Recent Research Progress and Potential Uses of the Amphibian Xenopus as a Biomedical and Immunological Model System
Abstract
:1. Introduction
2. Xenopus as a Model for Biomedical Research
2.1. Xenopus as a Model to Study Immune Tolerance
| Young tadpoles | Stage 35–40, 5–8 mm long, 2–7 days post-fertilization |
|
| Pre-metamorphic tadpoles | Stage 50–57, 30–50 mm long, 2–6 weeks post-fertilization |
|
| Metamorph | Stage 58–66, 40–60 mm long, 6–8 weeks post-fertilization |
|
| Young adults | 40–60 mm long, 3–12 months-old |
|
2.1.1. Xenopus as a Model to Study Skin Graft Tolerance and Rejection
| Definition of X. laevis strains | Name (MHC genotype) | Cell lines |
|---|---|---|
| Partially inbred, MHC homozygous strains | F (f/f) | Thymic lymphoid tumor cell lines |
| ||
| J (j/j) | Fibroblast cell lines | |
| ||
| Isogenetic laevis/gilli (LG) clones. | LG-15 (a/c) | Thymic lymphoid tumor cell line |
| Same heterozygous MHC but different minor H loci |
| |
| LG-6 (a/c) | Fibroblast cell line | |
|

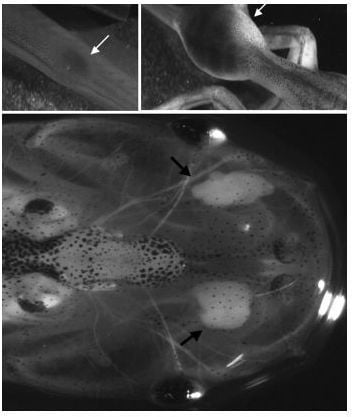
2.1.1.1. The Xenopus Immuno-Cancer Model
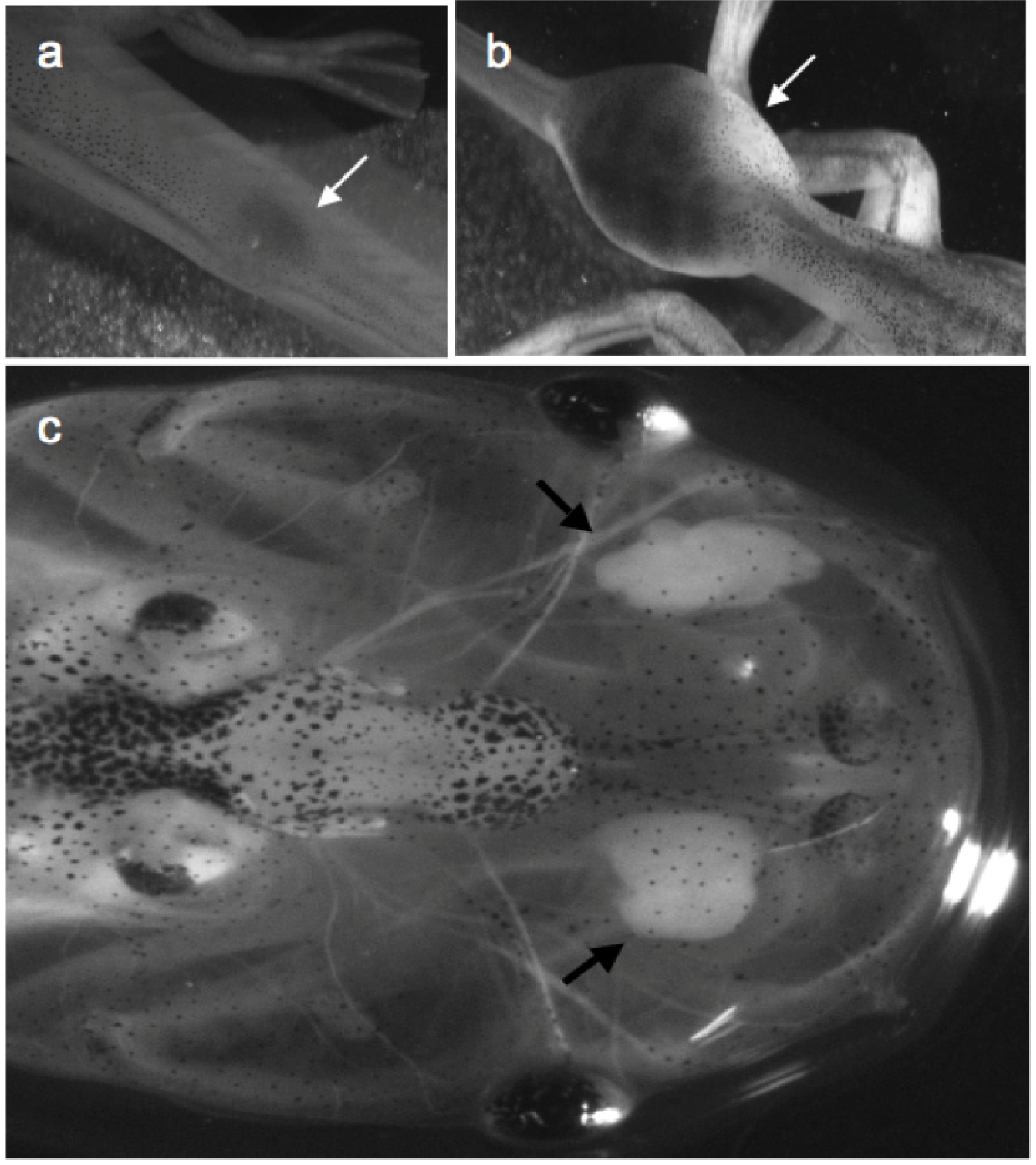
3. Reverse Genetics
3.1. Optimized Transgenesis for Immunological Studies in Xenopus

3.1.1. Recent Advances in Loss-of-Function Approaches
4. The Xenopus Genus: Genetic Resources and Polyploidy
4.1. Genomic Resources
4.1.1. Natural and Artificial Polyploidy in the Xenopus Genus
5. Concluding Remarks
Acknowledgments
Conflict of Interest
References
- Robert, J.; Ohta, Y. Comparative and developmental study of the immune system in Xenopus. Dev. Dyn. 2009, 238, 1249–1270. [Google Scholar] [CrossRef]
- Du Pasquier, L.; Schwager, J.; Flajnik, M.F. The immune system of Xenopus. Annu. Rev. Immunol. 1989, 7, 251–275. [Google Scholar] [CrossRef]
- Khokha, M.K. Xenopus white papers and resources: Folding functional genomics and genetics into the frog. Genesis 2012, 50, 133–142. [Google Scholar] [CrossRef]
- Hellsten, U.; Harland, R.M.; Gilchrist, M.J.; Hendrix, D.; Jurka, J.; Kapitonov, V.; Ovcharenko, I.; Putnam, N.H.; Shu, S.; Taher, L.; et al. The genome of the western clawed frog Xenopus tropicalis. Science 2010, 328, 633–636. [Google Scholar] [CrossRef]
- Xenopus laevis Genome Resources Web Page. Available online: http://www.xenbase.org/genomes/static/laevis.jsp (accessed on 9 July 2013).
- University of Rochester Medical Center Web Page. Xenopus laevis Research Resource for Immunobiology. Available online: http://www.urmc.rochester.edu/mbi/resources/Xenopus (accessed on 9 July 2013).
- Hsu, E. Mutation, selection, and memory in B lymphocytes of exothermic vertebrates. Immunol. Rev. 1998, 162, 25–36. [Google Scholar] [CrossRef]
- Du Pasquier, L.; Robert, J.; Courtet, M.; Mussmann, R. B-cell development in the amphibian Xenopus. Immunol. Rev. 2000, 175, 201–213. [Google Scholar] [CrossRef]
- Barlow, E.H.; Cohen, N. The thymus dependency of transplantation allotolerance in the metamorphosing frog Xenopus laevis. Transplantation 1983, 35, 612–619. [Google Scholar] [CrossRef]
- Nedelkovska, H.; Robert, J. Comparative Study of Skin Graft Tolerance and Rejection in the Frog Xenopus laevis. Skin Graft–Indications, Applications and Current Research; Spear, S., Ed.; InTech: New York, NY, USA, 2011. Available online: http://www.intechopen.com/books/skin-grafts-indications-applications-and-current-research/comparative-study-of-skin-graft-tolerance-and-rejection-in-the-frog-Xenopus-laevis (accessed on 10 July 2013).
- Chardonnens, X.; Du Pasquier, L. Induction of skin allograft tolerance during metamorphosis of the toad Xenopus laevis: A possible model for studying generation of self tolerance to histocompatibility antigens. Eur. J. Immunol. 1973, 3, 569–573. [Google Scholar] [CrossRef]
- Horton, J.D.; Horton, T.L.; Ritchie, P. Incomplete tolerance induced in Xenopus by larval tissue allografting: Evidence from immunohistology and mixed leucocyte culture. Dev. Comp. Immunol. 1993, 17, 249–262. [Google Scholar] [CrossRef]
- Turpen, J.B.; Smith, P.B. Precursor immigration and thymocyte succession during larval development and metamorphosis in Xenopus. J. Immunol. 1989, 142, 41–47. [Google Scholar]
- Flajnik, M.F.; Kaufman, J.F.; Hsu, E.; Manes, M.; Parisot, R.; Du Pasquier, L. Major histocompatibility complex-encoded class I molecules are absent in immunologically competent Xenopus before metamorphosis. J. Immunol. 1986, 137, 3891–3899. [Google Scholar]
- Salter-Cid, L.; Nonaka, M.; Flajnik, M.F. Expression of MHC class Ia and class Ib during ontogeny: High expression in epithelia and coregulation of class Ia and lmp7 genes. J. Immunol. 1998, 160, 2853–2861. [Google Scholar]
- Rollins-Smith, L.A.; Flajnik, M.F.; Blair, P.J.; Davis, A.T.; Green, W.F. Involvement of thyroid hormones in the expression of MHC class I antigens during ontogeny in Xenopus. Dev. Immunol. 1997, 5, 133–144. [Google Scholar] [CrossRef]
- Du Pasquier, L.; Flajnik, M.F. Expression of MHC class II antigens during Xenopus development. Dev. Immunol. 1990, 1, 85–95. [Google Scholar] [CrossRef]
- Horton, J.D.; Horton, T.L.; Dzialo, R.; Gravenor, I.; Minter, R.; Ritchie, P.; Gartland, L.; Watson, M.D.; Cooper, M.D. T-cell and natural killer cell development in thymectomized Xenopus. Immunol. Rev. 1998, 166, 245–258. [Google Scholar] [CrossRef]
- Robert, J.; Gantress, J.; Cohen, N.; Maniero, G.D. Xenopus as an experimental model for studying evolution of hsp—Immune system interactions. Methods 2004, 32, 42–53. [Google Scholar] [CrossRef]
- Du Pasquier, L.; Chardonnens, C. Genetic aspects of the tolerance to allografts induced at metamorphosis in the toad Xenopus laevis. Immunogenetics 1975, 2, 431–440. [Google Scholar] [CrossRef]
- Tochinai, S.K.; Katagiri, C. Complete abrogation of immune responses to skin allografts and rabbit erythrocytes in the early thymectomized Xenopus. Dev. Growth Differ. 1975, 17, 383–394. [Google Scholar]
- Xenbase Home Page. Available online: http://www.xenbase.org (accessed on 9 July 2013).
- Kobel, H.R.; Du Pasquier, L. Production of large clones of histocompatible, fully identical clawed toads (Xenopus). Immunogenetics 1975, 2, 7–91. [Google Scholar]
- Nedelkovska, H.; Cruz-Luna, T.; McPherson, P.; Robert, J. Comparative in vivo study of gp96 adjuvanticity in the frog Xenopus laevis. J. Vis. Exp. 2010, 43, 2026:1–2026:5. [Google Scholar]
- Robert, J.; Cohen, N. Evolution of immune surveillance and tumor immunity: Studies in Xenopus. Immunol. Rev. 1998, 166, 231–243. [Google Scholar] [CrossRef]
- Goyos, A.; Robert, J. Tumorigenesis and anti-tumor immune responses in Xenopus. Front. Biosci. 2009, 14, 167–176. [Google Scholar] [CrossRef]
- Du Pasquier, L.; Robert, J. In vitro growth of thymic tumor cell lines from Xenopus. Dev. Immunol. 1992, 2, 295–307. [Google Scholar] [CrossRef]
- Robert, J.; Guiet, C.; Du Pasquier, L. Lymphoid tumors of Xenopus laevis with different capacities for growth in larvae and adults. Dev. Immunol. 1994, 3, 297–307. [Google Scholar] [CrossRef]
- Robert, J.; Cohen, N. Ontogeny of CTX expression in Xenopus. Dev. Comp. Immunol. 1998, 22, 605–612. [Google Scholar] [CrossRef]
- Robert, J.; Guiet, C.; Du Pasquier, L. Ontogeny of the alloimmune response against a transplanted tumor in Xenopus laevis. Differentiation 1995, 59, 135–144. [Google Scholar] [CrossRef]
- Goyos, A.; Guselnikov, S.; Chida, A.S.; Sniderhan, L.F.; Maggirwar, S.B.; Nedelkovska, H.; Robert, J. Involvement of nonclassical MHC class Ib molecules in heat shock protein-mediated anti-tumor responses. Eur. J. Immunol. 2007, 37, 1494–1501. [Google Scholar] [CrossRef]
- Goyos, A.; Ohta, Y.; Guselnikov, S.; Robert, J. Novel nonclassical MHC class Ib genes associated with CD8 T cell development and thymic tumors. Mol. Immunol. 2009, 46, 1775–1786. [Google Scholar] [CrossRef]
- Zitvogel, L.; Tesniere, A.; Kroemer, G. Cancer despite immunosurveillance: Immunoselection and immunosubversion. Nat. Rev. Immunol. 2006, 6, 715–727. [Google Scholar] [CrossRef]
- Yie, S.M.; Yang, H.; Ye, S.R.; Li, K.; Dong, D.D.; Lin, X.M. Expression of human leucocyte antigen G (HLA-G) is associated with prognosis in non-small cell lung cancer. Lung Cancer 2007, 58, 267–274. [Google Scholar]
- De Kruijf, E.M.; Sajet, A.; van Nes, J.G.; Natanov, R.; Putter, H.; Smit, V.T.; Liefers, G.J.; van den Elsen, P.J.; van de Velde, C.J.; Kuppen, P.J. HLA-E and HLA-G expression in classical HLA class I-negative tumors is of prognostic value for clinical outcome of early breast cancer patients. J. Immunol. 2010, 185, 7452–7459. [Google Scholar] [CrossRef]
- Chesneau, A.; Sachs, L.M.; Chai, N.; Chen, Y.; Du Pasquier, L.; Loeber, J.; Pollet, N.; Reilly, M.; Weeks, D.L.; Bronchain, O.J. Transgenesis procedures in Xenopus. Biol. Cell 2008, 100, 503–521. [Google Scholar] [CrossRef]
- Kroll, K.L.; Amaya, E. Transgenic Xenopus embryos from sperm nuclear transplantations reveal FGF signaling requirements during gastrulation. Development 1996, 122, 3173–3183. [Google Scholar]
- Amaya, E.; Kroll, K.L. A method for generating transgenic frog embryos. MethodsMol. Biol. 1999, 97, 393–414. [Google Scholar]
- Allen, B.G.; Weeks, D.L. Transgenic Xenopus laevis embryos can be generated using phiC31 integrase. Nat. Methods 2005, 2, 975–979. [Google Scholar] [CrossRef]
- Sinzelle, L.; Vallin, J.; Coen, L.; Chesneau, A.; Du Pasquier, D.; Pollet, N.; Demeneix, B.; Mazabraud, A. Generation of trangenic Xenopus laevis using the Sleeping Beauty transposon system. Transgenic Res. 2006, 15, 751–760. [Google Scholar] [CrossRef]
- Ogino, H.; McConnell, W.B.; Grainger, R.M. High-throughput transgenesis in Xenopus using I-SceI meganuclease. Nat. Protoc. 2006, 1, 1703–1710. [Google Scholar] [CrossRef]
- Ogino, H.; McConnell, W.B.; Grainger, R.M. Highly efficient transgenesis in Xenopus tropicalis using I-SceI meganuclease. Mech. Dev. 2006, 123, 103–113. [Google Scholar] [CrossRef]
- Pan, F.C.; Chen, Y.; Loeber, J.; Henningfeld, K.; Pieler, T. I-SceI meganuclease-mediated transgenesis in Xenopus. Dev. Dyn. 2006, 235, 247–252. [Google Scholar] [CrossRef]
- Allen, B.G.; Weeks, D.L. Bacteriophage phiC31 integrase mediated transgenesis in Xenopus laevis for protein expression at endogenous levels. Methods Mol. Biol. 2009, 518, 113–122. [Google Scholar] [CrossRef]
- Groth, A.C.; Olivares, E.C.; Thyagarajan, B.; Calos, M.P. A phage integrase directs efficient site-specific integration in human cells. Proc. Natl. Acad. Sci. USA 2000, 97, 5995–6000. [Google Scholar] [CrossRef]
- Yergeau, D.A.; Kelley, C.M.; Zhu, H.; Kuliyev, E.; Mead, P.E. Transposon transgenesis in Xenopus. Methods 2010, 51, 92–100. [Google Scholar] [CrossRef]
- Hamlet, M.R.; Yergeau, D.A.; Kuliyev, E.; Takeda, M.; Taira, M.; Kawakami, K.; Mead, P.E. Tol2 transposon-mediated transgenesis in Xenopus tropicalis. Genesis 2006, 44, 438–445. [Google Scholar] [CrossRef]
- Miskey, C.; Izsvak, Z.; Plasterk, R.H.; Ivics, Z. The Frog Prince: A reconstructed transposon from Rana pipiens with high transpositional activity in vertebrate cells. Nucleic Acids Res. 2003, 31, 6873–6881. [Google Scholar] [CrossRef]
- Nedelkovska, H.; Robert, J. Optimized transgenesis in Xenopus laevis/gilli isogenetic clones for immunological studies. Genesis 2012, 50, 300–306. [Google Scholar] [CrossRef]
- Sifuentes-Romero, I.; Milton, S.L.; Garcia-Gasca, A. Post-transcriptional gene silencing by RNA interference in non-mammalian vertebrate systems: Where do we stand? Mutat. Res. 2011, 728, 158–171. [Google Scholar] [CrossRef]
- Lei, Y.; Guo, X.; Liu, Y.; Cao, Y.; Deng, Y.; Chen, X.; Cheng, C.H.; Dawid, I.B.; Chen, Y.; Zhao, H. Efficient targeted gene disruption in Xenopus embryos using engineered transcription activator-like effector nucleases (TALENs). Proc. Natl. Acad. Sci. USA 2012, 109, 17484–17489. [Google Scholar]
- Ishibashi, S.; Cliffe, R.; Amaya, E. Highly efficient bi-allelic mutation rates using TALENs in Xenopus tropicalis. Biol. Open 2012, 1, 1273–1276. [Google Scholar] [CrossRef]
- Young, J.J.; Cherone, J.M.; Doyon, Y.; Ankoudinova, I.; Faraji, F.M.; Lee, A.H.; Ngo, C.; Guschin, D.Y.; Paschon, D.E.; Miller, J.C.; et al. Efficient targeted gene disruption in the soma and germ line of the frog Xenopus tropicalis using engineered zinc-finger nucleases. Proc. Natl. Acad. Sci 2011, 108, 7052–7057. [Google Scholar] [CrossRef]
- Nedelkovska, H.; Edholm, E.S.; Haynes, N.; Robert, J. Effective RNAi-mediated beta2-microglobulin loss of function by transgenesis in Xenopus laevis. Biol. Open 2013, 2, 335–342. [Google Scholar] [CrossRef]
- National Center for Biotechnology Information (NCBI) Web Page. UniGene. Available online: http://www.ncbi.nlm.nih.gov/unigene (accessed on 9 July 2013).
- Kobel, H.R.; Tinsley, R.C. Xenopus Species and Ecology 2: The Extant Species. In The Biology of Xenopus; Clarendon: Oxford, UK, 1996; pp. 9–31. [Google Scholar]
- Evans, B.J. Genome evolution and speciation genetics of clawed frogs (Xenopus and Silurana). Front. Biosci. 2008, 13, 4687–4706. [Google Scholar] [CrossRef]
- Kobel, H.R.; Du Pasquier, L. Genetics of polyploidy Xenopus. Trends Genet. 1986, 2, 310–315. [Google Scholar] [CrossRef]
- Flajnik, M.F.; Kasahara, M. Comparative genomics of the MHC: Glimpses into the evolution of the adaptive immune system. Immunity 2001, 15, 351–362. [Google Scholar] [CrossRef]
- Hellsten, U.; Khokha, M.K.; Grammer, T.C.; Harland, R.M.; Richardson, P.; Rokhsar, D.S. Accelerated gene evolution and subfunctionalization in the pseudotetraploid frog Xenopus laevis. BMC Biol 2007, 5. [Google Scholar] [CrossRef]
- Hughes, M.K.; Hughes, A.L. Evolution of duplicate genes in a tetraploid animal, Xenopus laevis. Mol. Biol. Evol. 1993, 10, 1360–1369. [Google Scholar]
- Shum, B.P.; Avila, D.; Du Pasquier, L.; Kasahara, M.; Flajnik, M.F. Isolation of a classical MHC class I cDNA from an amphibian. Evidence for only one class I locus in the Xenopus MHC. J. Immunol. 1993, 151, 5376–5386. [Google Scholar]
- Courtet, M.; Flajnik, M.; Du Pasquier, L. Major histocompatibility complex and immunoglobulin loci visualized by in situ hybridization on Xenopus chromosomes. Dev. Comp. Immunol. 2001, 25, 149–157. [Google Scholar] [CrossRef]
© 2013 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Edholm, E.-S.; Robert, J. Recent Research Progress and Potential Uses of the Amphibian Xenopus as a Biomedical and Immunological Model System. Resources 2013, 2, 167-183. https://doi.org/10.3390/resources2030167
Edholm E-S, Robert J. Recent Research Progress and Potential Uses of the Amphibian Xenopus as a Biomedical and Immunological Model System. Resources. 2013; 2(3):167-183. https://doi.org/10.3390/resources2030167
Chicago/Turabian StyleEdholm, Eva-Stina, and Jacques Robert. 2013. "Recent Research Progress and Potential Uses of the Amphibian Xenopus as a Biomedical and Immunological Model System" Resources 2, no. 3: 167-183. https://doi.org/10.3390/resources2030167
APA StyleEdholm, E.-S., & Robert, J. (2013). Recent Research Progress and Potential Uses of the Amphibian Xenopus as a Biomedical and Immunological Model System. Resources, 2(3), 167-183. https://doi.org/10.3390/resources2030167