Rai1 Haploinsufficiency Is Associated with Social Abnormalities in Mice
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animals
2.2. General Health and Neurological Behavior
2.3. Self-Grooming in A Novel Environment
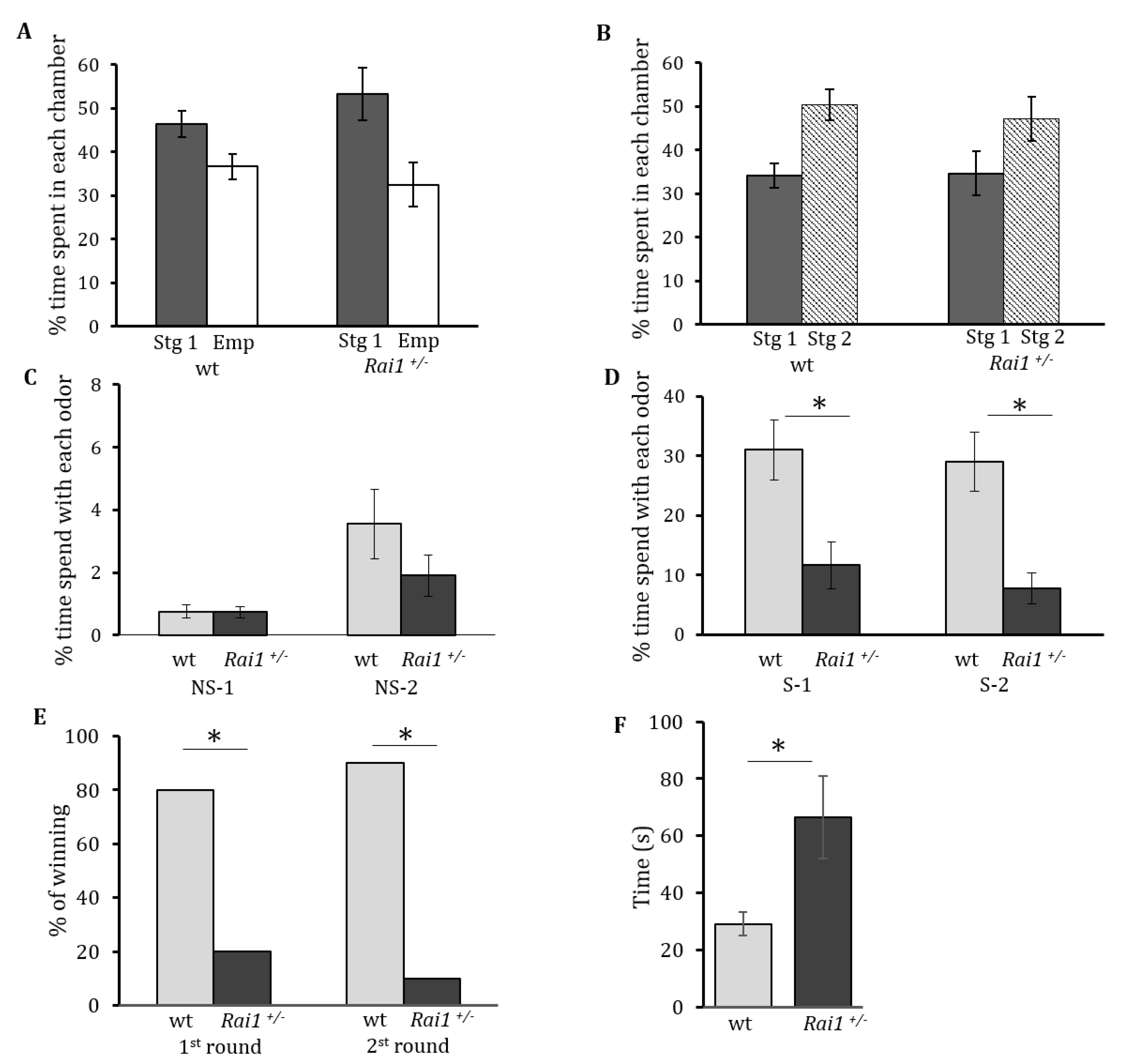
2.4. Sociability and Social Preference
2.5. Interest in Non-Social and Social Odors
2.6. Dominance Tube Test
2.7. Statistical Tests
3. Results
3.1. Abnormal Social Behaviors in Rai1+/− Mice
3.2. Repetitive Behaviors Were Augmented in Rai1+/− Mice
4. Discussion
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Casanova, E.L.; Sharp, J.L.; Chakraborty, H.; Sumi, N.S.; Casanova, M.F. Genes with high penetrance for syndromic and non-syndromic autism typically function within the nucleus and regulate gene expression. Mol. Autism 2016, 7. [Google Scholar] [CrossRef] [PubMed]
- Greenberg, F.; Guzzetta, V.; Montes de Oca-Luna, R.; Magenis, R.E.; Smith, A.C.; Richter, S.F.; Kondo, I.; Dobyns, W.B.; Patel, P.I.; Lupski, J.R. Molecular analysis of the Smith-Magenis syndrome: A possible contiguous-gene syndrome associated with del(17)(p11.2). Am. J. Hum. Genet. 1991, 49, 1207–1218. [Google Scholar] [PubMed]
- Edelman, E.A.; Girirajan, S.; Finucane, B.; Patel, P.I.; Lupski, J.R.; Smith, A.C.; Elsea, S.H. Gender, genotype, and phenotype differences in Smith-Magenis syndrome: A meta-analysis of 105 cases. Clin. Genet. 2007, 71, 540–550. [Google Scholar] [CrossRef] [PubMed]
- Slager, R.E.; Newton, T.L.; Vlangos, C.N.; Finucane, B.; Elsea, S.H. Mutations in RAI1 associated with Smith-Magenis syndrome. Nat. Genet. 2003, 33, 466–468. [Google Scholar] [CrossRef] [PubMed]
- Bi, W.; Saifi, G.M.; Shaw, C.J.; Walz, K.; Fonseca, P.; Wilson, M.; Potocki, L.; Lupski, J.R. Mutations of RAI1, a PHD-containing protein, in nondeletion patients with Smith-Magenis syndrome. Hum. Genet. 2004, 115, 515–524. [Google Scholar] [CrossRef] [PubMed]
- Girirajan, S.; Elsas, L.J.; Devriendt, K.; Elsea, S.H. RAI1 variations in Smith-Magenis syndrome patients without 17p11.2 deletions. J. Med. Genet. 2005, 42, 820–828. [Google Scholar] [CrossRef] [PubMed]
- Bi, W.; Saifi, G.M.; Girirajan, S.; Shi, X.; Szomju, B.; Firth, H.; Magenis, R.E.; Potocki, L.; Elsea, S.H.; Lupski, J.R. RAI1 point mutations, CAG repeat variation, and SNP analysis in non-deletion Smith-Magenis syndrome. Am. J. Med. Genet. A 2006, 140, 2454–2463. [Google Scholar] [CrossRef] [PubMed]
- Girirajan, S.; Vlangos, C.N.; Szomju, B.B.; Edelman, E.; Trevors, C.D.; Dupuis, L.; Nezarati, M.; Bunyan, D.J.; Elsea, S.H. Genotype-phenotype correlation in Smith-Magenis syndrome: Evidence that multiple genes in 17p11.2 contribute to the clinical spectrum. Genet. Med. 2006, 8, 417–427. [Google Scholar] [CrossRef] [PubMed]
- Truong, H.T.; Dudding, T.; Blanchard, C.L.; Elsea, S.H. Frameshift mutation hotspot identified in Smith-Magenis syndrome: Case report and review of literature. BMC Med. Genet. 2010, 11. [Google Scholar] [CrossRef] [PubMed]
- Vilboux, T.; Ciccone, C.; Blancato, J.K.; Cox, G.F.; Deshpande, C.; Introne, W.J.; Gahl, W.A.; Smith, A.C.; Huizing, M. Molecular analysis of the retinoic acid induced 1 gene (RAI1) in patients with suspected Smith-Magenis syndrome without the 17p11.2 deletion. PLoS ONE 2011, 6, e22861. [Google Scholar] [CrossRef] [PubMed]
- Vieira, G.H.; Rodriguez, J.D.; Carmona-Mora, P.; Cao, L.; Gamba, B.F.; Carvalho, D.R.; De Rezende Duarte, A.; Santos, S.R.; De Souza, D.H.; DuPont, B.R.; et al. Detection of classical 17p11.2 deletions, an atypical deletion and RAI1 alterations in patients with features suggestive of Smith-Magenis syndrome. Eur. J. Hum. Genet. 2012, 20, 148–154. [Google Scholar] [CrossRef] [PubMed]
- Gropman, A.L.; Duncan, W.C.; Smith, A.C. Neurologic and developmental features of the Smith-Magenis syndrome (del 17p11.2). Pediatr. Neurol. 2006, 34, 337–350. [Google Scholar] [CrossRef] [PubMed]
- Laje, G.; Morse, R.; Richter, W.; Ball, J.; Pao, M.; Smith, A.C. Autism spectrum features in Smith-Magenis syndrome. Am. J. Med. Genet. C Semin. Med. Genet. 2010, 154C, 456–462. [Google Scholar] [CrossRef] [PubMed]
- Wolters, P.L.; Gropman, A.L.; Martin, S.C.; Smith, M.R.; Hildenbrand, H.L.; Brewer, C.C.; Smith, A.C. Neurodevelopment of children under 3 years of age with Smith-Magenis syndrome. Pediatr. Neurol. 2009, 41, 250–258. [Google Scholar] [CrossRef] [PubMed]
- Wilde, L.; Silva, D.; Oliver, C. The nature of social preference and interactions in Smith-Magenis syndrome. Res. Dev. Disabil. 2013, 34, 4355–4365. [Google Scholar] [CrossRef] [PubMed]
- Osorio, A.; Sampaio, A.; Martínez Regueiro, R.; Garayzábal Heinze, E.; Carracedo, A.; Fernández Prieto, M. Autism spectrum symptoms in Smith-Magenis syndrome and Williams syndrome: Comparisons and contrasts. Int. J. Dev. Disabil. 2015, 61, 49–55. [Google Scholar] [CrossRef]
- Walz, K.; Caratini-Rivera, S.; Bi, W.; Fonseca, P.; Mansouri, D.L.; Lynch, J.; Vogel, H.; Noebels, J.L.; Bradley, A.; Lupski, J.R. Modeling del(17)(p11.2p11.2) and dup(17)(p11.2p11.2) contiguous gene syndromes by chromosome engineering in mice: Phenotypic consequences of gene dosage imbalance. Mol. Cell. Biol. 2003, 23, 3646–3655. [Google Scholar] [CrossRef] [PubMed]
- Walz, K.; Spencer, C.; Kaasik, K.; Lee, C.C.; Lupski, J.R.; Paylor, R. Behavioral characterization of mouse models for Smith-Magenis syndrome and dup(17)(p11.2p11.2). Hum. Mol. Genet. 2004, 13, 367–378. [Google Scholar] [CrossRef] [PubMed]
- Ricard, G.; Molina, J.; Chrast, J.; Gu, W.; Gheldof, N.; Pradervand, S.; Schutz, F.; Young, J.I.; Lupski, J.R.; Reymond, A.; et al. Phenotypic consequences of copy number variation: Insights from Smith-Magenis and Potocki-Lupski syndrome mouse models. PLoS Biol. 2010, 8, e1000543. [Google Scholar] [CrossRef] [PubMed]
- Walz, K.; Paylor, R.; Yan, J.; Bi, W.; Lupski, J.R. Rai1 duplication causes physical and behavioral phenotypes in a mouse model of dup(17)(p11.2p11.2). J. Clin. Invest. 2006, 116, 3035–3041. [Google Scholar] [CrossRef] [PubMed]
- Bi, W.; Yan, J.; Shi, X.; Yuva-Paylor, L.A.; Antalffy, B.A.; Goldman, A.; Yoo, J.W.; Noebels, J.L.; Armstrong, D.L.; Paylor, R.; et al. Rai1 deficiency in mice causes learning impairment and motor dysfunction, whereas Rai1 heterozygous mice display minimal behavioral phenotypes. Hum. Mol. Genet. 2007, 16, 1802–1813. [Google Scholar] [CrossRef] [PubMed]
- Bi, W.; Ohyama, T.; Nakamura, H.; Yan, J.; Visvanathan, J.; Justice, M.J.; Lupski, J.R. Inactivation of Rai1 in mice recapitulates phenotypes observed in chromosome engineered mouse models for Smith-Magenis syndrome. Hum. Mol. Genet. 2005, 14, 983–995. [Google Scholar] [CrossRef] [PubMed]
- Molina, J.; Carmona-Mora, P.; Chrast, J.; Krall, P.M.; Canales, C.P.; Lupski, J.R.; Reymond, A.; Walz, K. Abnormal social behaviors and altered gene expression rates in a mouse model for Potocki-Lupski syndrome. Hum. Mol. Genet. 2008, 17, 2486–2495. [Google Scholar] [CrossRef] [PubMed]
- Nadler, J.J.; Moy, S.S.; Dold, G.; Trang, D.; Simmons, N.; Perez, A.; Young, N.B.; Barbaro, R.P.; Piven, J.; Magnuson, T.R.; et al. Automated apparatus for quantitation of social approach behaviors in mice. Genes Brain Behav. 2004, 3, 303–314. [Google Scholar] [CrossRef] [PubMed]
- Rao, N.R.; Abad, C.; Perez, I.C.; Young, J.I.; Walz, K.; John, P.; Hussman Institute for Human Genomics. University of Miami: Miami, FL, USA, 2016; Unpublished data.
- Silverman, J.L.; Yang, M.; Lord, C.; Crawley, J.N. Behavioural phenotyping assays for mouse models of autism. Nat. Rev. Neurosci. 2010, 11, 490–502. [Google Scholar] [CrossRef] [PubMed]
- Goldman, A.M.; Potocki, L.; Walz, K.; Lynch, J.K.; Glaze, D.G.; Lupski, J.R.; Noebels, J.L. Epilepsy and chromosomal rearrangements in Smith-Magenis Syndrome [del(17)(p11.2p11.2)]. J. Child Neurol. 2006, 21, 93–98. [Google Scholar] [CrossRef] [PubMed]
- Shahbazian, M.; Young, J.; Yuva-Paylor, L.; Spencer, C.; Antalffy, B.; Noebels, J.; Armstrong, D.; Paylor, R.; Zoghbi, H. Mice with truncated MeCP2 recapitulate many Rett syndrome features and display hyperacetylation of histone H3. Neuron 2002, 35, 243–254. [Google Scholar] [CrossRef]
- Hayes, S.; Turecki, G.; Brisebois, K.; Lopes-Cendes, I.; Gaspar, C.; Riess, O.; Ranum, L.P.; Pulst, S.M.; Rouleau, G.A. CAG repeat length in RAI1 is associated with age at onset variability in spinocerebellar ataxia type 2 (SCA2). Hum. Mol. Genet. 2000, 9, 1753–1758. [Google Scholar] [CrossRef] [PubMed]
- Joober, R.; Benkelfat, C.; Toulouse, A.; Lafreniere, R.G.; Lal, S.; Ajroud, S.; Turecki, G.; Bloom, D.; Labelle, A.; Lalonde, P.; et al. Analysis of 14 CAG repeat-containing genes in schizophrenia. Am. J. Med. Genet. 1999, 88, 694–699. [Google Scholar] [CrossRef]
- Van der Zwaag, B.; Franke, L.; Poot, M.; Hochstenbach, R.; Spierenburg, H.A.; Vorstman, J.A.; Van Daalen, E.; De Jonge, M.V.; Verbeek, N.E.; Brilstra, E.H.; et al. Gene-network analysis identifies susceptibility genes related to glycobiology in autism. PLoS ONE 2009, 4, e5324. [Google Scholar] [CrossRef] [PubMed]
- Tsai, P.T.; Hull, C.; Chu, Y.; Greene-Colozzi, E.; Sadowski, A.R.; Leech, J.M.; Steinberg, J.; Crawley, J.N.; Regehr, W.G.; Sahin, M. Autistic-like behavior and cerebellar dysfunction in Purkinje cell Tsc1 mutant mice. Nature 2012, 488, 647–651. [Google Scholar] [CrossRef] [PubMed]
- Won, H.; Lee, H.; Gee, H.Y.; Mah, W.; Kim, J.; Lee, J.; Ha, S.; Chung, C.; Jung, E.S.; Park, S.; et al. Autistic-like behavior in Shank2-mutant mice improved by restoring NMDA receptor function. Nature 2014, 486, 261–265. [Google Scholar] [CrossRef] [PubMed]
- Etherton, M.R.; Blaiss, C.A.; Powell, C.M.; Sudhof, T.C. Mouse neurexin-1α deletion causes correlated electrophysiological and behavioral changes consistent with cognitive impairments. Proc. Natl. Acad. Sci. USA 2009, 106, 17998–18003. [Google Scholar] [CrossRef] [PubMed]
- Cheh, M.A.; Millonig, J.H.; Roselli, L.M.; Ming, X.; Jacobsen, E.; Kamdar, S.; Wagner, G.C. En2 knockout mice display neurobehavioral and neurochemical alterations relevant to autism spectrum disorder. Brain Res. 2006, 1116, 166–176. [Google Scholar] [CrossRef] [PubMed]
- Gallagher, D.; Voronova, A.; Zander, M.A.; Cancino, G.I.; Bramall, A.; Krause, M.P.; Abad, C.; Tekin, M.; Neilsen, P.M.; Callen, D.F.; et al. Ankrd11 is a chromatin regulator involved in autism that is essential for neural development. Dev. Cell. 2015, 32, 31–42. [Google Scholar] [CrossRef] [PubMed]
- Spencer, C.M.; Alekseyenko, O.; Serysheva, E.; Yuva-Paylor, L.A.; Paylor, R. Altered anxiety-related and social behaviors in the Fmr1 knockout mouse model of fragile X syndrome. Genes Brain Behav. 2005, 4, 420–430. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.; Guenthner, C.J.; Xu, J.; Nguyen, T.; Schwarz, L.A.; Wilkinson, A.W.; Gozani, O.; Chang, H.Y.; Shamloo, M.; Luo, L. Molecular and neural functions of Rai1, the casual gene for smith-magenis syndrome. Neuron 2016, 92, 392–406. [Google Scholar] [CrossRef] [PubMed]
- Chadman, K.K.; Gong, S.; Scattoni, M.L.; Boltuck, S.E.; Gandhy, S.U.; Heintz, N.; Crawley, J.N. Minimal aberrant behavioral phenotypes of neuroligin-3 R451C knockin mice. Autism Res. 2008, 1, 147–158. [Google Scholar] [CrossRef] [PubMed]
- McNaughton, C.H.; Moon, J.; Strawderman, M.S.; Maclean, K.N.; Evans, J.; Strupp, B.J. Evidence for social anxiety and impaired social cognition in a mouse model of fragile X syndrome. Behav. Neurosci. 2008, 122, 293–300. [Google Scholar] [CrossRef] [PubMed]
- Spencer, C.M.; Alekseyenko, O.; Hamilton, S.M.; Thomas, A.M.; Serysheva, E.; Yuva-Paylor, L.A.; Paylor, R. Modifying behavioral phenotypes in Fmr1 KO mice: Genetic background differences reveal autistic-like responses. Autism Res. 2011, 4, 40–56. [Google Scholar] [CrossRef] [PubMed]
- Fairless, A.H.; Katz, J.M.; Vijayvargiya, N.; Dow, H.C.; Kreibich, A.S.; Berrettini, W.H.; Abel, T.; Brodkin, E.S. Development of home cage social behaviors in BALB/cJ vs C57BL/6J mice. Behav. Brain Res. 2013, 237, 338–347. [Google Scholar] [CrossRef] [PubMed]
- Crawley, J.N.; Belknap, J.K.; Collins, A.; Crabbe, J.C.; Frankel, W.; Henderson, N.; Hitzemann, R.; Maxson, S.C.; Miner, L.L.; Silva, A.J.; et al. Behavioral phenotypes of inbred mouse strains: Implications and recommendations for molecular studies. Psychopharmacology 1997, 132, 107–124. [Google Scholar] [CrossRef] [PubMed]
- Ohashi, R.; Takao, K.; Miyakawa, T.; Shiina, N. Comprehensive behavioral analysis of RNG105 (Caprin 1) heterozygous mice: Reduced social interaction and attenuated response to novelty. Sci. Rep. 2016, 6. [Google Scholar] [CrossRef]
| SMS Patients | ||
|---|---|---|
| Phenotypes | % in Common 17p11.2 Deletion [3,8,11] | % in RAI1 Mutations [5,8,9,11] |
| Craniofacial Abnormalities | 100 | 100 |
| Skeletal Abnormalities | ||
| Short stature | 70–80 | 11 |
| Scoliosis/ Vertebral Abnormalities | 73 | 40–50 |
| Short broad hands/Brachydactyly | 85 | 88 |
| Otorhinolaryngological | ||
| Hoarse Voice | 66–80 | 76–86 |
| Hearing loss | 60–68 | 11–33 |
| Neurological | ||
| Cognitive Impairment | 100 | 100 |
| Infantile hypotonia | <90 | 50–61 |
| Speech delay | >90 | 70 |
| Motor delay | >90 | 60-70 |
| Sleep disturbance | 90 | 100 |
| EEG abnormalities | 50–66 | 80 |
| Seizures | 11–30 | 16.6–50 |
| Behavioral | ||
| Self-hugging | 50–80 | 100 |
| Onychotillomania | 25–85 | 80–100 |
| Polyembolokoilamania | 25–85 | 75–80 |
| Head banging/face slapping | 70 | 90 |
| Hand biting | 80 | 60–71 |
| Attention seeking | 80–100 | 100 |
| Aggressive behavior | 55 | |
| Self-injurious behavior | 70–90 | >90 |
| Hyperactivity | 80 | 100 |
| Autistic features | 90 [7] | NR |
| Other features | ||
| Cardiac defects | 30 | 0 |
| Renal/urinary tract defect | 30 | 0 |
| Obesity | 18 | 78 |
| Overeating | 25 | 81 |
| SMS Mouse Models | ||
|---|---|---|
| Phenotypes | Common Deletion [17,18,27] | Rai1+/− [19,22] |
| Neurological | ||
| Overt seizures | Yes (~20%) | Subtle |
| EEG | Abnormal | Abnormal |
| Locomotor activity + | Decreased | Decreased |
| Anxiety + | Normal | Normal |
| Learning and memory | Normal | Normal |
| Social novelty recognition & | Abnormal | Normal * |
| Social (dominant behavior) # | Decreased | Decreased * |
| Repetitive behaviors ++ | NR | Increased * |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rao, N.R.; Abad, C.; Perez, I.C.; Srivastava, A.K.; Young, J.I.; Walz, K. Rai1 Haploinsufficiency Is Associated with Social Abnormalities in Mice. Biology 2017, 6, 25. https://doi.org/10.3390/biology6020025
Rao NR, Abad C, Perez IC, Srivastava AK, Young JI, Walz K. Rai1 Haploinsufficiency Is Associated with Social Abnormalities in Mice. Biology. 2017; 6(2):25. https://doi.org/10.3390/biology6020025
Chicago/Turabian StyleRao, Nalini R., Clemer Abad, Irene C. Perez, Anand K. Srivastava, Juan I. Young, and Katherina Walz. 2017. "Rai1 Haploinsufficiency Is Associated with Social Abnormalities in Mice" Biology 6, no. 2: 25. https://doi.org/10.3390/biology6020025
APA StyleRao, N. R., Abad, C., Perez, I. C., Srivastava, A. K., Young, J. I., & Walz, K. (2017). Rai1 Haploinsufficiency Is Associated with Social Abnormalities in Mice. Biology, 6(2), 25. https://doi.org/10.3390/biology6020025





