A Novel Two-Level Fusion Feature for Mixed ECG Identity Recognition
Abstract
:1. Introduction
- The simplified process of identification without denoising algorithm;
- Novel combination of manual features for Hilbert transform and power spectrum;
- The MaxFusion algorithm is proposed to reduce the dimension of PCANet learning features;
- The identification implementation of mixed data set with different sampling frequencies.
2. Materials and Methods
2.1. The Overall Process of Two-Level Fusion Feature Identification Network
2.2. Preprocessing
2.3. Elementary Fusion Feature Extraction
2.3.1. Hilbert Transform Feature
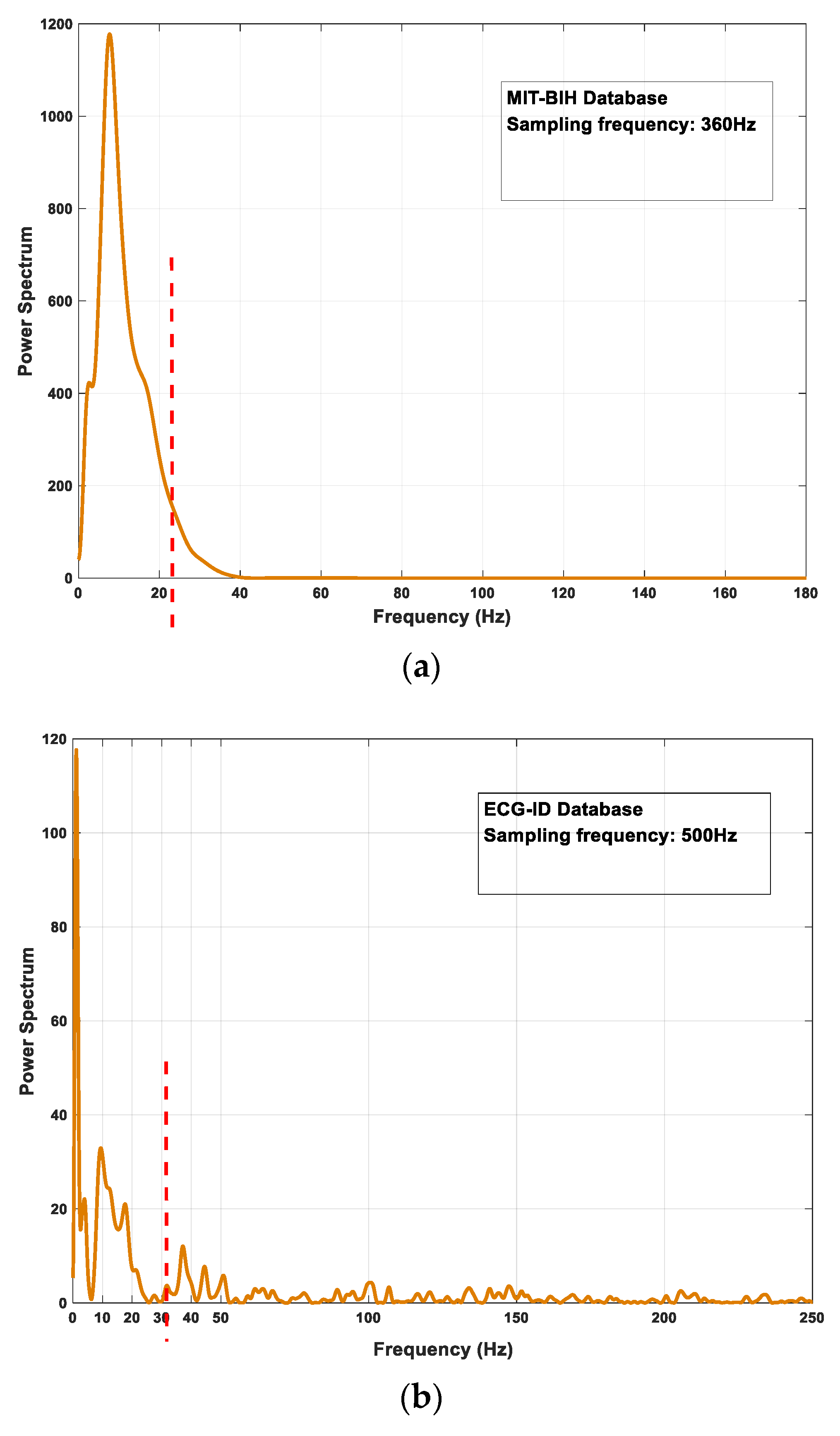
2.3.2. Power Spectrum Feature
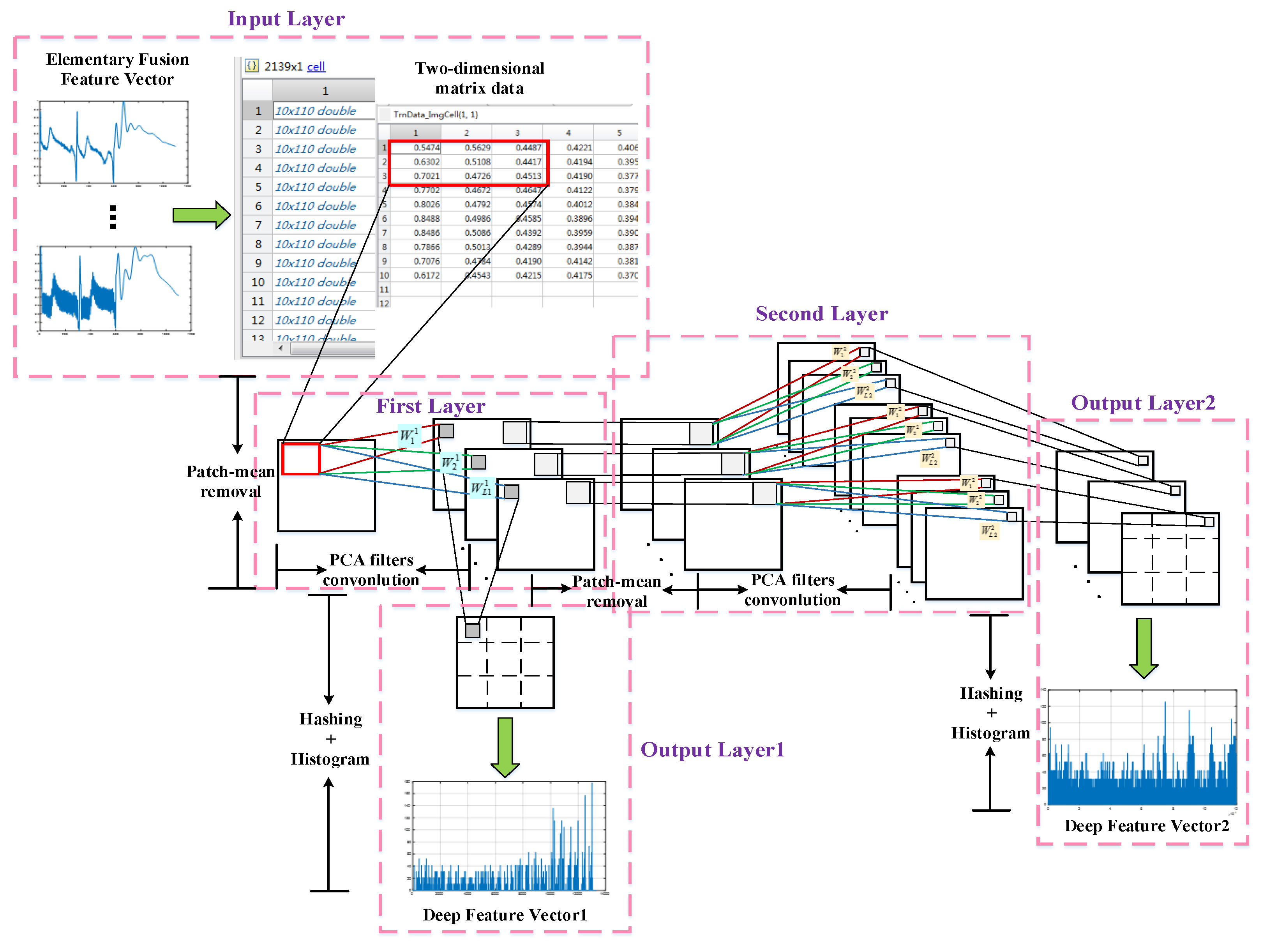
2.4. PCANet Deep Fusion Feature Extraction
2.4.1. PCANet
- Input Layer
- 2.
- First Convolutional Layer
- 3.
- Second Convolutional Layer
- 4.
- Output Layer
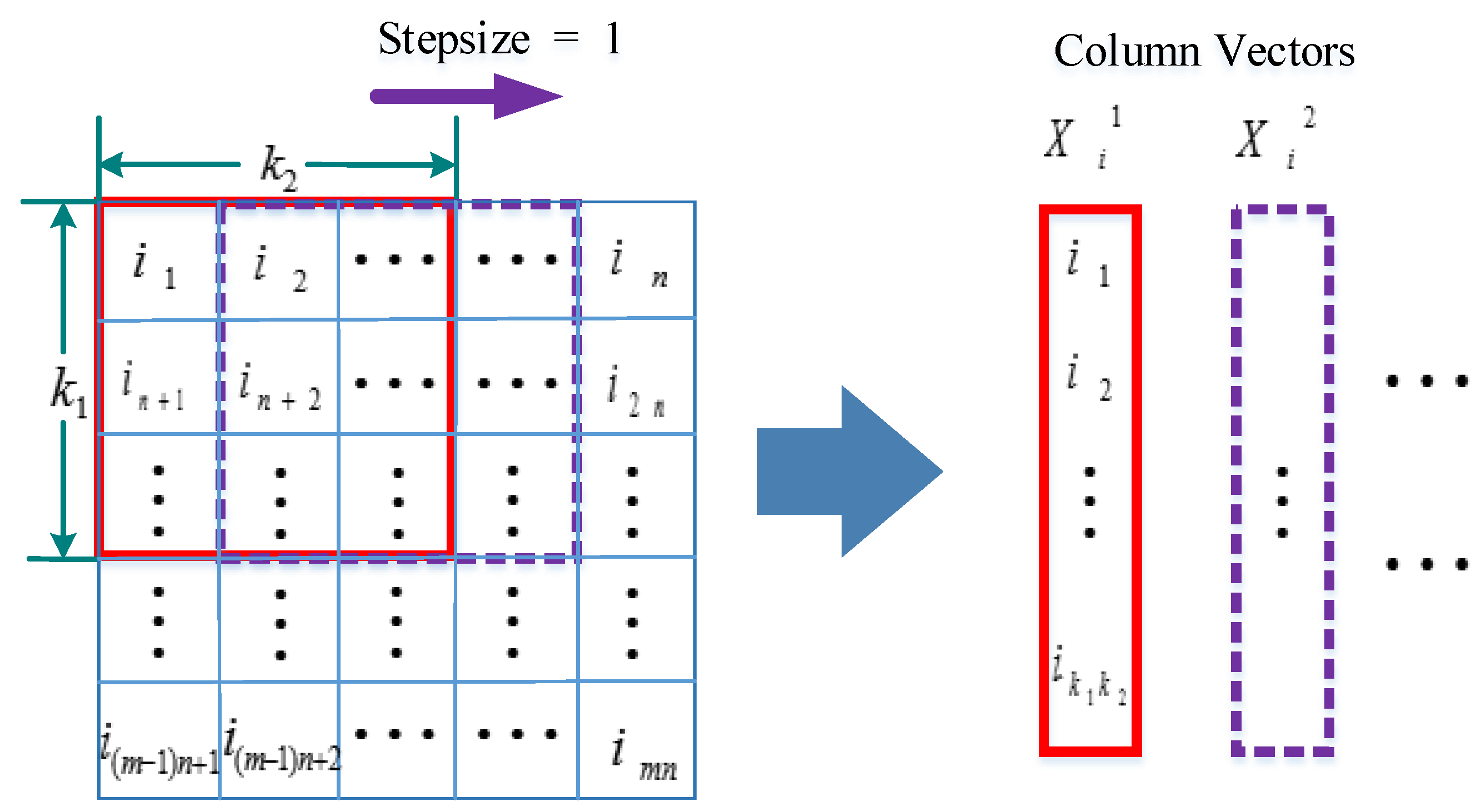
2.4.2. MaxFusion Algorithm
| Algorithm 1. MaxFusion algorithm procedure of two layers PCANet features |
| Input: Trainset samples: (The first layer feature of PCANet); (The second layer feature of PCANet); Testset samples: ; Output: MF Fusion feature: ; do Cascade two layers of training and testing features ; Set sliding window size for to or for to do Generate the window signal Find the maximum value of window signal Form the new single feature end for Form the new MF fusion data set ; end for |
2.5. Classifier
3. Results
3.1. Database
- ECG ID Database: 310 ECG recordings are collected in the ECG ID database, which comes from 90 healthy individuals. Each signal is recorded 20 s, the sampling frequency is 500 Hz. In the experiment, two data of each person are randomly selected as training and testing set, while the 47th individual is eliminated as only containing one recording of ECG signals. So the remaining 89 individuals are used as study subjects in all ECG-ID database experiments;
- MIT-BIH Arrhythmia Database: There are 48 ambulatory ECG signals with two-channel in the MIT-BIH arrhythmia database, among them, labels 201 and 202 come from the same subject. So the database records ECG data for 47 individuals with various arrhythmia diseases. Each signal lasts 30 min and the sampling frequency is 360 Hz. This database is used as abnormal ECG signals to test the proposed model;
- PTB Diagnostic ECG Database: It is composed of 549 data recordings for 290 subjects, 52 of which are healthy, and the sampling frequency is 1000 Hz. In the experiment, a total of 290 individuals of the same day signals are selected to detect model recognition ability of ECG signals with obvious baseline drift noise;
- The Mixed Data Set: The mixed three databases with different sampling frequencies contains 426 individuals which are applied to identify. The experiment chooses such a merged database to prove the feasibility of the model when the number of individuals increases and test generalization ability for ECG signals under different acquisition conditions.
3.2. Experimental Setting
3.3. Experimental Results
3.3.1. Comparison Experiment for Different Classifiers
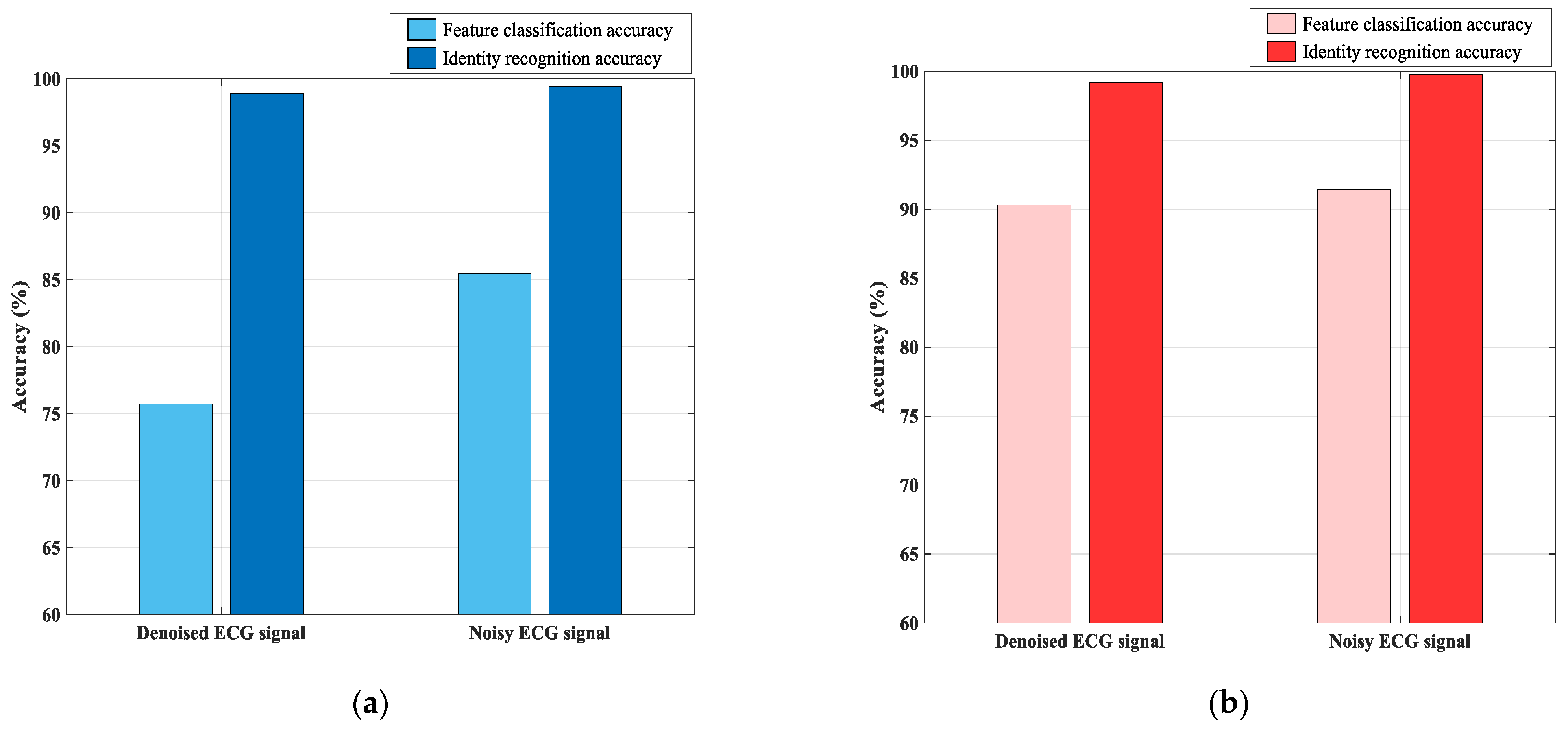
3.3.2. Comparison Experiment for Denoised and Original Signals
3.3.3. Comparison Experiment for Different Feature Extraction Algorithms
3.3.4. Comparison Experiment for Different PCANet Features
3.3.5. Comparison Experiment of the Related Researches
4. Discussion
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Kaplan Berkaya, S.; Uysal, A.K.; Sora Gunal, E.; Ergin, S.; Gunala, S.; Bilginer Gulmezogluc, M. A survey on ECG analysis. Biomed. Signal Process. Control 2018, 43, 216–235. [Google Scholar] [CrossRef]
- Karimian, N.; Guo, Z.M.; Tehranipoor, M.; Forte, D. Highly Reliable Key Generation from Electrocardiogram (ECG). IEEE Trans. Biomed. Eng. 2017, 64, 1400–1411. [Google Scholar] [CrossRef]
- Stegmayer, G.; Gerard, M.; Milone, D.H. Data mining over biological datasets: An integrated approach based on computational intelligence. Comput. Intell. Mag. 2012, 7, 22–34. [Google Scholar] [CrossRef]
- Agrafioti, F.; Gao, J.; Hatzinakos, D. Heart biometrics: Theory, methods and applications. Biometrics 2011, 3, 199–216. [Google Scholar]
- Pinto, J.R.; Cardoso, J.S.; Lourenco, A. Evolution, Current Challenges, and Future Possibilities in ECG Biometrics. IEEE Access 2018, 6, 34746–34776. [Google Scholar] [CrossRef]
- Khoma, V.; Pelc, M.; Khoma, Y. Artificial neural network capability for human being identification based on ECG. In Proceedings of the 2018 23rd International Conference on Methods & Models in Automation & Robotics (MMAR), Miedzyzdroje, Poland, 27–30 August 2018; pp. 479–482. [Google Scholar]
- Ferdinando, H.; Seppanen, T.; Alasaarela, E. Comparing features from ECG pattern and HRV analysis for emotion recognition system. In Proceedings of the 2016 IEEE Conference on Computational Intelligence in Bioinformatics and Computational Biology (CIBCB), Chiang Mai, Thailand, 5–7 October 2016; pp. 1–6. [Google Scholar]
- Rezgui, D.; Lachiri, Z. Integrating EMD attributes for person identification from electrocardiographic signals. In Proceedings of the 2016 2nd International Conference on Advanced Technologies for Signal and Image Processing (ATSIP), Monastir, Tunisia, 21–23 March 2016; pp. 478–482. [Google Scholar]
- Byeon, Y.-H.; Pan, S.-B.; Kwak, K.-C. Intelligent Deep Models Based on Scalograms of Electrocardiogram Signals for Biometrics. Sensors 2019, 19, 935. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kiranyaz, S.; Ince, T.; Gabbouj, M. Real-Time Patient-Specific ECG Classification by 1-D Convolutional Neural Networks. IEEE Trans. Biomed. Eng. 2016, 63, 664–675. [Google Scholar] [CrossRef] [PubMed]
- Ferdinando, H.; Seppänen, T.; Alasaarela, E. Bivariate empirical mode decomposition for ECG-based biometric identification with emotional data. In Proceedings of the 2017 39th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Jeju, Korea, 11–15 July 2017; pp. 450–453. [Google Scholar]
- Dar, M.N.; Akram, M.U.; Usman, A.; Khan, S.A. ECG biometric identification for general population using multiresolution analysis of DWT based features. In Proceedings of the Second International Conference on Information Security & Cyber Forensics, Cape Town, South Africa, 15–17 November 2015; pp. 5–10. [Google Scholar]
- Xu, J.; Li, T.; Chen, Y.; Chen, Y. Personal identification by convolutional neural network with ECG signal. In Proceedings of the 2018 International Conference on Information and Communication Technology Convergence (ICTC), Jeju, Korea, 17–19 October 2017; pp. 559–563. [Google Scholar]
- Wu, Y.; Wu, H.; Chang, W. Compressed Domain ECG Biometric Identification Using JPEG2000. In Proceedings of the International Joint Conference on E-business & Telecommunications, Colmar, France, 20–22 July 2015; pp. 5–13. [Google Scholar]
- Zhang, Q.; Zhou, D.; Zeng, X. HeartID: A multiresolution convolutional neural network for ECG-based biometric human identification in smart health applications. IEEE Access 2017, 5, 11805–11816. [Google Scholar] [CrossRef]
- Lin, S.L.; Chen, C.K.; Lin, C.L.; Yang, W.C.; Chiang, C.T. Individual identification based on chaotic electrocardiogram signals during muscular exercise. IET Biom. 2014, 3, 257–266. [Google Scholar] [CrossRef]
- AI Rahhal, M.M.; Bazi, Y.; Alhichri, H.; Alajlan, N.; Melgani, F.; Yager, F.F. Deep learning approach for active classification of electrocardiogram signals. Inf. Sci. 2016, 345, 340–354. [Google Scholar] [CrossRef]
- Kim, J.S.; Kim, S.H.; Pan, S.B. Personal recognition using convolutional neural network with ECG coupling image. J. Ambient. Intell. Humaniz. Comput. 2020, 11, 1923–1932. [Google Scholar] [CrossRef]
- Wieclaw, L.; Khoma, Y.; Falat, P.; Sabodashko, D.; Herasymenko, V. Biometrie identification from raw ECG signal using deep learning techniques. In Proceedings of the 2017 9th IEEE International Conference on Intelligent Data Acquisition and Advanced Computing Systems: Technology and Applications (IDAACS), Bucharest, Romania, 21–23 September 2017; pp. 129–133. [Google Scholar]
- Salloum, R.; Kuo, C.C.J. ECG-based biometrics using recurrent neural networks. In Proceedings of the IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), New Orleans, LA, USA, 5–9 March 2017; pp. 2062–2066. [Google Scholar]
- Acharya, U.R.; Fujita, H.; Lih, O.S.; Hagiwara, Y.; Tan, J.H.; Adam, M. Automated detection of arrhythmias using different intervals of tachycardia ECG segments with convolutional neural network. Inf. Sci. 2017, 405, 81–90. [Google Scholar] [CrossRef]
- Ramya, M.; Muthukumar, A.; Kannan, S. Multibiometric based authentication using feature level fusion. In Proceedings of the 2012 International Conference on Advances in Engineering, Science and Management (ICAESM), Nagapattinam, India, 30–31 March 2012; pp. 191–195. [Google Scholar]
- Shahram, T.; Nsen, T. Animal classification using facial images with score-level fusion. IET Comput. Vis. 2018, 12, 679–685. [Google Scholar]
- Vijayalakshmi, V.; Divya, R.; Jaganath, K. Finger print and Palm print based Multibiometric Authentication System with GUI Interface. In Proceedings of the 2013 International Conference on Communication and Signal Processing Melmaruvathur, Tamilnadu, India, 3–5 April 2013; pp. 738–742. [Google Scholar]
- Chen, X.; Xu, H.; Shen, H. ECG Identification Based on Fusion Features of Morphological Characteristics and KPCA. Electron. Technol. 2015, 3, 5–8. [Google Scholar]
- Golrizkhatami, Z.; Acan, A. ECG classification using three-level fusion of different feature descriptors. Expert Syst. Appl. 2018, 114, 54–64. [Google Scholar] [CrossRef]
- Ai, D.; Yang, J.; Wang, Z.; Fan, J. Fast multi-scale feature fusion for ECG heartbeat classification. Eurasip J. Adv. Signal Process. 2015, 2015, 46. [Google Scholar] [CrossRef] [Green Version]
- Meng, X.; Liang, C.; Polajnar, D.; Tong, W. Local binary pattern network: A deep learning approach for face recognition. In Proceedings of the IEEE International Conference on Image Processing, Phoenix, AZ, USA, 25–28 September 2016; pp. 3224–3228. [Google Scholar]
- Yang, X.; Liu, W.; Tao, D.; Cheng, J.; Li, S. Multiview canonical correlation analysis betworks for remote sensing image recognition. IEEE Geosci. Remote. Sens. Lett. 2017, 14, 1855–1859. [Google Scholar] [CrossRef]
- Yang, W.; Si, Y.; Wang, D.; Zhang, G. A novel method for identifying electrocardiograms using an independent component analysis and principal component analysis network. Measurement 2020, 152, 107363. [Google Scholar] [CrossRef]
- Yang, W.; Si, Y.; Wang, D.; Zhang, G. A novel approach for multi-Lead ECG classification using DL-CCANet and TL-CCANet. Sensors 2019, 19, 3214. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wei, W.; Zhang, C.X.; Lin, W. A QRS Wave Detection Algorithm Based on Complex Wavelet Transform. Appl. Mech. Mater. 2013, 239–240, 1284–1288. [Google Scholar] [CrossRef]
- Khazaee, A.; Zadeh, A.E. ECG beat classification using particle swarm optimization and support vector machine. Front. Comput. Sci. 2014, 8, 217–231. [Google Scholar] [CrossRef]
- Liu, W.; Wang, Z.; Liu, X.; Zeng, N.; Liu, Y.; Alsaadi, F.E. A survey of deep neural network architectures and their applications. Neurocomputing 2017, 234, 11–26. [Google Scholar] [CrossRef]
- Chan, T.H.; Jia, K.; Gao, S.; Lu, J.; Zeng, Z.; Ma, Y. PCANet: A Simple Deep Learning Baseline for Image Classification? IEEE Trans. Image Process. 2015, 24, 5017–5032. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, D.; Si, Y.; Yang, W.; Zhang, G.; Liu, T. A Novel Heart Rate Robust Method for Short-Term Electrocardiogram Biometric Identification. Appl. Sci. 2019, 9, 201. [Google Scholar] [CrossRef] [Green Version]
- Chu, Y.; Shen, H.; Huang, K. ECG Authentication Method Based on Parallel Multi-scale One-dimensional Residual Network with Center and Margin Loss. IEEE Access 2019, 7, 51598–51607. [Google Scholar] [CrossRef]
- Rui, L.A.; Gya, B.; Kw, A.; Huang, Y. Robust ECG biometrics using GNMF and sparse representation. Pattern Recognit. Lett. 2020, 129, 70–76. [Google Scholar]
- Gurkan, H.; Hanilci, A. ECG based biometric identification method using QRS images and convolutional neural network. Pamukkale Univ. J. Eng. Sci. 2020, 26, 318–327. [Google Scholar] [CrossRef]
- Dong, X.; Si, W.; Yu, W. Identity recognition based on the qrs complex dynamics of electrocardiogram. IEEE Access 2020, 99, 1. [Google Scholar] [CrossRef]
- Liu, F.; Si, Y.; Luo, T.; Wang, D.; Liu, Q.; Li, L. The ECG Identification Based on GRNN. In Proceedings of the 2018 IEEE International Conference on Communication Systems (ICCS), Chengdu, China, 19–21 December 2018; pp. 418–422. [Google Scholar]
- Pal, A.; Singh, Y.N. ECG Biometric Recognition. In International Conference on Mathematics & Computing; Springer: Singapore, 2018; pp. 61–73. [Google Scholar]
- Alotaiby, T.N.; Alshebeili, S.A.; Aljafar, L.M.; Alsabhan, W.M. ECGbased subject identification using common spatial pattern and SVM. J. Sens. 2019, 2019, 1–9. [Google Scholar]








| Layer | Parameters | Value |
|---|---|---|
| Input layer | Elementary fusion feature matrix size | |
| First layer | The size of patch | |
| The number of filters | 9 | |
| Output layer 1 | The size of histogram block | |
| The overlap ratio of histogram block | 0.5 | |
| Second layer | The size of patch | |
| The number of filters | 9 | |
| Output layer 2 | The size of histogram block | |
| The overlap ratio of histogram block | 0.5 | |
| Fusion layer | The length of fusion feature | 10,000 |
| Method 1 | Method 2 | Method 3 | Method 4 | Method 5 (The Proposed Method) | |
|---|---|---|---|---|---|
| Hilbert transform feature | Y | N | Y | N | Y |
| Power spectrum feature | Y | N | N | Y | Y |
| Two-layer PCANet fusion feature | N | Y | Y | Y | Y |
| Sensitivity | 49.88% | 88.90% | 88.80% | 89.55% | 93.84% |
| Precision | 50.02% | 89.78% | 89.73% | 89.87% | 94.58% |
| Feature classification accuracy | 59.04% | 87.20% | 86.97% | 85.33% | 91.45% |
| Number of Training Features | Training Feature Length | Training Time of Linear SVM | Feature Classification Accuracy | Identity Recognition Accuracy | |
|---|---|---|---|---|---|
| One-layer PCANet features | 7389 | 13,312 | 44.59 s | 89.59% | 99.53% |
| Two-layers PCANet features | 119,808 | 352.31 s | 90.97% | 99.53% | |
| The combined two layers PCANet features | 133,120 | 391.63 s | 92.17% | 99.77% | |
| The fused two layers PCANet features by MF algorithm | 10,000 | 159.28 s | 91.45% | 99.77% |
| Methods | Databases | Number of Subjects | Identity Recognition Accuracy | |
|---|---|---|---|---|
| Byeon (2019) [9] | AlexNet | PTB | 61 | 92.50% |
| Chu et al. (2019) [37] | Residual network | ECG-ID | 90 | 98.24% |
| MIT-BIH Arrhythmia | 47 | 95.99% | ||
| Rui et al. (2020) [38] | GNMF + sparse representation | ECG-ID | 89 | 98.03% |
| MIT-BIH Arrhythmia | 47 | 100% | ||
| Gurkan et al. (2020) [39] | QRS images + CNN | PTB | 50 | 83.95% |
| Dong et al. (2020) [40] | The QRS complex dynamics + radial basis function networks | PTB | 52 | 97.31% |
| Liu et al. (2018) [41] | SVD-LDA + GRNN | ECG-ID | 89 | 96.60% |
| Pal et al. (2018) [42] | Area fiducial features + PCA | PTB | 100 | 97.10% |
| Alotaiby et al. (2019) [43] | CSP features + SVM | PTB | 200 | 95.15% |
| The proposed model | Two-level fusion feature + SVM | ECG-ID | 89 | 99.44% |
| MIT-BIH Arrhythmia | 47 | 100% | ||
| PTB | 290 | 99.66% | ||
| Mixed Data Set | 426 | 99.77% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, X.; Si, Y.; Yang, W. A Novel Two-Level Fusion Feature for Mixed ECG Identity Recognition. Electronics 2021, 10, 2052. https://doi.org/10.3390/electronics10172052
Liu X, Si Y, Yang W. A Novel Two-Level Fusion Feature for Mixed ECG Identity Recognition. Electronics. 2021; 10(17):2052. https://doi.org/10.3390/electronics10172052
Chicago/Turabian StyleLiu, Xin, Yujuan Si, and Weiyi Yang. 2021. "A Novel Two-Level Fusion Feature for Mixed ECG Identity Recognition" Electronics 10, no. 17: 2052. https://doi.org/10.3390/electronics10172052
APA StyleLiu, X., Si, Y., & Yang, W. (2021). A Novel Two-Level Fusion Feature for Mixed ECG Identity Recognition. Electronics, 10(17), 2052. https://doi.org/10.3390/electronics10172052





