Spatial Phylogenetics, Biogeographical Patterns and Conservation Implications of the Endemic Flora of Crete (Aegean, Greece) under Climate Change Scenarios
Abstract
1. Introduction
2. Materials and Methods
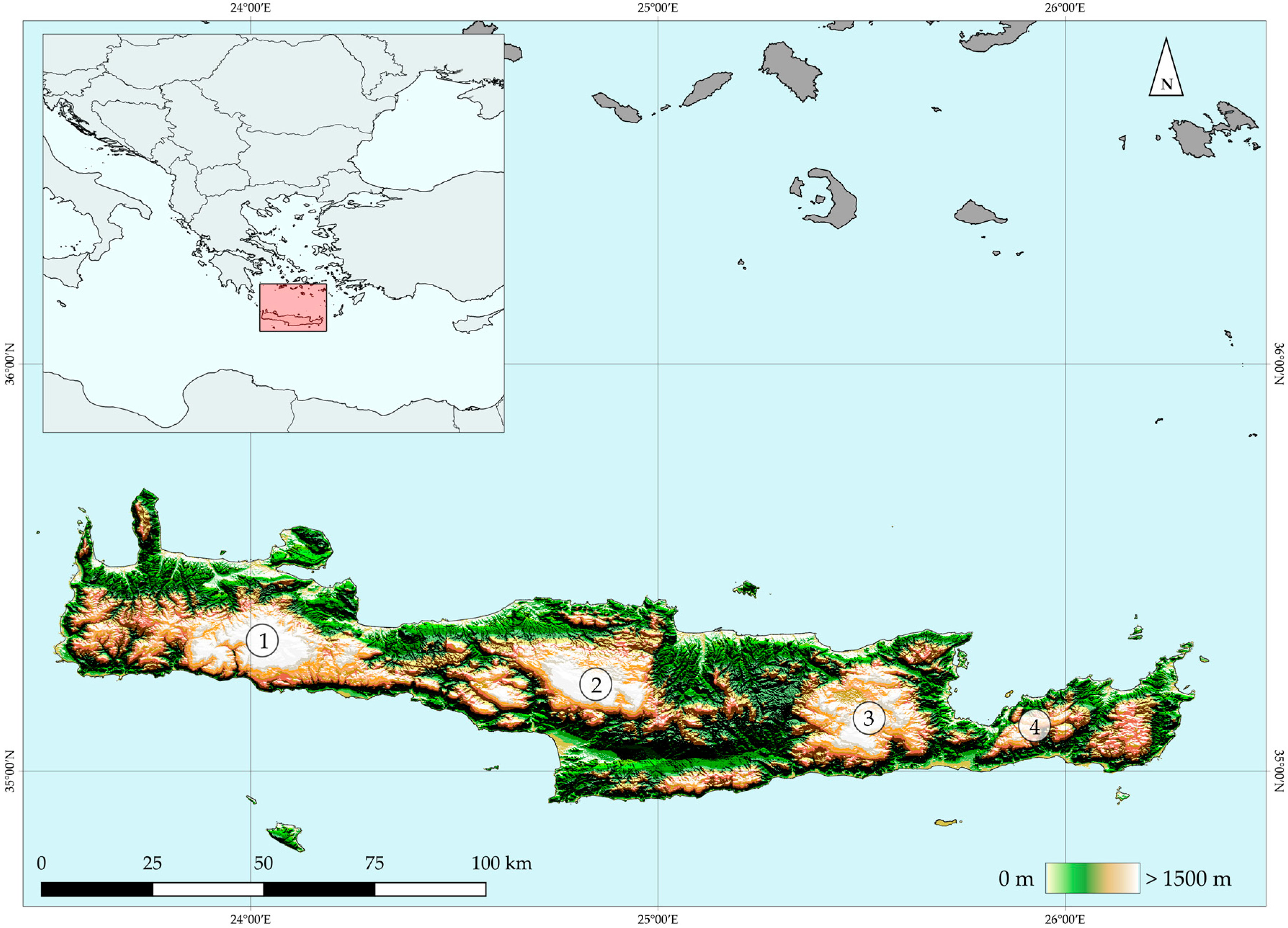
2.1. Study Area
2.2. Environmental Data
2.3. Species Occurrence Data
2.4. Biodiversity Analyses
2.5. Spatial Autoregressive Models
2.6. Future Diversity and Biogeographical Patterns
2.6.1. Changes in Species Richness (ΔSR)
2.6.2. Changes in Phylogenetic Diversity (ΔPD)
2.6.3. Changes in Ecological Generalism (ΔEG)
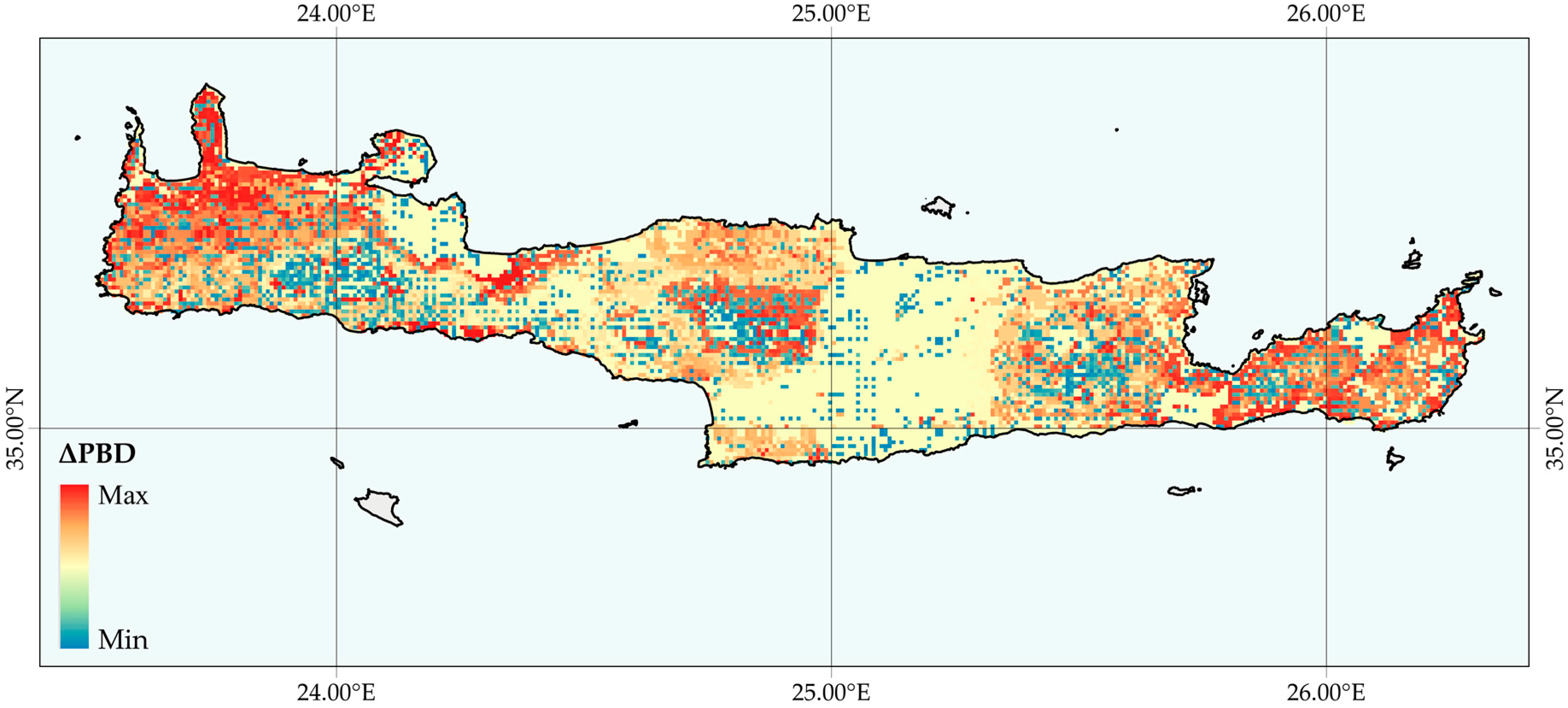
2.6.4. Changes in Phylogenetic Beta Diversity (ΔBD)
2.6.5. Changes in Biogeographical Patterns
3. Results
3.1. Biodiversity Indices
3.2. Future Diversity and Biogeographical Patterns
3.2.1. Changes in ΔEG, ΔPD, ΔSR and ΔBD
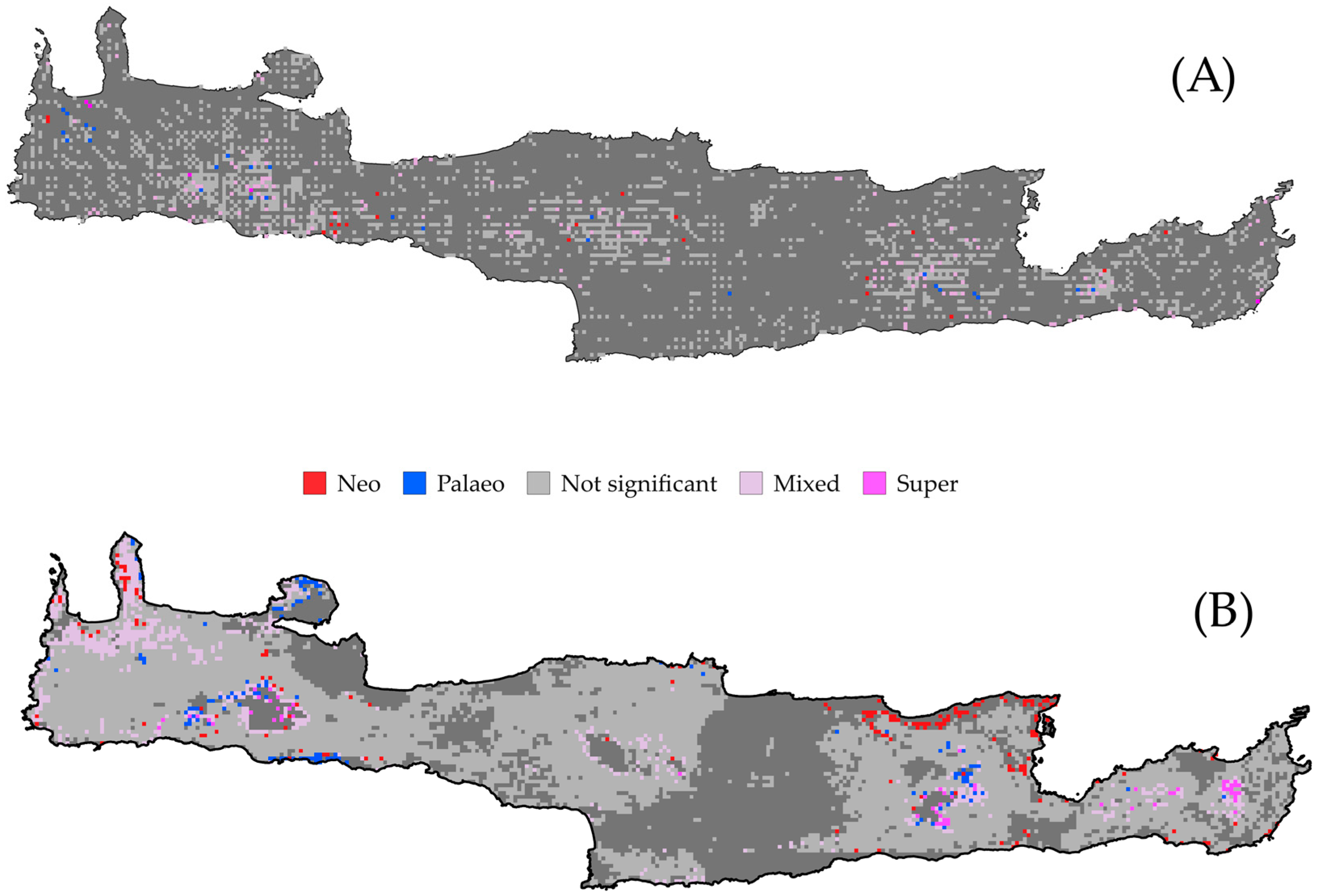
3.2.2. Changes in Biogeographical Patterns
3.3. Climatic Refugia and Protected Areas Network Overlap
4. Discussion
4.1. Centers of Endemism
4.2. Trends in Biogeographical Patterns
4.3. Conservation Prioritization
4.4. Conservation Considerations
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Le Roux, J.J.; Hui, C.; Castillo, M.L.; Iriondo, J.M.; Keet, J.H.; Khapugin, A.A.; Médail, F.; Rejmánek, M.; Theron, G.; Yannelli, F.A.; et al. Recent Anthropogenic Plant Extinctions Differ in Biodiversity Hotspots and Coldspots. Curr. Biol. 2019, 29, 2912–2918. [Google Scholar] [CrossRef] [PubMed]
- Gray, A. The ecology of plant extinction: Rates, traits and island comparisons. Oryx 2019, 53, 424–428. [Google Scholar] [CrossRef]
- Vellend, M.; Baeten, L.; Becker-Scarpitta, A.; Boucher-Lalonde, V.; McCune, J.L.; Messier, J.; Myers-Smith, I.H.; Sax, D.F. Plant Biodiversity Change Across Scales During the Anthropocene. Annu. Rev. Plant Biol. 2017, 68, 563–586. [Google Scholar] [CrossRef] [PubMed]
- Cronk, Q. Plant extinctions take time. Science 2016, 353, 446–447. [Google Scholar] [CrossRef]
- Wing, S.L. Mass Extinctions in Plant Evolution. In Extinctions in the History of Life; Cambridge University Press: Cambridge, UK, 2004. [Google Scholar]
- Enquist, B.J.; Feng, X.; Boyle, B.; Maitner, B.; Newman, E.A.; Jørgensen, P.M.; Roehrdanz, P.R.; Thiers, B.M.; Burger, J.R.; Corlett, R.T.; et al. The commonness of rarity: Global and future distribution of rarity across land plants. Sci. Adv. 2019, 5, eaaz0414. [Google Scholar] [CrossRef]
- Newbold, T. Future effects of climate and land-use change on terrestrial vertebrate community diversity under different scenarios. Proc. R. Soc. B Biol. Sci. 2018, 285. [Google Scholar] [CrossRef]
- Newbold, T.; Hudson, L.N.; Contu, S.; Hill, S.L.L.; Beck, J.; Liu, Y.; Meyer, C.; Phillips, H.R.P.; Scharlemann, J.P.W.; Purvis, A. Widespread winners and narrow-ranged losers: Land use homogenizes biodiversity in local assemblages worldwide. PLoS Biol. 2018, 16, e2006841. [Google Scholar] [CrossRef]
- Powers, R.P.; Jetz, W. Global habitat loss and extinction risk of terrestrial vertebrates under future land-use-change scenarios. Nat. Clim. Chang. 2019, 9, 323–329. [Google Scholar] [CrossRef]
- CBD (Convention on Biological Diversity) X/17. Consolidated Update of the Global Strategy for Plant Conservation 2011–2020; UNEP/CBD: Nagoya, Japan, 2010. [Google Scholar]
- Corlett, R.T. Safeguarding our future by protecting biodiversity. Plant Divers. 2020. [Google Scholar] [CrossRef]
- Busch, K.; Hermann, C.; Hinrichs, K.; Schulten, T. Euro Crisis, Austerity Policy and the European Social Model: How crisis policies in Southern Europe threaten the EU’s social dimension. Int. Policy Anal. Friedrich Ebert Found. Berl. 2013, 1–38. [Google Scholar]
- Arponen, A. Prioritizing species for conservation planning. Biodivers. Conserv. 2012, 21, 875–893. [Google Scholar] [CrossRef]
- Maes, J.; Teller, A.; Erhard, M.; Liquete, C.; Braat, L.; Berry, P.; Egoh, B.; Puydarrieus, P.; Fiorina, C.; Santos, F.; et al. Mapping and Assessment of Ecosystem and Their Services. An Analytical Framework for Ecosystem Assessments under Action 5 of the EU Biodiversity Strategy to 2020; European Union: Luxemburg, 2013. [Google Scholar]
- Kling, M.M.; Mishler, B.D.; Thornhill, A.H.; Baldwin, B.G.; Ackerly, D.D. Facets of phylodiversity: Evolutionary diversification, divergence and survival as conservation targets. Philos. Trans. R. Soc. B Biol. Sci. 2019, 374, 20170397. [Google Scholar] [CrossRef] [PubMed]
- Reece, J.S.; Noss, R.F. Prioritizing Species by Conservation Value and Vulnerability: A New Index Applied to Species Threatened by Sea-Level Rise and Other Risks in Florida. Nat. Areas J. 2014, 34, 31–45. [Google Scholar] [CrossRef]
- Keppel, G.; Ottaviani, G.; Harrison, S.; Wardell-Johnson, G.W.; Marcantonio, M.; Mucina, L. Towards an eco-evolutionary understanding of endemism hotspots and refugia. Ann. Bot. 2018, 122, 927–934. [Google Scholar] [CrossRef] [PubMed]
- Ashcroft, M.B. Identifying refugia from climate change. J. Biogeogr. 2010, 37, 1407–1413. [Google Scholar] [CrossRef]
- Hoffmann, S.; Irl, S.D.H.; Beierkuhnlein, C. Predicted climate shifts within terrestrial protected areas worldwide. Nat. Commun. 2019, 10, 1–10. [Google Scholar] [CrossRef]
- Le Saout, S.; Hoffmann, M.; Shi, Y.; Hughes, A.; Bernard, C.; Brooks, T.M.; Bertzky, B.; Butchart, S.H.M.; Stuart, S.N.; Badman, T.; et al. Protected Areas and Effective Biodiversity Conservation. Science 2013, 342, 803–805. [Google Scholar] [CrossRef]
- Crisp, M.D.; Laffan, S.; Linder, H.P.; Monro, A. Endemism in the Australian flora. J. Biogeogr. 2001, 28, 183–198. [Google Scholar] [CrossRef]
- Jetz, W.; Rahbek, C.; Colwell, R.K. The coincidence of rarity and richness and the potential signature of history in centres of endemism. Ecol. Lett. 2004, 7, 1180–1191. [Google Scholar] [CrossRef]
- Mishler, B.D.; Knerr, N.; González-Orozco, C.E.; Thornhill, A.H.; Laffan, S.W.; Miller, J.T. Phylogenetic measures of biodiversity and neo-and paleo-endemism in Australian acacia. Nat. Commun. 2014, 5, 4473. [Google Scholar] [CrossRef]
- Scherson, R.A.; Thornhill, A.H.; Urbina-Casanova, R.; Freyman, W.A.; Pliscoff, P.A.; Mishler, B.D. Spatial phylogenetics of the vascular flora of Chile. Mol. Phylogenetics Evol. 2017, 112, 88–95. [Google Scholar] [CrossRef] [PubMed]
- Laffan, S.W.; Rosauer, D.F.; Di Virgilio, G.; Miller, J.T.; González-Orozco, C.E.; Knerr, N.; Thornhill, A.H.; Mishler, B.D. Range-weighted metrics of species and phylogenetic turnover can better resolve biogeographic transition zones. Methods Ecol. Evol. 2016, 7, 580–588. [Google Scholar] [CrossRef]
- Thornhill, A.H.; Mishler, B.D.; Knerr, N.J.; González-Orozco, C.E.; Costion, C.M.; Crayn, D.M.; Laffan, S.W.; Miller, J.T. Continental-scale spatial phylogenetics of Australian angiosperms provides insights into ecology, evolution and conservation. J. Biogeogr. 2016, 43, 2085–2098. [Google Scholar] [CrossRef]
- Kier, G.; Kreft, H.; Lee, T.M.; Jetz, W.; Ibisch, P.L.; Nowicki, C.; Mutke, J.; Barthlott, W. A global assessment of endemism and species richness across island and mainland regions. Proc. Natl. Acad. Sci. USA 2009, 106, 9322–9327. [Google Scholar] [CrossRef] [PubMed]
- Gallagher, R.V.; Allen, S.; Rivers, M.C.; Allen, A.P.; Butt, N.; Keith, D.; Auld, T.D.; Enquist, B.J.; Wright, I.J.; Possingham, H.P.; et al. Global shortfalls in extinction risk assessments for endemic flora. bioRxiv 2020. [Google Scholar] [CrossRef]
- Barthlott, W.; Mutke, J.; Rafiqpoor, D.; Kier, G.; Kreft, H. Global Centers of Vascular Plant Diversity. Nova Acta Leopold. 2005, 92, 61–83. [Google Scholar]
- Myers, N.; Mittermeier, R.A.; Mittermeier, C.G.; Da Fonseca, G.A.B.; Kent, J. Biodiversity hotspots for conservation priorities. Nature 2000, 403, 853–858. [Google Scholar] [CrossRef]
- Médail, F.; Myers, N. Mediterranean Basin (in Mittermeier, C G Lamoreux, J Fonseca, G A B). Sierra 2004. [Google Scholar]
- CEP Fund. Mediterranean Basin Biodiversity Hotspot. 2017. Available online: https://www.cepf.net/sites/default/files/mediterranean-basin-2017-ecosystem-profile-english_0.pdf (accessed on 20 October 2019).
- Médail, F. The specific vulnerability of plant biodiversity and vegetation on Mediterranean islands in the face of global change. Reg. Environ. Chang. 2017, 17, 1775–1790. [Google Scholar] [CrossRef]
- Pacifici, M.; Foden, W.B.; Visconti, P.; Watson, J.E.M.; Butchart, S.H.M.; Kovacs, K.M.; Scheffers, B.R.; Hole, D.G.; Martin, T.G.; Akçakaya, H.R.; et al. Assessing species vulnerability to climate change. Nat. Clim. Chang. 2015, 5, 215–224. [Google Scholar] [CrossRef]
- Gentili, R.; Bacchetta, G.; Fenu, G.; Cogoni, D.; Abeli, T.; Rossi, G.; Salvatore, M.C.; Baroni, C.; Citterio, S. From cold to warm-stage refugia for boreo-alpine plants in southern European and Mediterranean mountains: The last chance to survive or an opportunity for speciation? Biodiversity 2015, 16, 247–261. [Google Scholar] [CrossRef]
- Domina, G.; Bazan, G.; Campisi, P.; Greuter, W. Taxonomy and conservation in Higher Plants and Bryophytes in the Mediterranean Area. Biodivers. J. 2015, 6, 197–204. [Google Scholar]
- Giorgi, F.; Lionello, P. Climate change projections for the Mediterranean region. Glob. Planet. Chang. 2008, 63, 90–104. [Google Scholar] [CrossRef]
- Fois, M.; Podda, L.; Médail, F.; Bacchetta, G. Endemic and alien vascular plant diversity in the small Mediterranean islands of Sardinia: Drivers and implications for their conservation. Biol. Conserv. 2020, 244, 108519. [Google Scholar] [CrossRef]
- Pressey, R.L.; Mills, M.; Weeks, R.; Day, J.C. The plan of the day: Managing the dynamic transition from regional conservation designs to local conservation actions. Biol. Conserv. 2013, 166, 155–169. [Google Scholar] [CrossRef]
- Beck, J.; Ballesteros-Mejia, L.; Nagel, P.; Kitching, I.J. Online solutions and the ‘Wallacean shortfall’: What does GBIF contribute to our knowledge of species’ ranges? Divers. Distrib. 2013, 19, 1043–1050. [Google Scholar] [CrossRef]
- Diniz-Filho, J.A.F.; Loyola, R.D.; Raia, P.; Mooers, A.O.; Bini, L.M. Darwinian shortfalls in biodiversity conservation. Trends Ecol. Evol. 2013, 28, 689–695. [Google Scholar] [CrossRef]
- Wilson, E.O. Biodiversity research requires more boots on the ground: Comment. Nat. Ecol. Evol. 2017, 1, 1590–1591. [Google Scholar] [CrossRef]
- Dimopoulos, P.; Raus, T.; Bergmeier, E.; Constantinidis, T.; Iatrou, G.; Kokkini, S.; Strid, A.; Tzanoudakis, D. Vascular plants of Greece: An annotated checklist. Englera 2013, 1–372. [Google Scholar] [CrossRef]
- Strid, A. Atlas of the Aegean Flora; Botanic Garden and Botanical Museum Berlin, Freie Universität Berlin: Berlin, Germany, 2016; ISBN 9783921800973. [Google Scholar]
- Kougioumoutzis, K.; Kokkoris, I.P.; Panitsa, M.; Trigas, P.; Strid, A.; Dimopoulos, P. Plant Diversity Patterns and Conservation Implications under Climate-Change Scenarios in the Mediterranean: The Case of Crete (Aegean, Greece). Diversity 2020, 12, 270. [Google Scholar] [CrossRef]
- Iliadou, E.; Bazos, I.; Kougioumoutzis, K.; Karadimou, E.; Kokkoris, I.; Panitsa, M.; Raus, T.; Strid, A.; Dimopoulos, P. Taxonomic and phylogenetic diversity patterns in the Northern Sporades islets complex (West Aegean, Greece). Plant Syst. Evol. 2020, 306, 1–17. [Google Scholar] [CrossRef]
- Mastrogianni, A.; Kallimanis, A.S.; Chytrý, M.; Tsiripidis, I. Phylogenetic diversity patterns in forests of a putative refugial area in Greece: A community level analysis. For. Ecol. Manag. 2019, 446, 226–237. [Google Scholar] [CrossRef]
- Fassou, G.; Kougioumoutzis, K.; Iatrou, G.; Trigas, P.; Papasotiropoulos, V. Genetic Diversity and Range Dynamics of Helleborus odorus subsp. cyclophyllus under Different Climate Change Scenarios. Forests 2020, 11, 620. [Google Scholar] [CrossRef]
- Stathi, E.; Kougioumoutzis, K.; Abraham, E.M.; Trigas, P.; Ganopoulos, I.; Avramidou, E.V.; Tani, E. Population genetic variability and distribution of the endangered Greek endemic Cicer graecum under climate change scenarios. AoB Plants 2020, 12, plaa007. [Google Scholar] [CrossRef] [PubMed]
- Tsiftsis, S.; Djordjević, V. Modelling sexually deceptive orchid species distributions under future climates: The importance of plant–pollinator interactions. Sci. Rep. 2020, 10, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Kougioumoutzis, K.; Simaiakis, S.M.; Tiniakou, A. Network biogeographical analysis of the central Aegean archipelago. J. Biogeogr. 2014, 41, 1848–1858. [Google Scholar] [CrossRef]
- Kougioumoutzis, K.; Valli, A.T.; Georgopoulou, E.; Simaiakis, S.M.; Triantis, K.A.; Trigas, P. Network biogeography of a complex island system: The Aegean Archipelago revisited. J. Biogeogr. 2017, 44, 651–660. [Google Scholar] [CrossRef]
- Simaiakis, S.M.; Rijsdijk, K.F.; Koene, E.F.M.; Norder, S.J.; Van Boxel, J.H.; Stocchi, P.; Hammoud, C.; Kougioumoutzis, K.; Georgopoulou, E.; Van Loon, E.; et al. Geographic changes in the Aegean Sea since the Last Glacial Maximum: Postulating biogeographic effects of sea-level rise on islands. Palaeogeogr. Palaeoclim. Palaeoecol. 2017, 471, 108–119. [Google Scholar] [CrossRef]
- Kagiampaki, A.; Triantis, K.; Vardinoyannis, K.; Mylonas, M. Factors affecting plant species richness and endemism in the South Aegean (Greece). J. Biol. Res. 2011, 16, 282–295. [Google Scholar]
- Trigas, P.; Iatrou, G.; Panitsa, M. Vascular plant species diversity, biogeography and vulnerability in the Aegean islands as exemplified by Evvia island (W Aegean, Greece). Fresenius Environ. Bull. 2008, 17, 48–57. [Google Scholar]
- Kallimanis, A.S.; Bergmeier, E.; Panitsa, M.; Georghiou, K.; Delipetrou, P.; Dimopoulos, P. Biogeographical determinants for total and endemic species richness in a continental archipelago. Biodivers. Conserv. 2010, 19, 1225–1235. [Google Scholar] [CrossRef]
- Panitsa, M.; Trigas, P.; Iatrou, G.; Sfenthourakis, S. Factors affecting plant species richness and endemism on land-bridge islands—An example from the East Aegean archipelago. Acta Oecologica 2010, 36, 431–437. [Google Scholar] [CrossRef]
- Strid, A. Phytogeographia Aegaea and the flora Hellenica database. Ann. Naturhistorischen Mus. Wien. Ser. B Bot. Zool. 1996, 279–289. [Google Scholar]
- Legakis, A.; Kypriotakis, Z. A Biogeographical Analysis of the Island of Crete, Greece. J. Biogeogr. 1994, 21, 441. [Google Scholar] [CrossRef]
- Panitsa, M.; Kagiampaki, A.; Kougioumoutzis, K. Plant diversity and biogeography of the Aegean Archipelago: A New Synthesis. In Biogeography and Biodiversity of the Aegean. In Honour of Prof. Moysis Mylonas; Sfenthourakis, S., Pafilis, P., Parmakelis, A., Poulakakis, N., Triantis, K., Eds.; Broken Hill Publishers Ltd: Nicosia, Cyprus, 2018; pp. 223–244. ISBN 9789925563784. [Google Scholar]
- Sfenthourakis, S.; Triantis, K.A. The Aegean archipelago: A natural laboratory of evolution, ecology and civilisations. J. Biol. Res. 2017, 24, 4. [Google Scholar] [CrossRef]
- Rundel, P.W.; Arroyo, M.T.K.; Cowling, R.M.; Keeley, J.E.; Lamont, B.B.; Vargas, P. Mediterranean Biomes: Evolution of Their Vegetation, Floras, and Climate. Annu. Rev. Ecol. Evol. Syst. 2016, 47, 383–407. [Google Scholar] [CrossRef]
- Dimopoulos, P.; Raus, T.; Bergmeier, E.; Constantinidis, T.; Iatrou, G.; Kokkini, S.; Strid, A.; Tzanoudakis, D. Vascular plants of Greece: An annotated checklist. Supplement. Willdenowia 2016, 46, 301–348. [Google Scholar] [CrossRef]
- Higgins, M. Greek islands, geology. In Encyclopedia of Islands; Gillespie, R.G., Clague, D.A., Eds.; University of California Press: Berkeley, CA, USA, 2009; pp. 392–396. ISBN 9780520256491. [Google Scholar]
- Sakellariou, D.; Galanidou, N. Pleistocene submerged landscapes and Palaeolithic archaeology in the tectonically active Aegean region. Geol. Soc. Lond. Spec. Publ. 2016, 411, 145–178. [Google Scholar] [CrossRef]
- Fassoulas, C. The geodynamic and paleogeographic evolution of the Aegean in the Tertiary and Quaternary: A review. In Biogeography and Biodiversity of the Aegean. In Honour of Prof. Moysis Mylonas; Sfenthourakis, S., Pafilis, P., Parmakelis, A., Poulakakis, N., Triantis, K.A., Eds.; Broken Hill Publishers Ltd: Nicosia, Cyprus, 2018; pp. 25–46. ISBN 9789925563784. [Google Scholar]
- Manzi, V.; Gennari, R.; Hilgen, F.; Krijgsman, W.; Lugli, S.; Roveri, M.; Sierro, F.J. Age refinement of the Messinian salinity crisis onset in the Mediterranean. Terra Nova 2013, 25, 315–322. [Google Scholar] [CrossRef]
- Hijmans, R.J.; Cameron, S.E.; Parra, J.L.; Jones, P.G.; Jarvis, A. Very high resolution interpolated climate surfaces for global land areas. Int. J. Clim. 2005, 25, 1965–1978. [Google Scholar] [CrossRef]
- Title, P.O.; Bemmels, J.B. ENVIREM: An expanded set of bioclimatic and topographic variables increases flexibility and improves performance of ecological niche modeling. Ecography 2017, 49, 291–307. [Google Scholar] [CrossRef]
- McSweeney, C.F.; Jones, R.G.; Lee, R.W.; Rowell, D.P. Selecting CMIP5 GCMs for downscaling over multiple regions. Clim. Dyn. 2015, 44, 3237–3260. [Google Scholar] [CrossRef]
- Hengl, T.; de Jesus, J.M.; Heuvelink, G.B.M.; Gonzalez, M.R.; Kilibarda, M.; Blagotić, A.; Shangguan, W.; Wright, M.N.; Geng, X.; Bauer-Marschallinger, B.; et al. SoilGrids250m: Global gridded soil information based on machine learning. PLoS ONE 2017, 12, e0169748. [Google Scholar] [CrossRef] [PubMed]
- Jarvis, A.; Reuter, H.I.; Nelson, A.; Guevara, E. Hole-Filled SRTM for the Globe Version 4. Available online: http//srtm.csi.cgiar.org (accessed on 20 October 2019).
- Hijmans, R.J. Package ‘raster’—Geographic Data Analysis and Modeling. CRAN Repos. 2019, 517, 2–12. [Google Scholar]
- Li, D.; Trotta, L.; Marx, H.E.; Allen, J.M.; Sun, M.; Soltis, D.E.; Soltis, P.S.; Guralnick, R.P.; Baiser, B. For common community phylogenetic analyses, go ahead and use synthesis phylogenies. Ecology 2019. [Google Scholar] [CrossRef]
- Swenson, N.G. Phylogenetic Resolution and Quantifying the Phylogenetic Diversity and Dispersion of Communities. PLoS ONE 2009, 4, e4390. [Google Scholar] [CrossRef]
- Faith, D.P. Conservation evaluation and phylogenetic diversity. Biol. Conserv. 1992, 61, 1–10. [Google Scholar] [CrossRef]
- Laffan, S.W.; Lubarsky, E.; Rosauer, D.F. Biodiverse, a tool for the spatial analysis of biological and related diversity. Ecography. 2010, 33, 643–647. [Google Scholar] [CrossRef]
- Rosauer, D.; Laffan, S.W.; Crisp, M.D.; Donnellan, S.C.; Cook, L.G. Phylogenetic endemism: A new approach for identifying geographical concentrations of evolutionary history. Mol. Ecol. 2009, 18, 4061–4072. [Google Scholar] [CrossRef]
- Allen, J.M.; Germain-Aubrey, C.C.; Barve, N.; Neubig, K.M.; Majure, L.C.; Laffan, S.W.; Mishler, B.D.; Owens, H.L.; Smith, S.A.; Whitten, W.M.; et al. Spatial Phylogenetics of Florida Vascular Plants: The Effects of Calibration and Uncertainty on Diversity Estimates. iScience 2019, 11, 57–70. [Google Scholar] [CrossRef]
- Kissling, W.D.; Carl, G. Spatial autocorrelation and the selection of simultaneous autoregressive models. Glob. Ecol. Biogeogr. 2008, 17, 59–71. [Google Scholar] [CrossRef]
- Naimi, B.; Hamm, N.A.S.; Groen, T.A.; Skidmore, A.K.; Toxopeus, A.G. Where is positional uncertainty a problem for species distribution modelling? Ecography 2014, 37, 191–203. [Google Scholar] [CrossRef]
- Ver Hoef, J.M.; Peterson, E.E.; Hooten, M.B.; Hanks, E.M.; Fortin, M.J. Spatial autoregressive models for statistical inference from ecological data. Ecol. Monogr. 2018, 88, 36–59. [Google Scholar] [CrossRef]
- Bivand, R.S.; Wong, D.W.S. Comparing implementations of global and local indicators of spatial association. TEST 2018, 27, 716–748. [Google Scholar] [CrossRef]
- McKnight, M.W.; White, P.S.; McDonald, R.I.; Lamoreux, J.F.; Sechrest, W.; Ridgely, R.S.; Stuart, S.N. Putting beta-diversity on the map: Broad-scale congruence and coincidence in the extremes. PLoS Biol. 2007, 5, e272. [Google Scholar] [CrossRef] [PubMed]
- Levins, R. Evolution in Changing Environments: Some Theoretical Explorations; Princeton University Press: Princeton, NJ, USA, 1968; ISBN 0691080623. [Google Scholar]
- Brown, J.H. On the relationship beween abundance and distribution of species. Am. Nat. 1984, 124, 255–279. [Google Scholar] [CrossRef]
- Menéndez-Guerrero, P.A.; Green, D.M.; Davies, T.J. Climate change and the future restructuring of Neotropical anuran biodiversity. Ecography 2020, 43, 222–235. [Google Scholar] [CrossRef]
- Baselga, A. The relationship between species replacement, dissimilarity derived from nestedness, and nestedness. Glob. Ecol. Biogeogr. 2012, 21, 1223–1232. [Google Scholar] [CrossRef]
- Baselga, A. Partitioning the turnover and nestedness components of beta diversity. Glob. Ecol. Biogeogr. 2010, 19, 134–143. [Google Scholar] [CrossRef]
- Legendre, P. Interpreting the replacement and richness difference components of beta diversity. Glob. Ecol. Biogeogr. 2014, 23, 1324–1334. [Google Scholar] [CrossRef]
- Baselga, A.; Orme, C.D.L. betapart: An R package for the study of beta diversity. Methods Ecol. Evol. 2012, 3, 808–812. [Google Scholar] [CrossRef]
- Baselga, A.; Gómez-Rodríguez, C.; Lobo, J.M. Historical Legacies in World Amphibian Diversity Revealed by the Turnover and Nestedness Components of Beta Diversity. PLoS ONE 2012, 7, e32341. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Su, G.; Xiong, Y.; Akasaka, M.; Molinos, J.G.; Matsuzaki, S.S.; Zhang, M. Complimentary analysis of metacommunity nestedness and diversity partitioning highlights the need for a holistic conservation strategy for highland lake fish assemblages. Glob. Ecol. Conserv. 2015, 3, 288–296. [Google Scholar] [CrossRef]
- Wood, S.N. Generalized Additive Models: An Introduction with R, 2nd ed.; CRC Press: Boca Raton, FL, USA, 2017; ISBN 9781498728348. [Google Scholar]
- Burnham, K.P.; Anderson, D.R. Model Selection and Multimodel Inference: A Practical Information-Theoretic Approach; Springer Science & Business Media: Berlin, Germany, 2003. [Google Scholar]
- Fitzpatrick, M.C.; Sanders, N.J.; Ferrier, S.; Longino, J.T.; Weiser, M.D.; Dunn, R. Forecasting the future of biodiversity: A test of single- and multi-species models for ants in North America. Ecography 2011, 34, 836–847. [Google Scholar] [CrossRef]
- Rousseeuw, P.J. Silhouettes: A graphical aid to the interpretation and validation of cluster analysis. J. Comput. Appl. Math. 1987, 20, 53–65. [Google Scholar] [CrossRef]
- Nowosad, J.; Stepinski, T.F. Spatial association between regionalizations using the information-theoretical V -measure. Int. J. Geogr. Inf. Sci. 2018, 32, 2386–2401. [Google Scholar] [CrossRef]
- Rosenberg, A.; Hirschberg, J. V-Measure: A Conditional Entropy-Based External Cluster Evaluation Measure. In Proceedings of the 2007 Joint Conference on Empirical Methods in Natural Language Processing and Computational Natural Language Learning (EMNLP-CoNLL), Prague, Czech Republic, 28–30 June 2007. [Google Scholar]
- Maechler, M.; Rousseeuw, P.; Struyf, A.; Hubert, M.; Hornik, K. Cluster, R package version 2.1.0; 2019. Available online: https://svn.r-project.org/R-packages/trunk/cluster (accessed on 20 October 2019).
- Desgraupes, B. clusterCrit, R package version 1.2.8; 2018. Available online: https://cran.r-project.org/web/packages/clusterCrit/index.html (accessed on 20 October 2019).
- Parmakelis, A.; Pfenninger, M.; Spanos, L.; Papagiannakis, G.; Louis, C.; Mylonas, M. Inference of a radiation in Mastus (Gastropoda, Pulmonata, Enidae) on the island of Crete. Evolution 2005, 59, 991. [Google Scholar] [CrossRef]
- Harrison, S.; Noss, R. Endemism hotspots are linked to stable climatic refugia. Ann. Bot. 2017, 119, 207–214. [Google Scholar] [CrossRef] [PubMed]
- Lazarina, M.; Kallimanis, A.S.; Dimopoulos, P.; Psaralexi, M.; Michailidou, D.E.; Sgardelis, S.P. Patterns and drivers of species richness and turnover of neo-endemic and palaeo-endemic vascular plants in a Mediterranean hotspot: The case of Crete, Greece. J. Biol. Res. 2019, 26, 12–13. [Google Scholar] [CrossRef]
- Trigas, P.; Panitsa, M.; Tsiftsis, S. Elevational Gradient of Vascular Plant Species Richness and Endemism in Crete—The Effect of Post-Isolation Mountain Uplift on a Continental Island System. PLoS ONE 2013, 8, e59425. [Google Scholar] [CrossRef]
- Spalink, D.; Kriebel, R.; Li, P.; Pace, M.C.; Drew, B.T.; Zaborsky, J.G.; Rose, J.; Drummond, C.P.; Feist, M.A.; Alverson, W.S.; et al. Spatial phylogenetics reveals evolutionary constraints on the assembly of a large regional flora. Am. J. Bot. 2018, 105, 1938–1950. [Google Scholar] [CrossRef] [PubMed]
- Molina-Venegas, R.; Aparicio, A.; Lavergne, S.; Arroyo, J. Climatic and topographical correlates of plant palaeo- and neoendemism in a Mediterranean biodiversity hotspot. Ann. Bot. 2017, 119, 229–238. [Google Scholar] [CrossRef] [PubMed]
- Dagallier, L.P.M.J.; Janssens, S.B.; Dauby, G.; Blach-Overgaard, A.; Mackinder, B.A.; Droissart, V.; Svenning, J.C.; Sosef, M.S.M.; Stévart, T.; Harris, D.J.; et al. Cradles and museums of generic plant diversity across tropical Africa. New Phytol. 2020, 225, 2196–2213. [Google Scholar] [CrossRef] [PubMed]
- Kokkoris, I.P.; Drakou, E.G.; Maes, J.; Dimopoulos, P. Ecosystem services supply in protected mountains of Greece: Setting the baseline for conservation management. Int. J. Biodivers. Sci. Ecosyst. Serv. Manag. 2018, 14, 45–59. [Google Scholar] [CrossRef]
- Chown, S.L.; Gaston, K.J. Areas cradles and museums: The latitudinal gradient in species richness. Trends Ecol. Evol. 2000, 15, 311–315. [Google Scholar] [CrossRef]
- Meulenkamp, J.E. Aspects of the Late Cenozoic Evolution of the Aegean Region. In Geological Evolution of the Mediterranean Basin; Springer: New York, NY, USA, 1985. [Google Scholar]
- Cellinese, N.; Smith, S.A.; Edwards, E.J.; Kim, S.-T.; Haberle, R.C.; Avramakis, M.; Donoghue, M.J. Historical biogeography of the endemic Campanulaceae of Crete. J. Biogeogr. 2009, 36, 1253–1269. [Google Scholar] [CrossRef]
- Greuter, W. The relict element of the flora of Crete and its evolutionary significance. In Taxonomy, Phytogeography and Evolution; Valentine, D.H., Ed.; Academic Press: London, UK, 1972; pp. 161–177. ISBN 0127102507. [Google Scholar]
- Edh, K.; Widén, B.; Ceplitis, A. Nuclear and chloroplast microsatellites reveal extreme population differentiation and limited gene flow in the Aegean endemic Brassica cretica (Brassicaceae). Mol. Ecol. 2007, 16, 4972–4983. [Google Scholar] [CrossRef]
- Trichas, A.; Smirli, M.; Papadopoulou, A.; Anastasiou, I.; Keskin, B.; Poulakakis, N. Dispersal versus vicariance in the Aegean: Combining molecular and morphological phylogenies of eastern Mediterranean Dendarus (Coleoptera: Tenebrionidae) sheds new light on the phylogeography of the Aegean area. Zool. J. Linn. Soc. 2020, 1–20. [Google Scholar] [CrossRef]
- Heenan, P.B.; Millar, T.R.; Smissen, R.D.; McGlone, M.S.; Wilton, A.D. Phylogenetic measures of neo- and palaeo-endemism in the indigenous vascular flora of the New Zealand archipelago. Aust. Syst. Bot. 2017, 30, 124–133. [Google Scholar] [CrossRef]
- Sosa, V.; De-Nova, J.A.; Vásquez-Cruz, M. Evolutionary history of the flora of Mexico: Dry forests cradles and museums of endemism. J. Syst. Evol. 2018, 56, 523–536. [Google Scholar] [CrossRef]
- Wang, S.W.; Boru, B.H.; Njogu, A.W.; Ochola, A.C.; Hu, G.W.; Zhou, Y.D.; Wang, Q.F. Floristic composition and endemism pattern of vascular plants in Ethiopia and Eritrea. J. Syst. Evol. 2020, 58, 33–42. [Google Scholar] [CrossRef]
- Urban, M.C. Escalator to extinction. Proc. Natl. Acad. Sci. USA 2018, 115, 11871–11873. [Google Scholar] [CrossRef] [PubMed]
- González-Orozco, C.E.; Pollock, L.J.; Thornhill, A.H.; Mishler, B.D.; Knerr, N.; Laffan, S.W.; Miller, J.T.; Rosauer, D.F.; Faith, D.P.; Nipperess, D.A.; et al. Phylogenetic approaches reveal biodiversity threats under climate change. Nat. Clim. Chang. 2016, 6, 1110–1114. [Google Scholar] [CrossRef]
- Azevedo, J.A.R.; Guedes, T.B.; de Nogueira, C.; Passos, P.; Sawaya, R.J.; Prudente, A.L.C.; Barbo, F.E.; Strüssmann, C.; Franco, F.L.; Arzamendia, V.; et al. Museums and cradles of diversity are geographically coincident for narrowly distributed Neotropical snakes. Ecography 2019. [Google Scholar] [CrossRef]
- Cabral, J.S.; Weigelt, P.; Kissling, W.D.; Kreft, H. Biogeographic, climatic and spatial drivers differentially affect α-, β-and γ-diversities on oceanic archipelagos. Proc. R. Soc. Lond. B Biol. Sci. 2014, 281, 20133246. [Google Scholar] [CrossRef] [PubMed]
- Kallimanis, A.S.; Panitsa, M.; Bergmeier, E.; Dimopoulos, P. Examining the relationship between total species richness and single island palaeo- and neo-endemics. Acta Oecol. 2011, 37, 65–70. [Google Scholar] [CrossRef]
- Wiens, J.J.; Graham, C.H. Niche Conservatism: Integrating Evolution, Ecology, and Conservation Biology. Annu. Rev. Ecol. Evol. Syst. 2005, 36, 519–539. [Google Scholar] [CrossRef]
- Rangel, T.F.; Edwards, N.R.; Holden, P.B.; Diniz-Filho, J.A.F.; Gosling, W.D.; Coelho, M.T.P.; Cassemiro, F.A.S.; Rahbek, C.; Colwell, R.K. Modeling the ecology and evolution of biodiversity: Biogeographical cradles, museums, and graves. Science 2018, 361. [Google Scholar] [CrossRef]
- Carta, A.; Pierini, B.; Roma-Marzio, F.; Bedini, G.; Peruzzi, L. Phylogenetic measures of biodiversity uncover pteridophyte centres of diversity and hotspots in Tuscany. Plant Biosyst. 2018, 152, 831–839. [Google Scholar] [CrossRef]
- Lykousis, V. Sea-level changes and shelf break prograding sequences during the last 400 ka in the Aegean margins: Subsidence rates and palaeogeographic implications. Cont. Shelf Res. 2009, 29, 2037–2044. [Google Scholar] [CrossRef]
- Kundrata, R.; Baalbergen, E.; Bocak, L.; Schilthuizen, M. The origin and diversity of Drilus Olivier, 1790 (Elateridae: Agrypninae: Drilini) in Crete based on mitochondrial phylogeny. Syst. Biodivers. 2015, 13, 52–75. [Google Scholar] [CrossRef]
- Schule, W. Mammals, Vegetation and the Initial Human Settlement of the Mediterranean Islands: A Palaeoecological Approach. J. Biogeogr. 1993, 20, 399. [Google Scholar] [CrossRef]
- Thompson, J.D. Plant Evolution in the Mediterranean; Oxford University Press: Oxford, UK, 2005; ISBN 0198515332. [Google Scholar]
- Harrison, S.; Spasojevic, M.J.; Li, D. Climate and plant community diversity in space and time. Proc. Natl. Acad. Sci. USA 2020, 117, 4464–4470. [Google Scholar] [CrossRef] [PubMed]
- Olden, J.D.; Poff, N.L.R.; Douglas, M.R.; Douglas, M.E.; Fausch, K.D. Ecological and evolutionary consequences of biotic homogenization. Trends Ecol. Evol. 2004, 19, 18–24. [Google Scholar] [CrossRef] [PubMed]
- Cardinale, B.J.; Duffy, J.E.; Gonzalez, A.; Hooper, D.U.; Perrings, C.; Venail, P.; Narwani, A.; MacE, G.M.; Tilman, D.; Wardle, D.A.; et al. Biodiversity loss and its impact on humanity. Nature 2012, 486, 59–67. [Google Scholar] [CrossRef] [PubMed]
- Cantalapiedra, J.L.; Aze, T.; Cadotte, M.W.; Riva, G.V.D.; Huang, D.; Mazel, F.; Pennell, M.W.; Ríos, M.; Mooers, A. Conserving evolutionary history does not result in greater diversity over geological time scales. Proc. R. Soc. B Biol. Sci. 2019, 286, 20182896. [Google Scholar] [CrossRef]
- Bottrill, M.C.; Joseph, L.N.; Carwardine, J.; Bode, M.; Cook, C.; Game, E.T.; Grantham, H.; Kark, S.; Linke, S.; McDonald-Madden, E.; et al. Is conservation triage just smart decision making? Trends Ecol. Evol. 2008, 23, 649–654. [Google Scholar] [CrossRef]
- Allan, J.R.; Possingham, H.P.; Atkinson, S.C.; Waldron, A.; Marco, M.D.; Adams, V.M.; Butchart, S.H.M.; Venter, O.; Maron, M.; Williams, B.A.; et al. Conservation attention necessary across at least 44% of Earth’s terrestrial area to safeguard biodiversity. bioRxiv 2019, 839977. [Google Scholar] [CrossRef]
- Müller, A.; Schneider, U.A.; Jantke, K. Evaluating and expanding the European Union’s protected-area network toward potential post-2020 coverage targets. Conserv. Biol. 2020, 34, 654–665. [Google Scholar] [CrossRef]
- Pollock, L.J.; Thuiller, W.; Jetz, W. Large conservation gains possible for global biodiversity facets. Nature 2017, 546, 141–144. [Google Scholar] [CrossRef]
- Heikkinen, R.K.; Leikola, N.; Aalto, J.; Aapala, K.; Kuusela, S.; Luoto, M.; Virkkala, R. Fine-grained climate velocities reveal vulnerability of protected areas to climate change. Sci. Rep. 2020, 10, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Humphreys, A.M.; Govaerts, R.; Ficinski, S.Z.; Nic Lughadha, E.; Vorontsova, M.S. Global dataset shows geography and life form predict modern plant extinction and rediscovery. Nat. Ecol. Evol. 2019, 3, 1043–1047. [Google Scholar] [CrossRef] [PubMed]
- Tucker, C.M.; Cadotte, M.W. Unifying measures of biodiversity: Understanding when richness and phylogenetic diversity should be congruent. Divers. Distrib. 2013, 19, 845–854. [Google Scholar] [CrossRef]
- Dimitrakopoulos, P.G.; Memtsas, D.; Troumbis, A.Y. Questioning the effectiveness of the Natura 2000 Special Areas of Conservation strategy: The case of Crete. Glob. Ecol. Biogeogr. 2004, 13, 199–207. [Google Scholar] [CrossRef]
- Trigas, P.; Tsiftsis, S.; Tsiripidis, I.; Iatrou, G. Distribution Patterns and Conservation Perspectives of the Endemic Flora of Peloponnese (Greece). Folia Geobot. 2012, 47, 421–439. [Google Scholar] [CrossRef]
- Feng, G.; Ma, Z.; Sandel, B.; Mao, L.; Normand, S.; Ordonez, A.; Svenning, J.C. Species and phylogenetic endemism in angiosperm trees across the Northern Hemisphere are jointly shaped by modern climate and glacial–interglacial climate change. Glob. Ecol. Biogeogr. 2019, 28, 1393–1402. [Google Scholar] [CrossRef]
- García-Ruiz, J.M.; López-Moreno, J.I.; Vicente-Serrano, S.M.; Lasanta–Martínez, T.; Beguería, S. Mediterranean water resources in a global change scenario. Earth-Science Rev. 2011, 105, 121–139. [Google Scholar] [CrossRef]
- Monsarrat, S.; Jarvie, S.; Svenning, J.C. Anthropocene refugia: Integrating history and predictive modelling to assess the space available for biodiversity in a human-dominated world. Philos. Trans. R. Soc. B. Biol. Sci. 2019, 374, 20190219. [Google Scholar] [CrossRef]
- Li, D.; Olden, J.D.; Lockwood, J.L.; Record, S.; McKinney, M.L.; Baiser, B. Changes in taxonomic and phylogenetic diversity in the Anthropocene. Proc. R. Soc. B Biol. Sci. 2020, 287, 20200777. [Google Scholar] [CrossRef]
- Dornelas, M.; Gotelli, N.J.; Shimadzu, H.; Moyes, F.; Magurran, A.E.; McGill, B.J. A balance of winners and losers in the Anthropocene. Ecol. Lett. 2019, 22, 847–854. [Google Scholar] [CrossRef]
- Olden, J.D.; Comte, L.; Giam, X. The Homogocene: A research prospectus for the study of biotic homogenisation. NeoBiota 2018, 37, 23–36. [Google Scholar] [CrossRef]
- Thuiller, W.; Lavergne, S.; Roquet, C.; Boulangeat, I.; Lafourcade, B.; Araujo, M.B. Consequences of climate change on the tree of life in Europe. Nature 2011, 470, 531–534. [Google Scholar] [CrossRef] [PubMed]
- D’Agata, C.D.C.; Skoula, M.; & Brundu, G. A preliminary inventory of the alien flora of Crete (Greece). Bocconea 2009, 23, 301–315. [Google Scholar]
- Panitsa, M.; Iliadou, E.; Kokkoris, I.; Kallimanis, A.; Patelodimou, C.; Strid, A.; Raus, T.; Bergmeier, E.; Dimopoulos, P. Distribution patterns of ruderal plant diversity in Greece. Biodivers. Conserv. 2020, 29, 869–891. [Google Scholar] [CrossRef]
- Geijzendorffer, I.R.; Cohen-Shacham, E.; Cord, A.F.; Cramer, W.; Guerra, C.; Martín-López, B. Ecosystem services in global sustainability policies. Environ. Sci. Policy 2017, 74, 40–48. [Google Scholar] [CrossRef]
- Kokkoris, I.P.; Bekri, E.S.; Skuras, D.; Vlami, V.; Zogaris, S.; Maroulis, G.; Dimopoulos, D.; Dimopoulos, P. Integrating MAES implementation into protected area management under climate change: A fine-scale application in Greece. Sci. Total Environ. 2019. [Google Scholar] [CrossRef]
- Van Der Plas, F.; Manning, P.; Soliveres, S.; Allan, E.; Scherer-Lorenzen, M.; Verheyen, K.; Wirth, C.; Zavala, M.A.; Ampoorter, E.; Baeten, L.; et al. Biotic homogenization can decrease landscape-scale forest multifunctionality. Proc. Natl. Acad. Sci. USA 2016. [Google Scholar] [CrossRef]
- Kokkoris, I.P.; Mallinis, G.; Bekri, E.S.; Vlami, V.; Zogaris, S.; Chrysafis, I.; Mitsopoulos, I.; Dimopoulos, P. National Set of MAES Indicators in Greece: Ecosystem Services and Management Implications. Forests 2020, 11, 595. [Google Scholar] [CrossRef]


© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kougioumoutzis, K.; Kokkoris, I.P.; Panitsa, M.; Trigas, P.; Strid, A.; Dimopoulos, P. Spatial Phylogenetics, Biogeographical Patterns and Conservation Implications of the Endemic Flora of Crete (Aegean, Greece) under Climate Change Scenarios. Biology 2020, 9, 199. https://doi.org/10.3390/biology9080199
Kougioumoutzis K, Kokkoris IP, Panitsa M, Trigas P, Strid A, Dimopoulos P. Spatial Phylogenetics, Biogeographical Patterns and Conservation Implications of the Endemic Flora of Crete (Aegean, Greece) under Climate Change Scenarios. Biology. 2020; 9(8):199. https://doi.org/10.3390/biology9080199
Chicago/Turabian StyleKougioumoutzis, Konstantinos, Ioannis P. Kokkoris, Maria Panitsa, Panayiotis Trigas, Arne Strid, and Panayotis Dimopoulos. 2020. "Spatial Phylogenetics, Biogeographical Patterns and Conservation Implications of the Endemic Flora of Crete (Aegean, Greece) under Climate Change Scenarios" Biology 9, no. 8: 199. https://doi.org/10.3390/biology9080199
APA StyleKougioumoutzis, K., Kokkoris, I. P., Panitsa, M., Trigas, P., Strid, A., & Dimopoulos, P. (2020). Spatial Phylogenetics, Biogeographical Patterns and Conservation Implications of the Endemic Flora of Crete (Aegean, Greece) under Climate Change Scenarios. Biology, 9(8), 199. https://doi.org/10.3390/biology9080199







