TIP_finder: An HPC Software to Detect Transposable Element Insertion Polymorphisms in Large Genomic Datasets
Abstract
1. Introduction
2. Materials and Methods
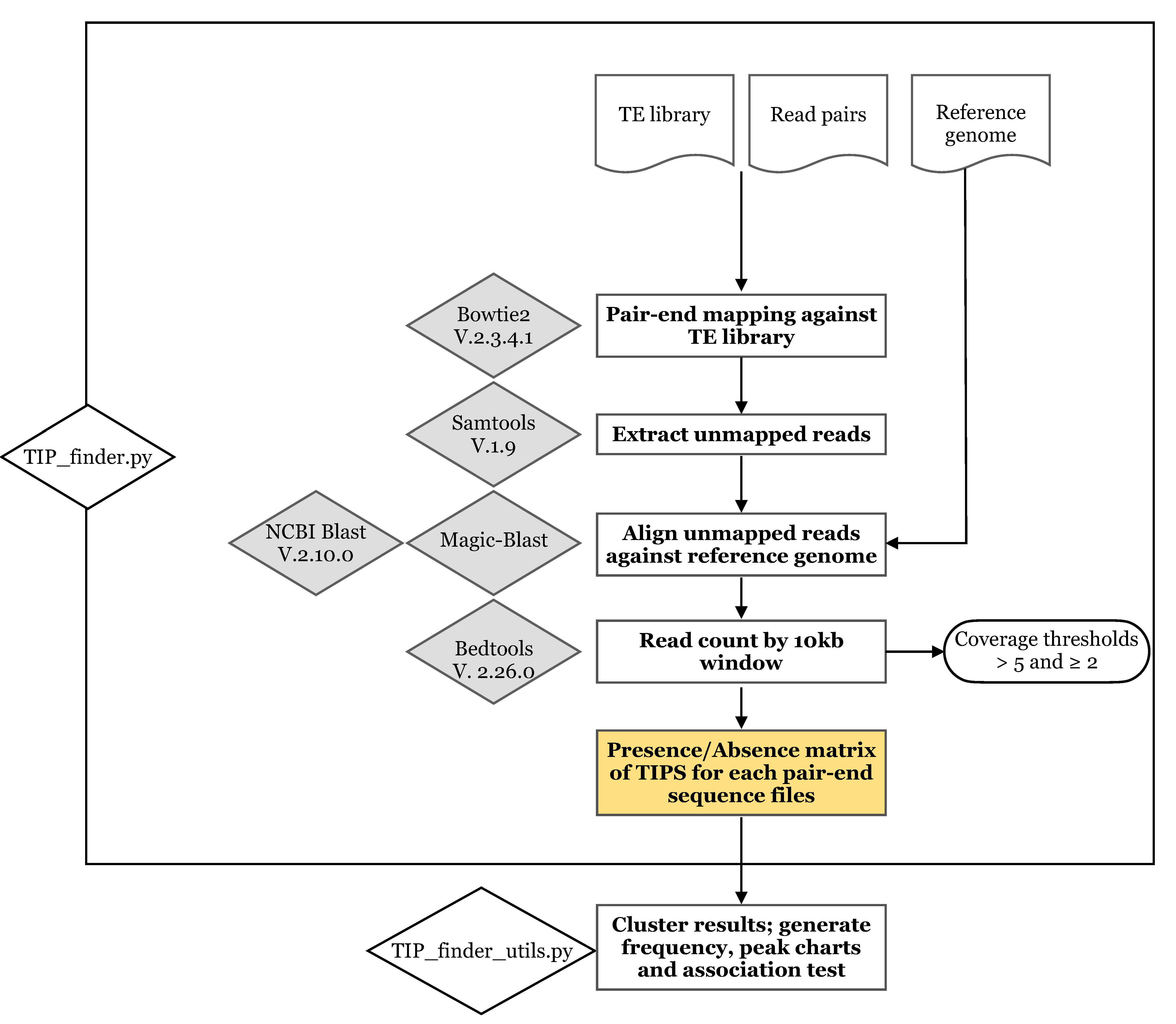
2.1. Implementation of TIP_finder
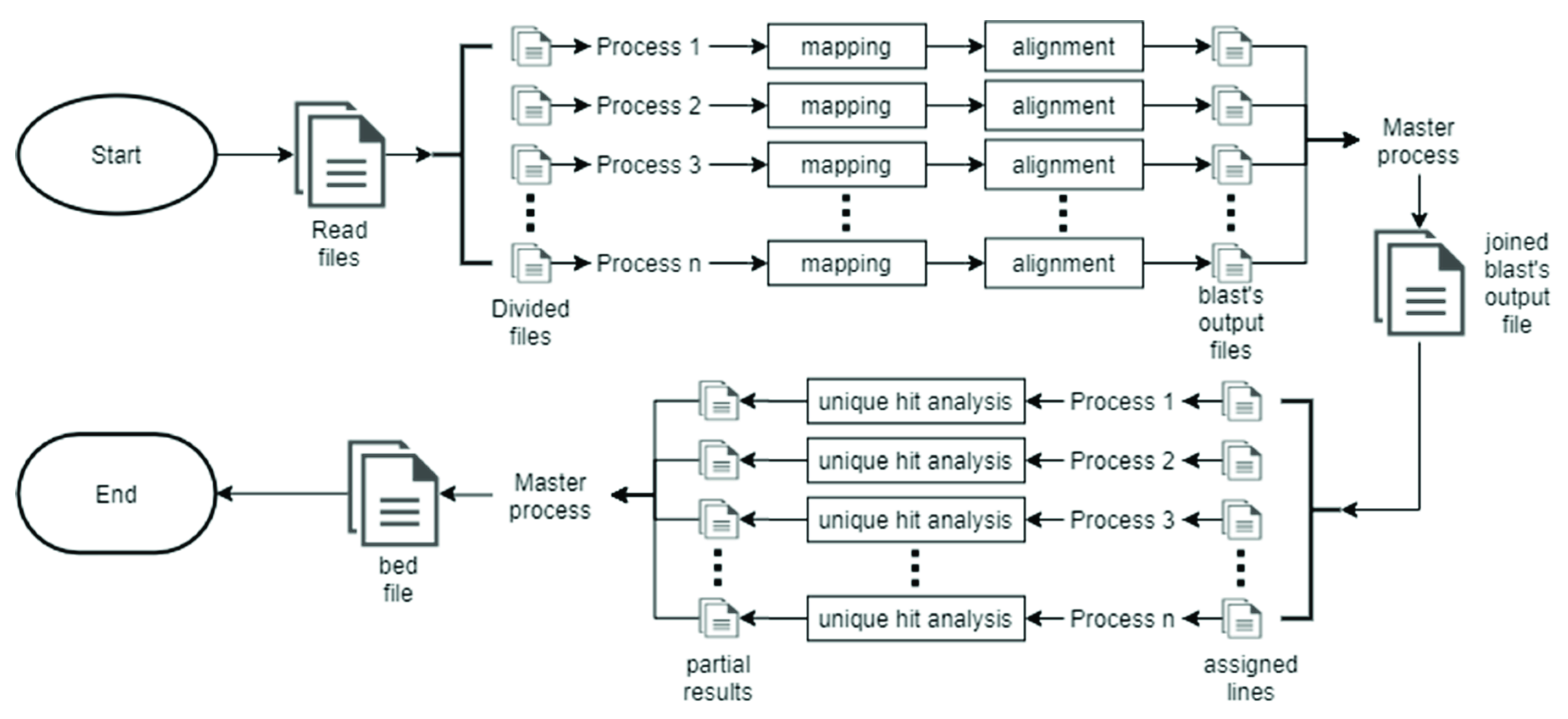
2.2. Parallel Strategy Implemented
2.3. Statistical Association Analysis
2.4. Genomic Data Used by TIP_finder
2.5. Computational Resources
3. Results
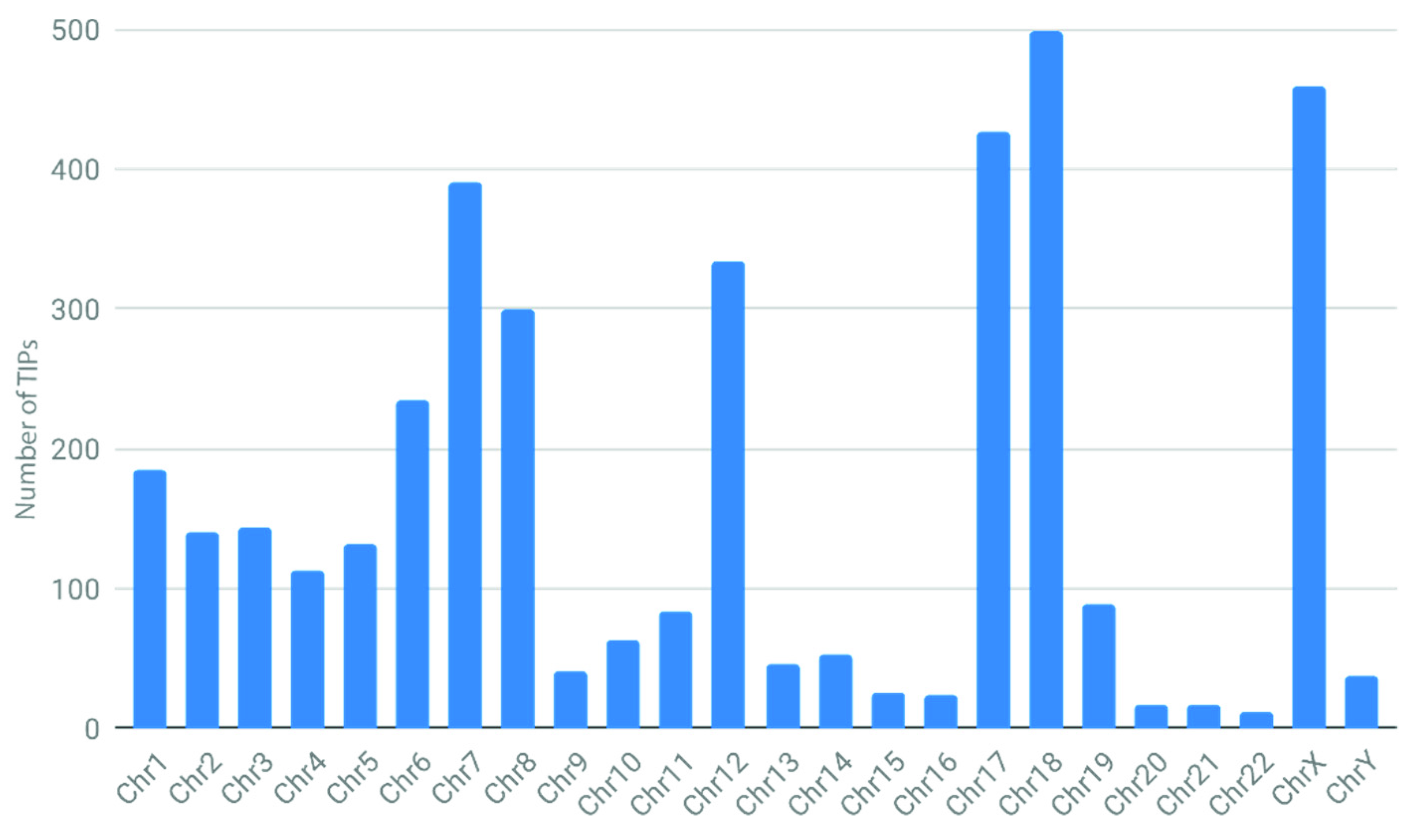
3.1. Problems Encountered with Large Genomes and Testing TIP_finder
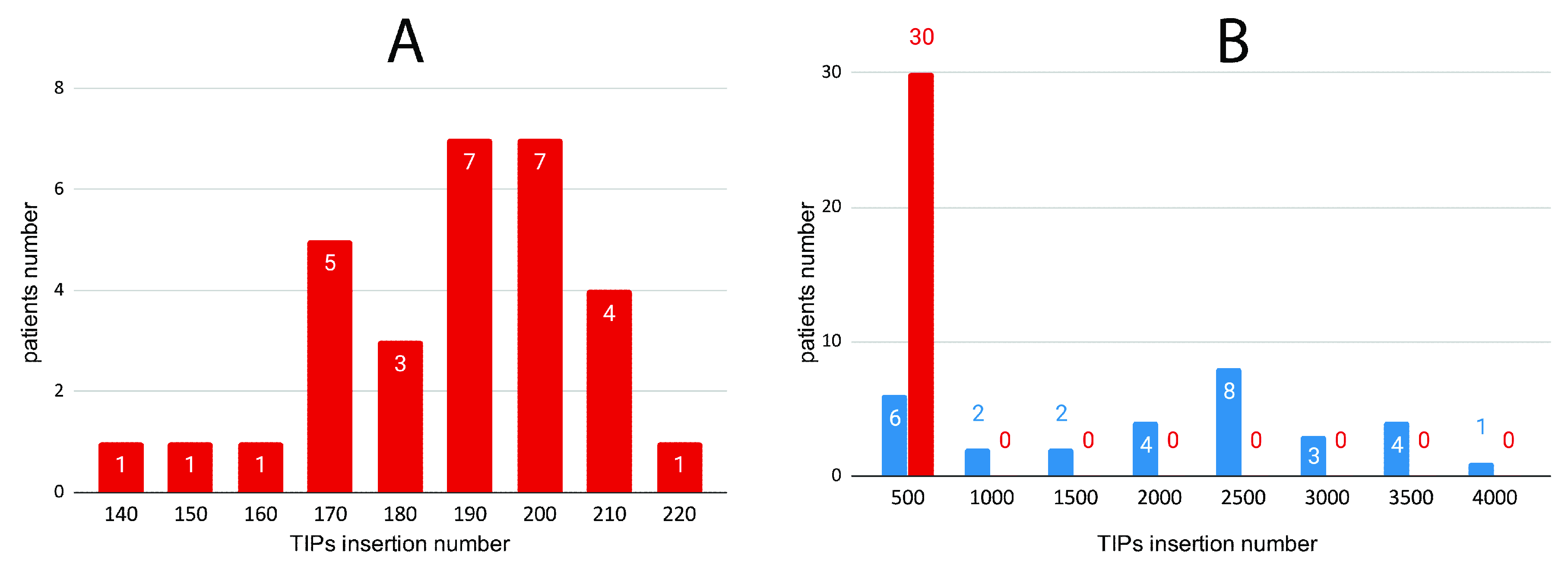
3.2. Correlation of HERV-K TIPs with Breast Cancer as a Proof of Concept
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- McClintock, B. The Origin and Behavior of Mutable Loci in Maize. Proc. Natl. Acad. Sci. USA 1950, 36, 344–355. [Google Scholar] [CrossRef] [PubMed]
- Orozco-Arias, S.; Liu, J.; Id, R.T.; Ceballos, D.; Silva, D.; Id, D.; Ming, R.; Guyot, R. Inpactor, Integrated and Parallel Analyzer and Classifier of LTR Retrotransposons and Its Application for Pineapple LTR Retrotransposons Diversity and Dynamics. Biology 2018, 7, 32. [Google Scholar] [CrossRef] [PubMed]
- Bourque, G.; Burns, K.H.; Gehring, M.; Gorbunova, V.; Seluanov, A.; Hammell, M.; Imbeault, M.; Izsvák, Z.; Levin, H.L.; Macfarlan, T.S.; et al. Ten things you should know about transposable elements. Genome Biol. 2018, 19, 199. [Google Scholar] [CrossRef] [PubMed]
- Rishishwar, L.; Mariño-Ramírez, L.; Jordan, I.K. Benchmarking computational tools for polymorphic transposable element detection. Brief. Bioinform. 2017, 18, 908–918. [Google Scholar] [CrossRef]
- Orozco-Arias, S.; Isaza, G.; Guyot, R.; Tabares-soto, R. A systematic review of the application of machine learning in the detection and classi fi cation of transposable elements. PeerJ 2019, 7, 18311. [Google Scholar] [CrossRef]
- Choulet, F.; Alberti, A.; Theil, S.; Glover, N.; Barbe, V.; Daron, J.; Pingault, L.; Sourdille, P.; Couloux, A.; Paux, E.; et al. Structural and functional partitioning of bread wheat chromosome 3B. Science 2014, 345, 1249721. [Google Scholar] [CrossRef]
- Su, W.; Gu, X.; Peterson, T. TIR-Learner, a New Ensemble Method for TIR Transposable Element Annotation, Provides Evidence for Abundant New Transposable Elements in the Maize Genome. Mol. Plant 2019, 12, 447–460. [Google Scholar] [CrossRef]
- De Koning, A.P.J.; Gu, W.; Castoe, T.A.; Batzer, M.A.; Pollock, D.D. Repetitive elements may comprise over Two-Thirds of the human genome. PLoS Genet. 2011, 7. [Google Scholar] [CrossRef]
- Orozco-Arias, S.; Isaza, G.; Guyot, R. Retrotransposons in Plant Genomes: Structure, Identification, and Classification through Bioinformatics and Machine Learning. Int. J. Mol. Sci. 2019, 20, 3837. [Google Scholar] [CrossRef]
- Todorovska, E. Retrotransposons and their role in plant—Genome evolution. Biotechnol. Biotechnol. Equip. 2014, 21, 294–305. [Google Scholar] [CrossRef]
- Casacuberta, E.; González, J. The impact of transposable elements in environmental adaptation. Mol. Ecol. 2013, 22, 1503–1517. [Google Scholar] [CrossRef]
- Zhang, M.; Liang, J.Q. Expressional activation and functional roles of human endogenous retroviruses in cancers. Rev. Med. Virol. 2019, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Lisch, D. How important are transposons for plant evolution? Nat. Rev. Genet. 2013, 14, 49–61. [Google Scholar] [CrossRef] [PubMed]
- Deininger, P.L.; Batzer, M.A. Alu repeats and human disease. Mol. Genet. Metab. 1999, 67, 183–193. [Google Scholar] [CrossRef]
- Hancks, D.C.; Kazazian, H.H. Active human retrotransposons: Variation and disease. Curr. Opin. Genet. Dev. 2012, 22, 191–203. [Google Scholar] [CrossRef] [PubMed]
- Beck, C.R.; Garcia-Perez, J.L.; Badge, R.M.; Moran, J.V. LINE-1 Elements in Structural Variation and Disease. Annu. Rev. Genom. Hum. Genet. 2011, 12, 187–215. [Google Scholar] [CrossRef] [PubMed]
- Chaparro, C.; Gayraud, T.; de Souza, R.F.; Domingues, D.S.; Akaffou, S.S.; Vanzela, A.L.L.; de Kochko, A.; Rigoreau, M.; Crouzillat, D.; Hamon, S.; et al. Terminal-repeat retrotransposons with GAG domain in plant genomes: A new testimony on the complex world of transposable elements. Genome Biol. Evol. 2015, 7, 493–504. [Google Scholar] [CrossRef]
- Wicker, T.; Sabot, F.; Hua-Van, A.; Bennetzen, J.L.; Capy, P.; Chalhoub, B.; Flavell, A.; Leroy, P.; Morgante, M.; Panaud, O.; et al. A unified classification system for eukaryotic transposable elements. Nat. Rev. Genet. 2007, 8, 973–982. [Google Scholar] [CrossRef]
- Neumann, P.; Novák, P.; Hoštáková, N.; MacAs, J. Systematic survey of plant LTR-retrotransposons elucidates phylogenetic relationships of their polyprotein domains and provides a reference for element classification. Mob. DNA 2019, 10, 1. [Google Scholar] [CrossRef]
- Laten, H.M.; Gaston, G.D. Plant Endogenous Retroviruses? A Case of Mysterious ORFs. In Plant Transposable Elements; Spriger: Berlin/Heidelberg, Germany, 2012; pp. 89–112. [Google Scholar]
- Grandbastien, M.-A.A. LTR retrotransposons, handy hitchhikers of plant regulation and stress response. Biochim. Biophys. Acta Gene Regul. Mech. 2015, 1849, 403–416. [Google Scholar] [CrossRef]
- Subramanian, R.P.; Wildschutte, J.H.; Russo, C.; Coffin, J.M. Identification, characterization, and comparative genomic distribution of the HERV-K (HML-2) group of human endogenous retroviruses. Retrovirology 2011, 1–22. [Google Scholar] [CrossRef] [PubMed]
- Havecker, E.R.; Gao, X.; Voytas, D.F. The diversity of LTR retrotransposons. Genome Biol. 2004, 5, 225. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Rishishwar, L.; Wang, L.; Clayton, E.A.; Mariño-Ramírez, L.; McDonald, J.F.; Jordan, I.K. Population and clinical genetics of human transposable elements in the (post) genomic era. Mob. Genet. Elem. 2017. [Google Scholar] [CrossRef] [PubMed]
- Asch, H.L.; Eliacin, E.; Fanning, T.G.; Connolly, J.L.; Bratthauer, G.; Asch, B.B. Comparative Expression of the LINE-1 p40 Protein in Human Breast Breast Carcinomas and Normal Breast Tissues. Oncol. Res. Featur. Preclin. Clin. Cancer Ther. 1996, 8, 239–247. [Google Scholar]
- Johanning, G.L.; Malouf, G.G.; Zheng, X.; Esteva, F.J.; Weinstein, J.N.; Wang-Johanning, F.; Su, X. Expression of human endogenous retrovirus-K is strongly associated with the basal-like breast cancer phenotype. Sci. Rep. 2017, 7, 41960. [Google Scholar] [CrossRef]
- Goering, W.; Schmitt, K.; Dostert, M.; Schaal, H.; Mayer, J.; Schulz, W.A. Human Endogenous Retrovirus HERV-K (HML-2) Activity in Prostate Cancer Is Dominated by a Few Loci. Prostate 2015, 1971, 1958–1971. [Google Scholar] [CrossRef]
- Roesch, A.; Meese, E.; Mayer, J.; Schmitt, K. Transcriptional Profiling of Human Endogenous Retrovirus Group HERV-K (HML-2) Loci in Melanoma. Genome Biol. Evol. 2013, 5, 307–328. [Google Scholar] [CrossRef]
- Bratthauer, G.L.; Fanning, T.G. LINE-1 retrotransposon expression in pediatric germ cell tumors. Cancer 1993, 71, 2383–2386. [Google Scholar] [CrossRef]
- Carpentier, M.C.; Manfroi, E.; Wei, F.J.; Wu, H.P.; Lasserre, E.; Llauro, C.; Debladis, E.; Akakpo, R.; Hsing, Y.I.; Panaud, O. Retrotranspositional landscape of Asian rice revealed by 3000 genomes. Nat. Commun. 2019, 10. [Google Scholar] [CrossRef]
- Martienssen, R. Epigenetic phenomena: Paramutation and gene silencing in plants. Curr. Biol. 1996, 6, 810–813. [Google Scholar] [CrossRef]
- Drongitis, D.; Aniello, F.; Fucci, L.; Donizetti, A. Roles of Transposable Elements in the Different Layers of Gene Expression Regulation. Int. J. Mol. Sci. 2019, 20, 5755. [Google Scholar] [CrossRef]
- Barrón, M.G.; Fiston-Lavier, A.-S.; Petrov, D.A.; González, J. Population Genomics of Transposable Elements in Drosophila. Annu. Rev. Genet. 2014, 48, 561–581. [Google Scholar] [CrossRef]
- Rigal, M.; Mathieu, O. A “mille-feuille” of silencing: Epigenetic control of transposable elements. Biochim. Biophys. Acta Gene Regul. Mech. 2011, 1809, 452–458. [Google Scholar] [CrossRef]
- Ewing, A.D. Transposable element detection from whole genome sequence data. Mob. DNA 2015, 6. [Google Scholar] [CrossRef]
- Vendrell-Mir, P.; Barteri, F.; Merenciano, M.; González, J.; Casacuberta, J.M.; Castanera, R. A benchmark of transposon insertion detection tools using real data. Mob. DNA 2019, 10, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Jiang, C.; Chen, C.; Huang, Z.; Liu, R.; Verdier, J. ITIS, a bioinformatics tool for accurate identification of transposon insertion sites using next-generation sequencing data. BMC Bioinform. 2015, 16. [Google Scholar] [CrossRef][Green Version]
- Hénaff, E.; Zapata, L.; Casacuberta, J.M.; Ossowski, S. Jitterbug: Somatic and germline transposon insertion detection at single-nucleotide resolution. BMC Genom. 2015, 16, 768. [Google Scholar] [CrossRef] [PubMed]
- Helman, E.; Lawrence, M.S.; Stewart, C.; Sougnez, C.; Getz, G.; Meyerson, M. Somatic retrotransposition in human cancer revealed by whole-genome and exome sequencing. Genome Res. 2014, 24, 1053–1063. [Google Scholar] [CrossRef] [PubMed]
- Mohiyuddin, M.; Mu, J.C.; Li, J.; Bani Asadi, N.; Gerstein, M.B.; Abyzov, A.; Wong, W.H.; Lam, H.Y.K. MetaSV: An accurate and integrative structural-variant caller for next generation sequencing. Bioinformatics 2015, 31, 2741–2744. [Google Scholar] [CrossRef] [PubMed]
- Kroon, M.; Lameijer, E.W.; Lakenberg, N.; Hehir-Kwa, J.Y.; Thung, D.T.; Slagboom, P.E.; Kok, J.N.; Ye, K. Detecting dispersed duplications in high-throughput sequencing data using a database-free approach. Bioinformatics 2016, 32, 505–510. [Google Scholar] [CrossRef]
- Tran, H.T.M.; Ramaraj, T.; Furtado, A.; Lee, L.S.; Henry, R.J. Use of a draft genome of coffee (Coffea arabica) to identify SNP s associated with caffeine content. Plant Biotechnol. J. 2018, 16, 1756–1766. [Google Scholar] [CrossRef] [PubMed]
- Mueller, L.; Strickler, S.; Domingues, D.; Pereira, L.; Andrade, A.; Marraccini, P.; Ming, R.; Wai, J.; Albert, V.; Giuliano, G.; et al. Towards a better understanding of the Coffea Arabica genome structure. In Proceedings of the Embrapa Café-Artigo em Anais de Congresso (ALICE), International Conference on Coffee Science, Armenia, Colombia, 8–13 September 2014. [Google Scholar]
- Wu, A.R.; Neff, N.F.; Kalisky, T.; Dalerba, P.; Treutlein, B.; Rothenberg, M.E.; Mburu, F.M.; Mantalas, G.L.; Sim, S.; Clarke, M.F.; et al. Quantitative assessment of single-cell RNA-sequencing methods. Nat. Methods 2014, 11, 41–46. [Google Scholar] [CrossRef]
- Berlin, K.; Koren, S.; Chin, C.S.; Drake, J.P.; Landolin, J.M.; Phillippy, A.M. Assembling large genomes with single-molecule sequencing and locality-sensitive hashing. Nat. Biotechnol. 2015, 33, 623–630. [Google Scholar] [CrossRef]
- Lander, E.S.; Linton, L.M.; Birren, B.; Nusbaum, C.; Zody, M.C.; Baldwin, J.; Devon, K.; Dewar, K.; Doyle, M.; FitzHugh, W.; et al. Initial sequencing and analysis of the human genome. Nature 2001, 409, 860–921. [Google Scholar] [PubMed]
- Cheng, S.; Melkonian, M.; Smith, S.A.; Brockington, S.; Archibald, J.M.; Delaux, P.-M.; Li, F.-W.; Melkonian, B.; Mavrodiev, E.V.; Sun, W.; et al. 10KP: A phylodiverse genome sequencing plan. Gigascience 2018, 7, giy013. [Google Scholar] [CrossRef] [PubMed]
- Lewin, H.A.; Robinson, G.E.; Kress, W.J.; Baker, W.J.; Coddington, J.; Crandall, K.A.; Durbin, R.; Edwards, S.V.; Forest, F.; Gilbert, M.T.P.; et al. Earth BioGenome Project: Sequencing life for the future of life. Proc. Natl. Acad. Sci. USA 2018, 115, 4325–4333. [Google Scholar] [CrossRef] [PubMed]
- Tatusova, T. Update on genomic databases and resources at the national center for biotechnology information. In Methods in Molecular Biology; Humana Press Inc.: Totowa, NJ, USA, 2016; Volume 1415, pp. 3–30. [Google Scholar]
- Tabares Soto, R. Programación Paralela Sobre Arquitecturas Heterogéneas. Master’s Thesis, Universidad Nacional de Colombia, Manizales, Colombia, November 2016. [Google Scholar]
- Orozco-Arias, S.; Tabares-Soto, R.; Ceballos, D.; Guyot, R. Parallel Programming in Biological Sciences, Taking Advantage of Supercomputing in Genomics. In Advances in Computing; Solano, A., Ordoñez, H., Eds.; Springer: Zurich, Switzerland, 2017; Volume 735, pp. 627–643. ISBN 9781457720819. [Google Scholar]
- Mikailov, M.; Luo, F.J.; Barkley, S.; Valleru, L.; Whitney, S.; Liu, Z.; Thakkar, S.; Tong, W.; Petrick, N. Scaling bioinformatics applications on HPC. BMC Bioinform. 2017, 18, 501. [Google Scholar] [CrossRef]
- Orozco-Arias, S.; Camargo-forero, L.; Correa, J.C.; Guyot, R.; Cristancho, M. BIOS-ParallelBlast: Paralelización optimizada de alineamiento de secuencias sobre Xeon Phi. Ing. Investig. Technol. 2017, 18, 423–432. [Google Scholar]
- Rodrigues, F.M.; von Mering, C. Sequence analysis HPC-CLUST: Distributed hierarchical clustering for large sets of nucleotide sequences. Bioinformatics 2014, 30, 287–288. [Google Scholar] [CrossRef]
- Sawyer, S.; Horton, M.; Burdyshaw, C.; Brook, G. HPC-BLAST: Distributed BLAST for Modern HPC Clusters. In Proceedings of the 11th International Conference on Bioinformatics and Computational Biology, Honolulu, HI, USA, 18–20 March 2019. [Google Scholar]
- Gropp, W.; Lusk, E. Fault Tolerance in Message Passing Interface Programs. Int. J. High Perform. Comput. Appl. 2004, 18, 363–372. [Google Scholar] [CrossRef]
- Gropp, W.; Lusk, E.; Doss, N.; Skjellum, A. A high-performance, portable implementation of the MPI message passing interface standard. Parallel Comput. 1996, 22, 789–828. [Google Scholar] [CrossRef]
- Aguilar Castro, J.L.; Leiss, E. Introducción a la Computación Paralela; Universidad de los Andes: Mérida, Venezuela, 2004; ISBN 9801207523. [Google Scholar]
- Chen, W.; Feng, P.M.; Deng, E.Z.; Lin, H.; Chou, K.C. iTIS-PseTNC: A sequence-based predictor for identifying translation initiation site in human genes using pseudo trinucleotide composition. Anal. Biochem. 2014, 462, 76–83. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R.; Data, G.P.; et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [PubMed]
- Quinlan, A.R.; Hall, I.M. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 2010, 26, 841–842. [Google Scholar] [CrossRef]
- Altschup, S.F.; Gish, W.; Pennsylvania, T.; Park, U. Basic Local Alignment Search Tool 2Department of Computer Science. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Boratyn, G.M.; Thierry-Mieg, J.; Thierry-Mieg, D.; Busby, B.; Madden, T.L. Magic-BLAST, an accurate RNA-seq aligner for long and short reads. BMC Bioinform. 2019, 20, 1–19. [Google Scholar] [CrossRef]
- Dalcín, L.; Paz, R.; Storti, M.; D’Elía, J. MPI for Python: Performance improvements and MPI-2 extensions. J. Parallel Distrib. Comput. 2008, 68, 655–662. [Google Scholar] [CrossRef]
- McKinney, W. Data Structures for Statistical Computing in Python. In Proceedings of the 9th Python in Science Conference, Austin, TX, USA, 28 June–3 July 2010; pp. 51–56. [Google Scholar]
- Hunter, J.D. Matplotlib: A 2D graphics environment. Comput. Sci. Eng. 2007, 9, 90–95. [Google Scholar] [CrossRef]
- Waskom, M.; Botvinnik, O.; O’Kane, D.; Hobson, P.; Lukauskas, S.; Gemperline, D.C.; Augspurger, T.; Halchenko, Y.; Cole, J.B.; Warmenhoven, J.; et al. Mwaskom/seaborn: v0.8.1 (September 2017). Zenodo 2017. [Google Scholar] [CrossRef]
- Virtanen, P.; Gommers, R.; Oliphant, T.E.; Haberland, M.; Reddy, T.; Cournapeau, D.; Burovski, E.; Peterson, P.; Weckesser, W.; Bright, J.; et al. SciPy 1.0: Fundamental algorithms for scientific computing in Python. Nat. Methods 2020, 17, 261–272. [Google Scholar] [CrossRef]
- Maurya, V.N.; Singh, V.V.; Yusuf, M.U. Statistical Analysis on the Rate of Kidney (Renal) Failure. Am. J. Appl. Math. Stat. 2014, 2, 6–12. [Google Scholar] [CrossRef]
- Edition, S. Smooth Tests of Goodness of Fit; John Wiley & Sons: Hoboken, NJ, USA, 2009; ISBN 9780470824429. [Google Scholar]
- Cochran, W.G. Some methods for strenghthening the commom χ^2 tests. Biometrics 2012, 10, 417–451. [Google Scholar] [CrossRef]
- Denoeud, F.; Carretero-Paulet, L.; Dereeper, A.; Droc, G.; Guyot, R.; Pietrella, M.; Zheng, C.; Alberti, A.; Anthony, F.; Aprea, G.; et al. The coffee genome provides insight into the convergent evolution of caffeine biosynthesis. Science 2014, 345, 1181–1184. [Google Scholar] [CrossRef]
- Huang, L.; Wang, X.; Dong, Y.; Long, Y.; Hao, C.; Yan, L.; Shi, T. Resequencing 93 accessions of coffee unveils independent and parallel selection during Coffea species divergence. Plant Mol. Biol. 2020, 103, 1–11. [Google Scholar] [CrossRef]
- Li, M.; Radvanyi, L.; Yin, B.; Rycaj, K.; Li, J.; Chivukula, R.; Lin, K.; Lu, Y.; Shen, J.; Chang, D.Z.; et al. Downregulation of Human Endogenous Retrovirus Type K (HERV-K) Viral env RNA in Pancreatic Cancer Cells Decreases Cell Proliferation and Tumor Growth. Clin. Cancer Res. 2017, 23. [Google Scholar] [CrossRef] [PubMed]
- Cegolon, L.; Salata, C.; Weiderpass, E.; Vineis, P.; Palù, G.; Mastrangelo, G. Human endogenous retroviruses and cancer prevention: Evidence and prospects. BMC Cancer 2013, 13, 4. [Google Scholar] [CrossRef] [PubMed]
- Desantis, C.E.; Ma, J.; Goding Sauer, A.; Newman, L.A.; Jemal, A. Breast Cancer Statistics, 2017, Racial Disparity in Mortality by State. CA Cancer J. Clin. 2017, 67, 439–448. [Google Scholar] [CrossRef] [PubMed]
- Chen, X. Understanding the Genetic Architecture of Schizophrenia in Chinese Population; University of Nevada Las Vegas: Las Vegas, NV, USA, 2016. [Google Scholar]
- Sherry, S.; Xiao, C.; Durbrow, K.; Kimelman, M.; Rodarmer, K.; Shumway, M.; Yaschenko, E. NCBI SRA Toolkit Technology for Next Generation Sequence Data. In Proceedings of the Plant and Animal Genome XX Conference, San Diego, CA, USA, 14–18 January 2012. [Google Scholar]
- Jurka, J.; Kapitonov, V.V.; Pavlicek, A.; Klonowski, P.; Kohany, O.; Walichiewicz, J. Repbase Update, a database of eukaryotic repetitive elements. Cytogenet. Genome Res. 2005, 110, 462–467. [Google Scholar] [CrossRef]
- Furlani, J.L.; Osel, P.W. Abstract Yourself With Modules. In Proceedings of the 10th USENIX Conference on System Administration, San Jose, CA, USA, 7–12 November1996; USENIX Association: Berkley, CA, USA, 1996; pp. 193–204. [Google Scholar]
- Leinonen, R.; Sugawara, H.; Shumway, M.; International Nucleotide Sequence Database Collaboration. The sequence read archive. Nucleic Acids Res. 2010, 39, D19–D21. [Google Scholar] [CrossRef]
- Lynch, V.J.; Leclerc, R.D.; May, G.; Wagner, G.P. Transposon-mediated rewiring of gene regulatory networks contributed to the evolution of pregnancy in mammals. Nat. Genet. 2011, 43, 1154–1159. [Google Scholar] [CrossRef]
- Chuong, E.B. Retroviruses facilitate the rapid evolution of the mammalian placenta. Bioessays 2013, 35, 853–861. [Google Scholar] [CrossRef] [PubMed]
- Hermann, D.; Egue, F.; Tastard, E.; Nguyen, D.-H.; Casse, N.; Caruso, A.; Hiard, S.; Marchand, J.; Chenais, B.; Morant-Manceau, A.; et al. An introduction to the vast world of transposable elements—what about the diatoms? DIATOM Res. 2014, 29, 91–104. [Google Scholar] [CrossRef]
- Pericay, C.; Díez, O.; Campos, B.; Balmaña, J.; Domènech, M.; Lerma, E.; Baena, M.; Sabaté, J.M.; Gómez, A.; López, J.J.; et al. Características clinicopatológicas y evolución clínica de pacientes con cáncer de mama y mutaciones en los genes BRCA1 o BRCA2. Med. Clin. 2001, 117, 161–166. [Google Scholar] [CrossRef]
- The 3000 Rice Genomes Project. The 3000 rice genomes project. Gigascience 2014, 3, 1–6. [Google Scholar] [CrossRef]
- McDowell, J.M.; Meyers, B.C. A transposable element is domesticated for service in the plant immune system. Proc. Natl. Acad. Sci. USA 2013, 110, 14821–14822. [Google Scholar] [CrossRef]





| Y/X | X = 0 | X = 1 |
|---|---|---|
| Y = 0 | O00 | O01 |
| Y = 1 | O10 | O11 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Orozco-Arias, S.; Tobon-Orozco, N.; Piña, J.S.; Jiménez-Varón, C.F.; Tabares-Soto, R.; Guyot, R. TIP_finder: An HPC Software to Detect Transposable Element Insertion Polymorphisms in Large Genomic Datasets. Biology 2020, 9, 281. https://doi.org/10.3390/biology9090281
Orozco-Arias S, Tobon-Orozco N, Piña JS, Jiménez-Varón CF, Tabares-Soto R, Guyot R. TIP_finder: An HPC Software to Detect Transposable Element Insertion Polymorphisms in Large Genomic Datasets. Biology. 2020; 9(9):281. https://doi.org/10.3390/biology9090281
Chicago/Turabian StyleOrozco-Arias, Simon, Nicolas Tobon-Orozco, Johan S. Piña, Cristian Felipe Jiménez-Varón, Reinel Tabares-Soto, and Romain Guyot. 2020. "TIP_finder: An HPC Software to Detect Transposable Element Insertion Polymorphisms in Large Genomic Datasets" Biology 9, no. 9: 281. https://doi.org/10.3390/biology9090281
APA StyleOrozco-Arias, S., Tobon-Orozco, N., Piña, J. S., Jiménez-Varón, C. F., Tabares-Soto, R., & Guyot, R. (2020). TIP_finder: An HPC Software to Detect Transposable Element Insertion Polymorphisms in Large Genomic Datasets. Biology, 9(9), 281. https://doi.org/10.3390/biology9090281






