Based Feature Selection of Discrete and Numeric Class Machine Learning; University of Waikato, Department of Computer Science (Working paper 00/08); University of Waikato, Department of Computer Science: Hamilton, NZ, USA, 2000. [Google Scholar]
- Asuncion, A.; Newman, D. UCI Machine Learning Repository; Irvine University of California: Irvine, CA, USA, 2007. [Google Scholar]
- Kerber, R. Chimerge: Discretization of numeric attributes. In Proceedings of the Tenth National Conference on Artificial Intelligence, San Jose, CA, USA, 12–16 July 1992; pp. 123–128. [Google Scholar]
- Sachar, S.; Kumar, A. Survey of feature extraction and classification techniques to identify plant through leaves. Expert Syst. Appl. 2021, 167, 114181. [Google Scholar] [CrossRef]
- Langley, P.; Iba, W.; Thompson, K. An analysis of Bayesian classifiers. In Proceedings of the AAAI, San Jose, CA, USA, 12–16 July 1992; Volume 90, pp. 223–228. [Google Scholar]
- Nadeau, C.; Bengio, Y. Inference for the generalization error. Mach. Learn. 2003, 52, 239–281. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).