Abstract
Aromatic compounds are the second most abundant class of molecules on the earth and frequent environmental pollutants. They are difficult to metabolize due to an inert chemical structure, and of all living organisms, only microbes have evolved biochemical pathways that can open an aromatic ring and catabolize thus formed organic molecules. In bacterial genomes, the phenylacetate (PA) utilization pathway is abundant and represents the central route for degradation of a variety of organic compounds, whose degradation reactions converge at this pathway. The PA pathway is a hybrid pathway and combines the dual features of aerobic metabolism, i.e., usage of both oxygen to open the aromatic ring and of anaerobic metabolism—coenzyme A derivatization of PA. This allows the degradation process to be adapted to fluctuating oxygen conditions. In this review we focus on the structural and functional aspects of enzymes and their complexes involved in the PA degradation by the catabolic hybrid pathway. We discuss the ability of the central PaaABCE monooxygenase to reversibly oxygenate PA, the controlling mechanisms of epoxide concentration by the pathway enzymes, and the similarity of the PA utilization pathway to the benzoate utilization Box pathway and β-oxidation of fatty acids.
1. Introduction
Aromatic compounds are abundant in nature and diverse in their chemical structure. Together with carbohydrates they are one of the most abundant sources of organic carbon [1] in the world. Compounds derived from petroleum such as benzene, toluene, ethylbenzene and xylene (BTEX) and others such as polychlorinated biphenyls (PCBs), polycyclic aromatic hydrocarbons (PAHs) and pentachlorophenol are among common environmental pollutants that contain an aromatic ring [2]. Plants lack the pathway for degradation of aromatic compounds and in the animal kingdom, there exist limited capacities for their utilization [3].
The degradation/utilization pathways for aromatic compounds are found predominantly in microbes allowing them to use aromatic compounds as the sole source of carbon [3]. Due to the high resonance energy of the aromatic system, these compounds have limited reactivity and are quite resistant to degradation. Nevertheless, microbes have evolved enzymatic pathways to accomplish this task, both under aerobic [4] as well as anaerobic conditions [5]. To access a wide variety of aromatic compounds, microbes have developed a strategy of converting them through the so-called “peripheral pathways” to a few central intermediates, therefore reducing the need to develop numerous specialized pathways [4]. Two aerobic strategies have evolved for breaking down aromatic rings. The classical aerobic strategy employs two multicomponent monooxygenases or a dioxygenase to introduce two hydroxyl groups into the aromatic ring, converting it first into catechol and subsequently cleaving the bond adjacent to the hydroxyls in an oxygen-dependent reaction [6]. The second strategy, discovered more recently for phenylacetate and benzoate catabolism [7], proceeds through attachment of the aromatic compound to coenzyme A, followed by its oxidation and ring opening. The initial mechanism postulated that the aromatic character of the ring was destabilized by the addition of two hydroxyl groups in two successive oxygenase steps [8]. However, several years later the conversion of the aromatic ring was proven to occur through a non-aromatic epoxide intermediate, a reaction catalyzed by a multicomponent monooxygenase [9,10].
The breakdown of aromatic compounds also occurs under anaerobic conditions whereby two independent metabolic strategies have evolved, both requiring the formation of a benzoyl-CoA intermediate. The first one reduces benzoyl-CoA by a class I reductase in a reaction driven by ATP hydrolysis [11]. The second, more recently discovered strategy uses ATP-independent class II benzoyl-CoA reductases that are multisubunit enzymes containing several cofactors [12]. Several reasons were proposed as to why processing of phenylacetate and benzoate in the form of CoA esters may be beneficial. CoA-bound intermediates are membrane impermeable, which keeps the degradation products contained within the cell. The large attachment in the form of a CoA-moiety significantly improves the recognition of a relatively small hydrophobic molecule. Finally, facultative anaerobes can degrade CoA-derivatives through anaerobic pathways in conditions of oxygen shortage.
In this review we focus generally on the hybrid phenylacetate utilization pathway and in particular on the three-dimensional structures of the Paa enzymes and the spatial coordination of reactions within this pathway through the formation of multiprotein complexes.
2. Phenylacetate Hybrid Degradation Pathway
The best characterized of the aerobic aromatic compound degradation pathways utilizing CoA is the phenylacetate degradation pathway, also known as a hybrid pathway, as it contains elements of aerobic and anaerobic pathways. This hybrid pathway exists in ~1/6 of currently sequenced bacterial genomes [9]. A parallel benzoate-CoA degradation hybrid pathway has been identified in an additional 5% of the genomes [4]. In conjunction with the peripheral pathways, this hybrid pathway serves as the central route for degradation of a wide variety of aromatic compounds such as 2-phenylacetate, 2-phenylethylamine, tropic acid, and styrene [7]. Genes associated with phenylacetate catabolism in Escherichia coli are located within three transcription units, the main one being the paa operon. They encode twelve enzymes or enzymatic subunits: PaaA, PaaB, PaaC, PaaE, PaaF, PaaG, PaaH, PaaI, PaaJ, PaaK, PaaY and PaaZ, a transcription regulator PaaX and a protein with unknown function PaaD (Table 1).

Table 1.
General information about the enzymes from the phenylacetate degradation pathway.
| Protein | Function | Structure | Organism | Homologue | PDB ID | Organism | Family/fold |
|---|---|---|---|---|---|---|---|
| PaaK | Phenylacetate-CoA ligase | 2Y4O (PaaK2) 2Y27 (PaaK1) | Burkholderia cenopacia | Benzoate-CoA ligase | 2V7B | Burkholderia xenovorans | Adenylate-forming domain, class I |
| PaaI | Phenylacetyl-CoA Thioesterase | 1PSU | E. coli | PaaI-like protein | 4M20 | Staphylococcus aureus | Thioesterase group of hotdog superfamily |
| 1J1Y | Thermus thermophilus | 4-hydroxybenzoyl-CoA thioesterase | 3R37 | Arthrobacter sp. | |||
| PaaA | Catalytic subunit of 1,2-phenylacetyl-CoA epoxidase | 3PW1 | E. coli | BoxB | 3Q1G | Azoarcus evansii | Bacterial multicomponent monooxygenase |
| MMOH | 4GAM | Methylococcus capsulatus | |||||
| PaaB | Bridging subunit of 1,2-phenylacetyl-CoA epoxidase | 3EGR | Ralstonia eutropha | PaaB-like proteins | |||
| PaaC | Structural subunit of 1,2-phenylacetyl-CoA epoxidase | 3PW1 | E. coli | MMOH | 4GAM | Methylococcus capsulatus | Bacterial multicomponent monooxygenase |
| PaaE | Reductase subunit of 1,2-phenylacetyl-CoA epoxidase | Phtalate Dehydrogenase | 2PIA | Burkholderia cepacia | Class IA Reductase | ||
| PaaD | unknown | Duf59 | 3LNO | Bacillus Anthracis | Domain of the unknown function 59 | ||
| PaaG | 1,2-epoxyphenylacetyl-CoA isomerase | 4FZW | E. coli | BoxC | 2W3P | Burkholderia xenovorans | Enoyl-CoA isomerase; Crotonase superfamily |
| 3HRX | Thermus thermophilus | ||||||
| PaaZ | Oxepin-CoA hydrolase | BoxD | No structure | Azoarcus evansii | N-terminal: NAD(P)+-dependent aldehyde dehydrogenase C-terminal: (De)Hydratase group of hotdog superfamily | ||
| Aldehyde dehydrogenase | 2VRO | Burkholderia xenovorans | |||||
| PaaJ | 3-oxoadipyl-CoA/3-oxo-5,6-dehydrosuberyl-CoA thiolase | 1ULQ | Thermus thermophilus | Acetyl-CoA acetyltransferase | 4N44 | Clostridium acetobutylicum | 3-ketoacyl-CoA thiolase (thiolase I) |
| ThlA2 | 4E1L | Clostridium difficile | |||||
| PaaF | 2.3-dehydroadipyl-CoA hydratase | 4FZW | E. coli | BoxC | 2W3P | Burkholderia xenovorans | Enoyl-CoA hydratase; Crotonase superfamily |
| PaaH | 3-hydroxyadipyl-CoA dehydrogenase | 3MOG | E. coli | 3-hydroxyacyl-CoA dehydrogenase | |||
| PaaY | 2-hydroxycyclohepta-1,4,6-triene-1-carboxyl-CoA thioesterase | GK2848 | 3VNP | Geobacillus kaustophilus | |||
| PaaX | Transcriptional repressor | PaaX-like protein | 3LO9 | Jannaschia sp. | PaaX-like proteins containing helix-turn-helix motif |
All chemical reactions within this pathway have been defined, intermediates identified and enzyme functions assigned [9,13,14] (Schema 1). The pathway can be divided into two parts, the upper and the lower part. The lower part has enzymes that are homologous to well-characterized enzymes of fatty acid degradation found in the β-oxidation pathway. In the upper part of the pathway (Schema 1) the first enzyme is the phenyl-CoA acetate ligase (PaaK) [15], which is followed by a multicomponent monooxygenase complex consisting of four proteins (PaaABCE) [9,14,16]. PaaABCE converts PA-CoA (compound II) to the 1,2-epoxide derivative (compound III). This epoxide derivative is converted by the PaaG isomerase to an oxepin, an oxygen-containing heterocycle consisting of a seven-membered ring with three double bonds (compound IV), cleaved to 3-oxo-5,6-dehydrosuberyl-CoA semialdehyde (compound V) and oxidized by PaaZ to yield a β-keto C8-intermediate (compound VI).
Schema 1.
Paa phenylacetate degradation pathway. Comparison with Box pathway (top) and a fatty acid β-oxidation pathway (bottom). Compounds: I, phenylacetate; II, phenylacetyl-CoA; III, ring 1,2-epoxyphenylacetyl-CoA; IV, 2-oxepin-2(3H)-ylideneacetyl-CoA (oxepin-CoA); V, 3-oxo-5,6-dehydrosuberyl-CoA semialdehyde, VI, 3-oxo-5,6-dehydrosuberyl-CoA; VII, 3,4-dehydroadipyl-CoA; VIII, 2,3-dehydroadipyl-CoA; IX, 3-hydroxyadipyl-CoA; X, 3-oxoadipyl-CoA. Box pathway (top)—Compounds: (from left to right) benzoate; benzoyl-CoA; 2,3-epoxybenzoyl-CoA; the proposed oxepin-CoA is depicted in square brackets; 3,4-dehydroadipyl-CoA semialdehyde; 3,4-dehydroadipyl-CoA. Fatty acid β-oxidation pathway—Enzymes: ECI—Δ3-Δ2-enoyl-CoA isomerase; ECH—2-enoyl-CoA hydratase; HACD—3-hydroxyacyl-CoA dehydrogenase; KACT—3-ketoacyl-CoA thiolase.
In the lower part of the phenylacetate hybrid pathway, the C8-intermediate is cleaved by a thiolase (PaaJ), forming C6-dehydroadipyl-CoA (compound VII), which is isomerized to the α,β-unsaturated thioester 2,3-dehydroadipyl-CoA (compound VIII) by PaaG, hydrated by PaaF to 3-hydroxyadipyl-CoA (compound IX), and oxidized by NAD+-dependent dehydrogenase PaaH to 3-oxoadipyl-CoA (compound X) [9]. This final intermediate is cleaved by PaaJ to yield common metabolites, acetyl-CoA and succinyl-CoA [17].
3. The Upper Part of the Pathway
3.1. Activation of the Aromatic Compound
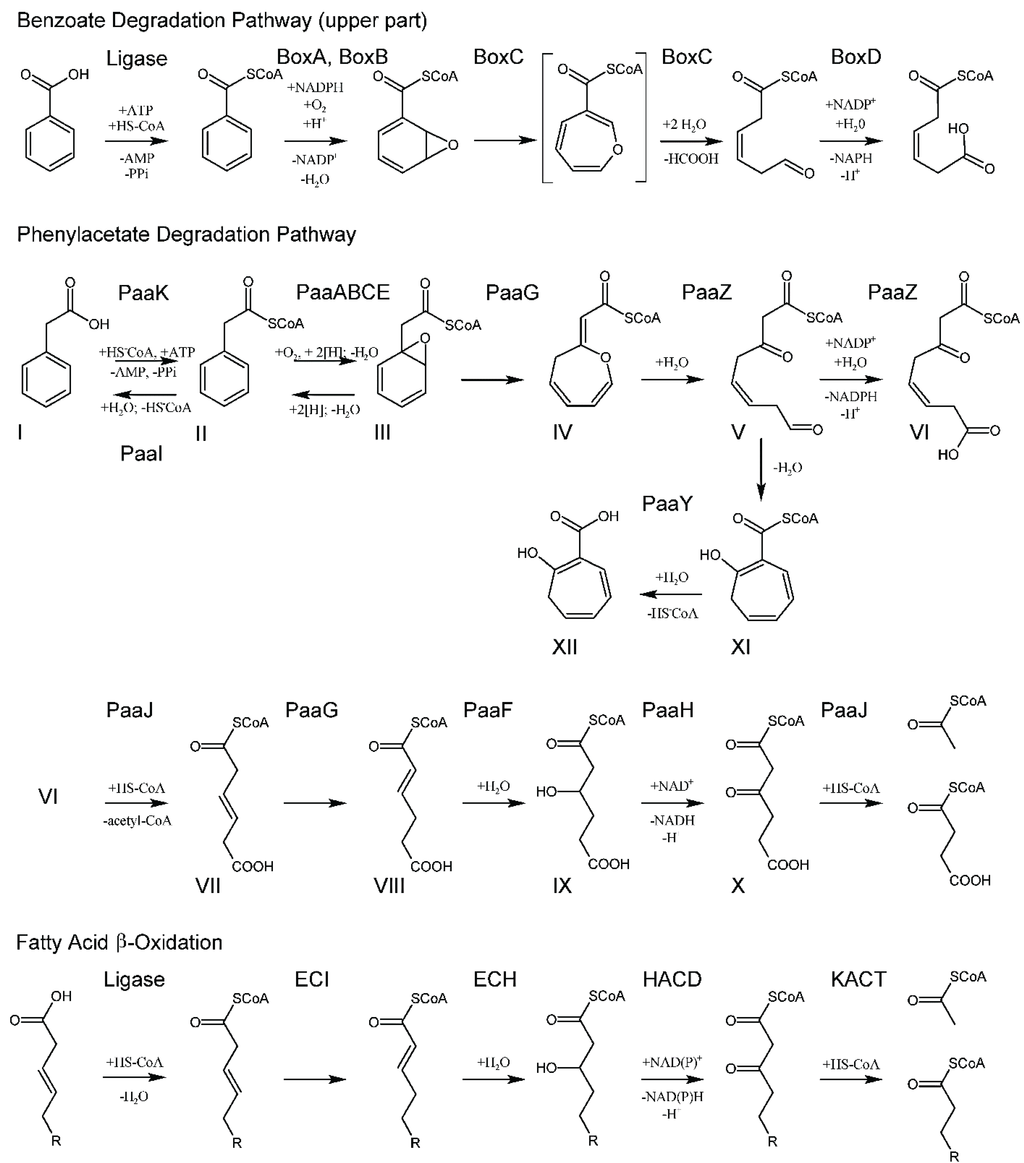
PaaK catalyzes ATP and Mg2+-dependent attachment of phenylacetate to CoA, with AMP and PPi as other reaction products. PaaK belongs to the adenylate-forming enzyme superfamily, which includes enzymes activating short-to-long fatty acids, aromatic compounds, biosynthesis of peptide antibiotics and siderophores [18]. PaaK was shown to be relatively heat-stable and very specific toward phenylacetate [15,19]. The structure of PaaK (Figure 1) was determined for the enzymes from Burkholderia cenocepacia, which unusually possesses two PaaK enzymes, PaaK1 and PaaK2, but only a single copy of all other enzymes of the phenylacetate pathway. PaaK1 and PaaK2 share 69% identity over the entire length of enzymes, while the corresponding genes are located among other genes of the hybrid pathway. PaaK1 has lower Km value for phenylacetate and a more relaxed substrate specificity than PaaK2, which likely arises from its extended aryl substrate pocket [20]. Nevertheless, the knockout of the paaK1 gene alone does not impair growth and pathogenicity of the microbe [21].
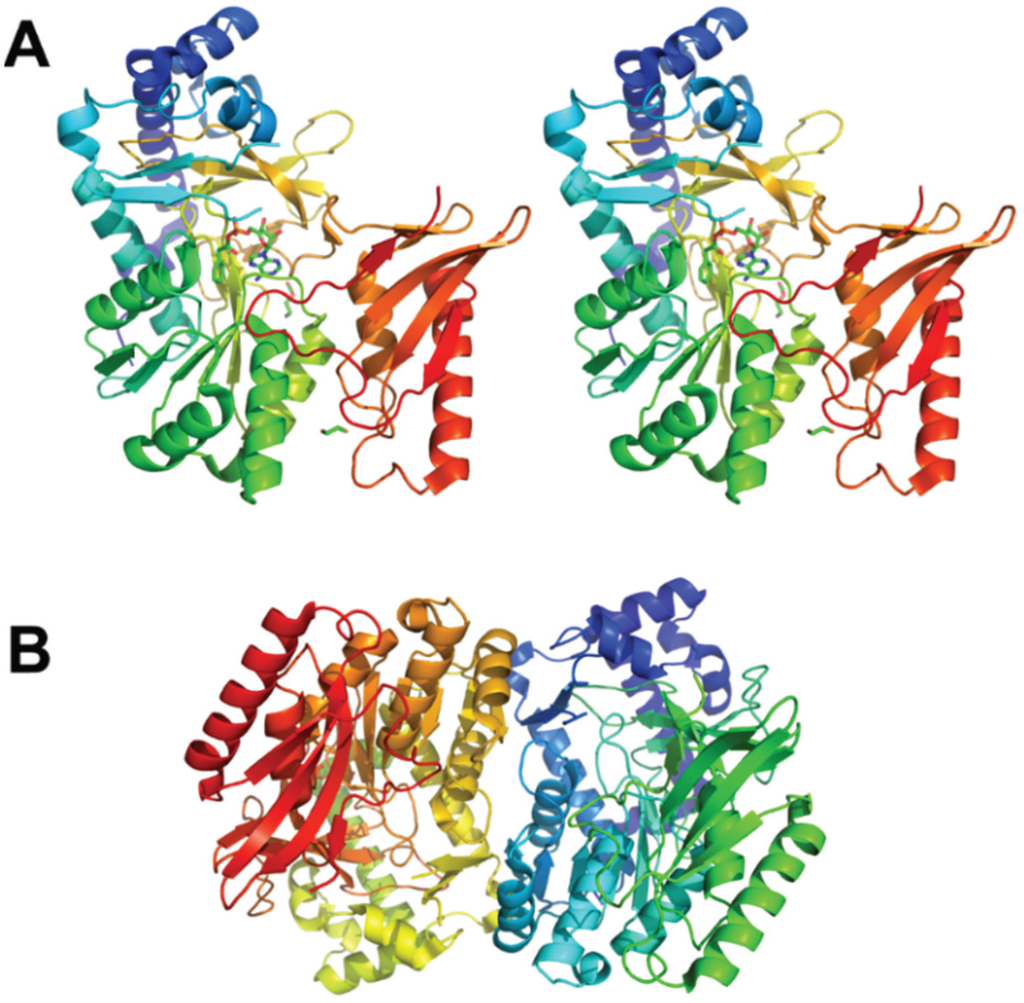
Figure 1.
The crystal structure of PaaK1 from Burkholderia cenocepacia. (A) The stereoview of the PaaK1 monomer with the phenylacetyl adenylate intermediate painted in rainbow colors from blue at the N-terminus to red at the C-terminus. (B) Biological unit of PaaK is a dimer. This and other figures were prepared using PyMol (www.pymol.org).
3.2. Epoxidation of the Aromatic Ring
Monooxygenation of the aromatic ring is the most crucial step of the pathway. Gene-knockout experiments have shown that when any of the five genes paaA,B,C,D or E is deleted, the pathway is inactivated at this step [22]. However, experiments with co-expression of recombinant proteins from Escherichia coli [16] and Pseudomonas sp. strain Y2 [9], as well as pull-down experiments showed that only PaaA, B, C and E components are necessary for the oxidation reaction and formation of the epoxide. Not only could the full complex PaaABCE be purified [9] but PaaAC, PaaBC and PaaABC could also form stable subcomplexes [16]. The binding of the reductase subunit PaaE, despite being essential for the overall reaction, is the weakest in the overall complex. Subsequent structural and biochemical studies revealed that the monooxygenase contains a catalytic subunit PaaA, a structural subunit PaaC, a bridging subunit PaaB and a reductase paaE with the overall complex composition of PaaA2B3-4C2E1 as judged by SDS-PAGE. The complex contained six iron atoms of which two belonged to the iron-sulfur cluster of PaaE and four to the two molecules of PaaA [14].
The crystals of PaaAC were obtained in complex with several CoA derivatives, including the phenylacetyl-CoA substrate and the structures were determined at resolutions in the range 2.03–2.25 Å [23]. The PaaAC complex forms a heterotetramer (dimer of heterodimers) (Figure 2A). Although the PaaA and PaaC subunits display only 17% sequence identity, their structures share the same fold and can be superimposed with the root-mean-squares deviation of 1.7 Å [16] (Figure 2B). The PaaA-PaaC heterodimer interface has a significantly hydrophobic character, particularly for PaaA, explaining the insolubility of PaaA alone. Additionally, hydrophilic/charged interactions at the edges of the interface add to the stability of the complex. The association of the two heterodimers into a tetramer occurs solely through the interactions of the PaaC molecules from each heterodimer and is less extensive than the PaaA-PaaC interface [16].
The catalytic subunit in this complex is PaaA, while PaaC is thought to play a structural role. No iron ions were present in the PaaA structure; however, the expected iron binding residues Glu 42, Glu 72, His 75, Asp 126, Glu 155 and His 158 are very similar to those found in other BMMs [24]. Indeed, soaking the crystals with iron, although it significantly increased crystal disorder and decreased resolution, showed that the ions were bound in the expected site [16]. The phenylacetate-CoA binds to the PaaA subunit in a hairpin conformation within a ~20Å deep pocket in the protein. The adenine and phenyl moieties at the two ends of the substrate point in the same direction and the phosphates are at the hairpin bend near the protein surface (Figure 2A). The entrance to the substrate-binding pocket is partially covered by surface segments containing Lys103 and Leu286, which likely provide a dynamic door for substrate entry. The interaction of the substrate with Phe108 helps to orient the phenylacetyl moiety so that the aromatic C1–C2 bond becomes located exactly above the putative di-iron center and in the proximity to the unusual constellation of Lys68 coordinating three acidic residues, Glu49, Glu72 and Asp126, named the lysine bridge [16].
The information about the structure of PaaACB complex came from the investigation of these proteins in Klebsiella pneumonia. Although the crystal diffracted only to 4.3 Å resolution, the structure could be determined using the known high resolution models for all three components [25]. The stoichiometry of the three subunits in the complex is 2:2:2. The PaaAC subcomplex is the same as observed previously on its own while also containing the phenylacetate-CoA molecule bound to the PaaA subunit. The PaaB subunit binds to both PaaA and PaaC, subunits of one heterodimer in a cleft formed near to the PaaA/PaaC interface (Figure 2C). The heterohexamer is assembled through dimerization of two PaaB subunits (Figure 2D). This arrangement into a hexamer with the PaaB dimer in the middle and two PaaAC heterodimers on the outside was confirmed by negative stain electron microscopy and small angle X-ray scattering in solution. The structure of the heterohexameric PaaACB complex with two PaaAC lobes connected by the PaaB dimer displays a concave surface close to the phenylacetate-CoA binding site. The iron-binding α-helices C, E and F of PaaA are exposed on that surface. There is sufficient space for the molecule of PaaE reductase to bind to this surface near the substrate and participate in electron transfer. The lysine bridge, identified in the PaaA subunit near the di-iron center, was suggested as being involved in this transfer [25].
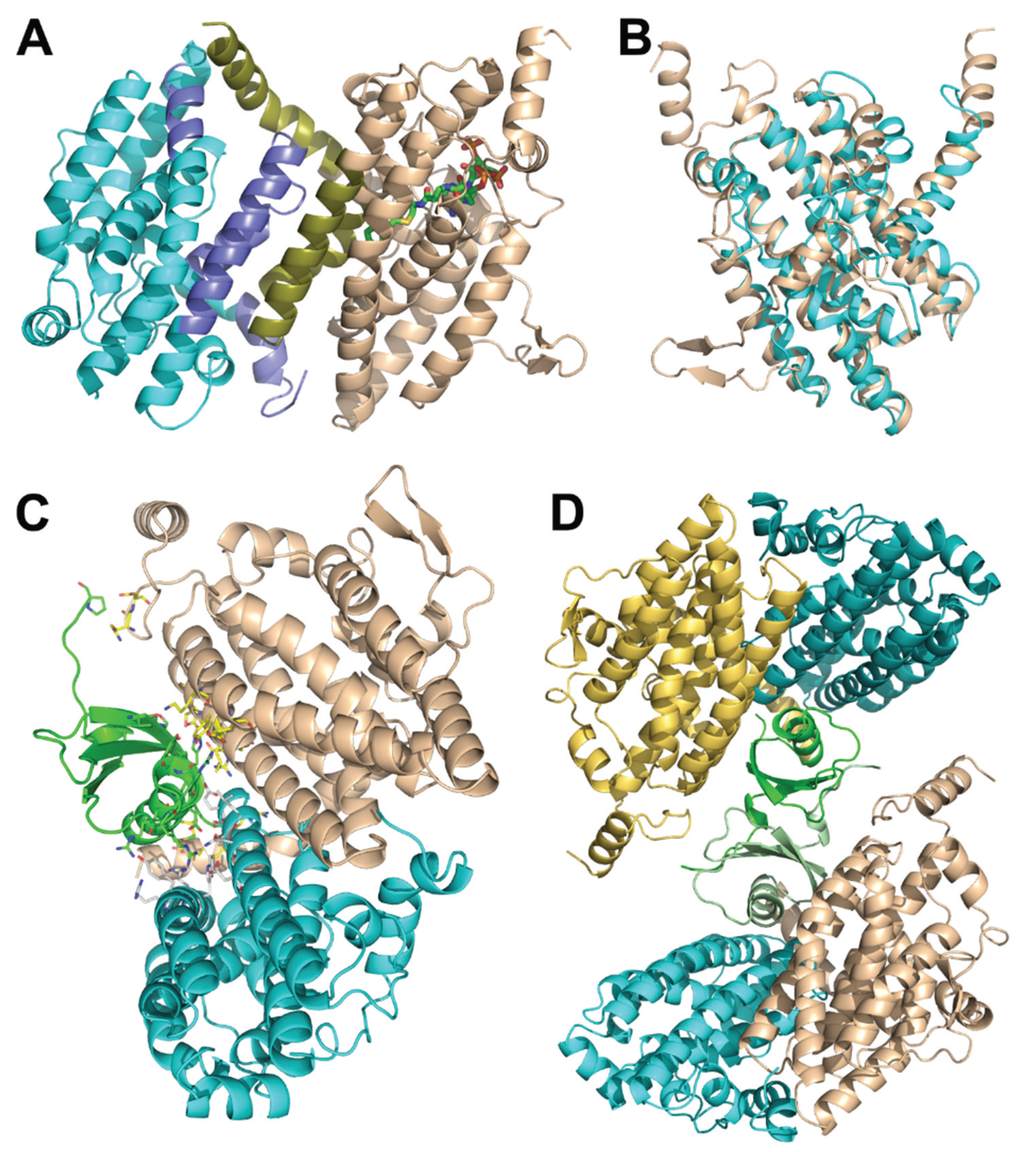
Figure 2.
Paa monooxygenase complex. (A) The PaaA-PaaC heterodimer with bound phenylacetyl-CoA. The catalytic PaaA subunit is painted in a wheat color, the structural PaaC subunit is painted in cyan. (B) Superposition of PaaA (wheat) and PaaC (cyan); (C) The PaaACB heterotrimer. PaaA is painted wheat, PaaC is cyan and PaaB is green. The residues at the interface of PaaB with PaaA and PaaC are shown in stick mode. (D) The (PaaACB)2 heterohexamer. PaaA is painted wheat and olive, PaaC is cyan and light blue, and PaaB is green and light green.
PaaE was characterized kinetically and spectroscopically [14,16]. It belongs to the class IA reductases [26], which are usually associated with dioxygenases, while classical monooxygenases, e.g., methane monooxygenase, utilize class IB or class III reductases. Indeed, advanced BLAST analysis identified the phthalate dioxygenase reductase from Burkholderia cepacia (PDB code 2PIA, [26]) as a structural homolog albeit with only a 22% sequence identity. PaaE contains an N-terminal NADPH- and FAD-binding domain and a C-terminal [2Fe-2S] ferredoxin-like domain and is thought to transfer electrons from NADPH through FAD and the iron-sulfur cluster to iron atoms in the active center of PaaA.
3.3. Ring Opening
In the next step of the degradation pathway, PaaG isomerizes 1,2-epoxyphenylacetyl-CoA epoxide (compound III), formed by the action of monooxygenase, to 2-oxepin-2(3H)-ylideneacetyl-CoA (compound IV) [9]. PaaG belongs to the crotonase fold superfamily, which also contains enoyl-CoA isomerases and enoyl-CoA hydratases involved in fatty acid β-oxidation. The structure of PaaG will be discussed below.
The next two steps, the hydrolysis of the oxepin and the oxidation of the terminal aldehyde group, are performed by the bifunctional enzyme PaaZ. It comprises two domains, the C-terminal R-specific hotdog fold hydratase (where R- and S- refer to the stereochemistry of the hydroxyacyl-CoA reaction product) and the N-terminal aldehyde dehydrogenase [13]. Despite different folds both R-specific hydratases, i.e., PaaZ, and S-specific hydratases, such as PaaF (see below), utilize an acid-base catalytic mechanism.
In many microorganisms the paaZ gene contains only an aldehyde dehydrogenase domain instead of two above-mentioned domains, raising questions about the enzyme responsible for the ring opening. Such an enzyme, which performs oxepin-CoA hydrolysis (ring-opening), has been identified in Aromatoleum aromaticum as a hotdog fold hydratase coded by a gene outside the paa gene cluster [13]. This enzyme was also capable of hydrating crotonyl-CoA with high activity, suggesting that the hydrolysis of the oxepin-CoA may be a side reaction [13].
3.4. Controlling the Fate of Toxic Epoxide
Epoxides are active towards DNA and proteins [27] and significantly inhibit cell growth [28]. It is possible that some pathogenic bacteria, such as Burkholderia cepacia, have evolved to use epoxide against their hosts to promote pathogen survival and replication [29]. The fate of the toxic epoxide is controlled by several means. The most prominent safeguard is the ability of the PaaABCE complex to catalyze the reverse reaction—deoxygenation (distinguish from dioxygenation), in which 2H+ equivalents are used to yield phenylacetyl-CoA and water [14]. This PaaABCE-catalyzed reverse deoxygenation reaction occurs when the epoxide accumulates due to inadequate processing by the downstream enzymes PaaG and PaaZ.
The second control mechanism involves the thioesterase PaaI. It belongs to the hotdog fold superfamily [30] and catalyzes the breakdown of the early-stage metabolites of the Paa pathway (PA-CoA), reversing the action of PaaK. PaaI has a very narrow substrate specificity limited to PA-CoA [14], suggesting that PaaI removes the excess of PA-CoA and prevents overloading the capacity of PaaABCE.
Exploring the reaction catalyzed by PaaZ, Teufel and colleagues have found that oxepin-CoA spontaneously rearranges to 2-hydroxycyclohepta-1,4,6-triene-1-formyl-CoA (compound XI), which inhibits ring cleavage by paaZ [13]. However, the inhibitory activity of the compound XI is mitigated by PaaY, which was found to be able to hydrolyze this product to yield compound XII with the highest specificity among all reagents tested to date. A homolog of PaaY from Geobacillus kaustophilus HTA426 with 33% sequence identity has been structurally characterized (PDB code 3VNP) providing information about the overall structure of PaaY. Further studies revealed that PaaY associates into trimers, contains Ca2+ and Zn2+ ions and is able to hydrolyse an even wider range of CoA derivatives, including acetoacetyl-CoA [31].
The Lower Part of the Paa Pathway
The lower part of the phenylacetate degradation pathway is similar to the fatty acid β-oxidation pathway with the rearrangement of a double bond from Δ3 to Δ2 position, i.e., Δ3-Δ2-enoyl-CoA isomerization, hydratation of the double bond, i.e., Δ2-enoyl-CoA hydratation, oxidation of the hydroxyl-group to the keto-group and cleavage of the acetyl-CoA.
The product of ring opening by the isomerase PaaG and the hydrolase PaaZ, 3-oxo-5,6-dehydrosuberyl-CoA (compound VI), is shortened by thiolase PaaJ by two carbon atoms to 2,3-dehydroadipyl-CoA (compound VII). The Δ3-double bond is rearranged to the Δ2-position by PaaG (compound VIII) and hydrated to 3-hydroxyadipyl-CoA (compound IX) by PaaF. Next, dehydrogenase PaaH oxidizes the hydroxyl-group to yield 3-oxoadipyl-CoA (compound X), which is finally cleaved by PaaJ to common metabolites acetyl-CoA and succinyl-CoA [9].
3.5. Protein-protein Interactions Among Enzymes of the Lower Part of the Paa Pathway
As enzymes of the fatty acid degradation pathway assemble in multiprotein complexes [32] and the reactions in the lower part of the Paa pathway are similar to fatty acid degradation, we suggested that the enzymes of the lower part may also associate into multiprotein complexes. Methods similar to those described above were used to identify stable complexes among enzymes PaaA, PaaB, PaaC, PaaF, PaaG, PaaH, PaaJ, and PaaZ. In numerous co-expression experiments of up to seven Paa enzymes in a single cell, only one stable complex was identified, that between PaaF and PaaG [33].
3.6. Structure of the PaaFG Complex
The crystal structure of the PaaF-PaaG complex was determined at 2.5 Å resolution [33]. Both proteins possess a crotonase fold and, as is common for proteins with this fold, they assemble into homotrimeric discs (Figure 3A,B). The complex is composed of a stack of four such discs, two PaaF discs in the center sandwiched between PaaG discs on each end (Figure 3C). In the stack the PaaF disc is positioned between another PaaF disc and a PaaG disc. The latter interacts only with PaaF through one of its flat surfaces, while the other surface is exposed to the solvent. This arrangement is also maintained in solutions, as visualized by negative stain electron microscopy [33].
PaaF and PaaG, like other members of the crotonase superfamily, are folded with repeating ββα-units, which form two almost perpendicular β-sheets, surrounded by α-helices [34]. Since the active sites of PaaF and PaaG are facing the solvent and are located far from each other, the tunnelling hypothesis is unlikely. However, assembling 12 active sites in close proximity within the complex likely speeds up two consecutive reactions along the pathway due to increased local concentration of active sites.
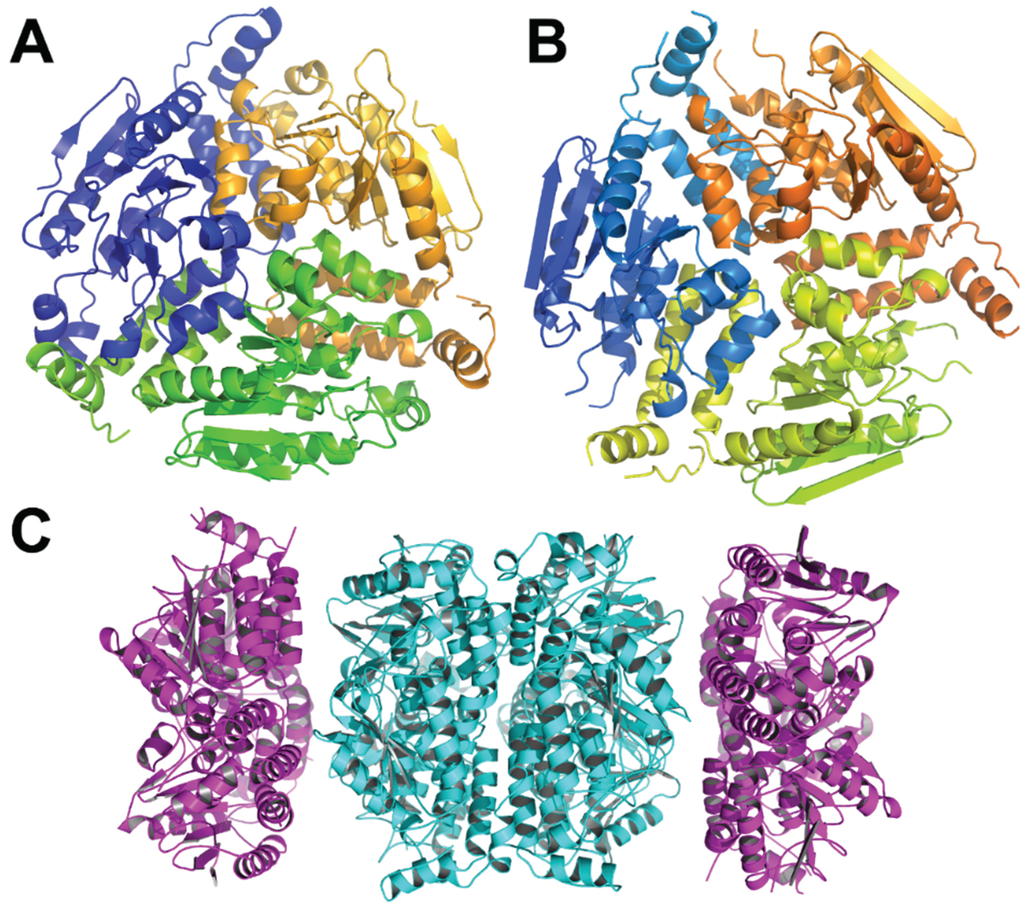
Figure 3.
The E. coli PaaF-PaaG complex forms a four-layered hetero-dodecamer. (A) The trimer of PaaF; (B) the trimer of PaaG; (C) the arrangement of trimeric rings within the dodecamer. The outside rings (magenta) are PaaG trimers, the inner rings (cyan) are PaaF trimers.
3.7. Other Enzymes Involved in the Lower Part of the Paa Pathway
The structures of β-ketoadipyl-CoA thiolase PaaJ from several bacteria have been determined (Thermus thermophilus, 1ULQ, 57% identity to E. coli PaaJ; Clostridium acetobutylicum EA 2018, PDB code 4N44, 45% identity; Clostridium difficile, PDB code 4E1L, 44% identity). The monomer has an α/β structure with the central layer of two α-helices sandwiched between four- and five-stranded β-sheets followed by α-helical layers (Figure 4). The monomers assemble into tetramers with two lobes and a central β-barrel composed of elements from four monomers holding them together.
The structure of the 3-hydroxyadipyl-CoA dehydrogenase PaaH from E. coli has been recently determined (PDB code 3MOG). This ~500-amino-acid enzyme contains three domains: the N-terminal Rossmann-fold domain, the “dimerization” domain comprising residues 188–280 and 388–475, and a reduced Rossmann-fold domain (residues 305–383) (Figure 5).
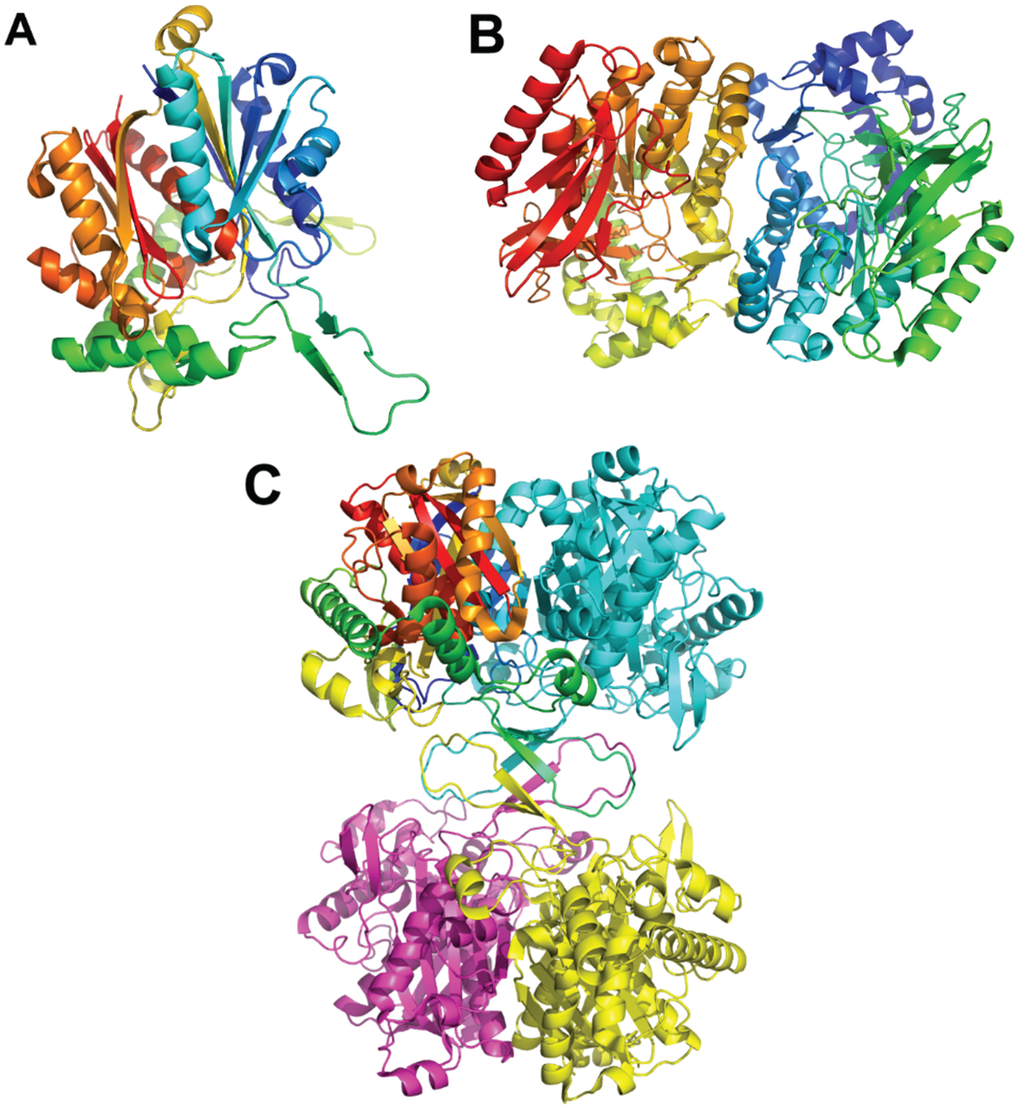
Figure 4.
The Thermus thermophilus HB8 PaaJ. The biological unit is a dimer of dimers. (A) The cartoon representation of the PaaJ monomer painted in rainbow color from blue at the N-terminus to red at the C-terminus. The structure is composed of five layers α/β/α/β/α. (B) The cartoon representation of a tightly bound dimer. The second monomer is in cyan. (C) The tetramer, third monomer is in magenta and fourth in yellow.
Other Proteins of the Paa Operon
PaaD is a small protein of around 170 residues, which was shown to be indispensable in vivo for the function of PaaABCE monooxygenase complex [8], although in vitro its presence was not required for catalysis [9,16]. It is predicted to contain a domain of unknown function DUF59 [35] and shows similarity to SufT, a protein involved in iron-sulfur cluster assembly. The similarity to SufT might indicate that PaaD is involved in the incorporation of iron ions in PaaE or PaaA.
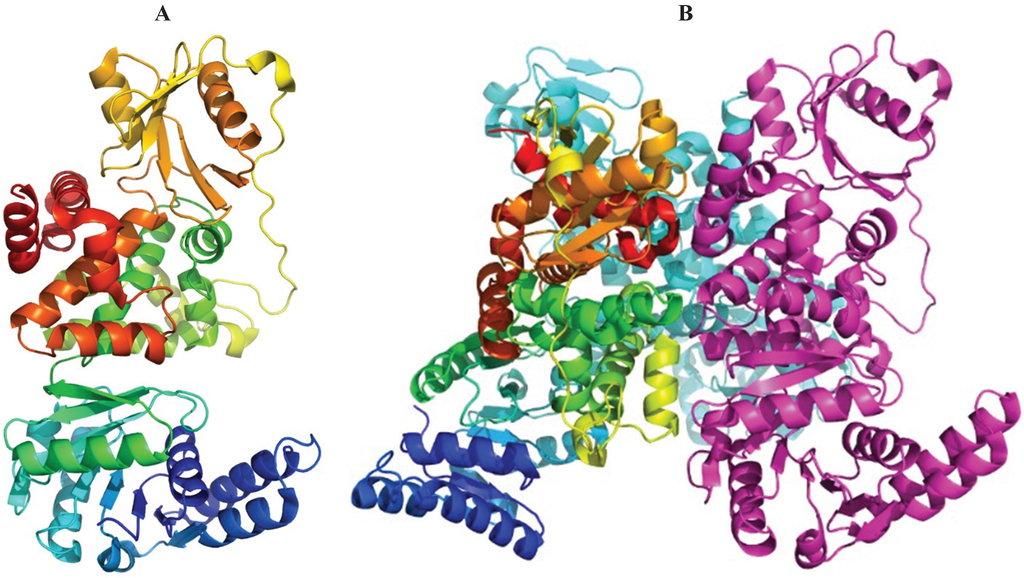
Figure 5.
The structure of E. coli PaaH. (A) The cartoon representation of the monomer painted rainbow colors. The dimerization domain is made of two segments, the one following the N-terminal domain (in green) and the C-terminus (in red); (B) the structure of a trimer. The inter-monomer contacts involve mainly the oligomerization domain.
PaaX is a transcriptional regulator to all three open-reading frames of the Paa operon. The binding to DNA is inhibited by phenylacetyl-CoA but not by phenylacetate alone [31,36]. It is also hypothesized that the intermediary CoA derivatives of the Paa catabolic pathway increase transcription inhibition by PaaX, while the inhibition is released by PaaY, which hydrolyses 2-hydroxycyclohepta-1,4,6-triene-1-formyl-CoA and PA-CoA [31].
PaaX has a distant homolog of known structure, that from Mycobacterium tuberculosis with 23% sequence identity (PDB code 3KFW), and which structure indicates the presence of the helix-turn-helix DNA-binding motif. PaaX belongs to the GntR family of regulators. Although crystallization of PaaX from E. coli was reported [37], no structural data are yet available [31,36].
4. Similarities with Other Metabolic Pathways
Steps in the upper part of the Paa pathway are very similar to the degradation of benzoic acid in the Box pathway, where a ligase first attaches the aromatic compound to CoA, the ring is then oxidized by the BoxAB complex to yield a 2,3-epoxybenzoyl-CoA, the ring is then opened by BoxC through a hypothesized formation on an oxepin and the terminal aldehyde group is oxidized by BoxD to yield 3,4-dehydroadipyl-CoA [10].
The action of BoxA and BoxB enzymes, which perform monooxygenation of the benzoyl-CoA, is similar to that of the PaaABCE complex. BoxB is a structural homolog of PaaA, but unlike other monooxygenases that are heteromeric, BoxB does not require a structural subunit and is monomeric [38]. While the di-iron center was empty in the high resolution structure of PaaA, the iron ions were observed in the structure of BoxB together with the substrate benzoic moiety. Both PaaA and BoxB are very specific toward their corresponding substrates and show no cross-reactivity for benzoyl-CoA and phenylacetyl-CoA, correspondingly. Indeed, PaaABCE was inhibited by the addition of benzoyl-CoA [14], while BoxB could not catalyze phenylacetyl-CoA monooxygenation [39].
BoxA is a reductase, which contains two [4Fe-4S] clusters and is specific for NADPH and FAD [38], and thus BoxA does not bear direct similarity to PaaE or to typical reductases of monooxygenases. BoxA does not co-purify with BoxB, and its concentration in living cells is much lower than that of BoxB. Epoxidation of the benzoic ring by BoxB requires only minor amounts of BoxA, suggesting a transient interaction between these two enzymes. However, the enzyme that catalyzes ring opening in the next step of the pathway, BoxC, co-purifies with BoxB suggesting that they form a complex. Moreover, BoxC is essential for BoxB to overcome auto-inhibition with its reaction product 2,3-epoxybenzoyl-CoA [10]. PaaABCE activity is similarly dependent on the downstream enzyme PaaZ [14].
BoxC (2,3-epoxybenzoyl-CoA dihydrolase, PaaF and PaaG homolog) and BoxD (3,4-dehydroadipyl-CoA semialdehyde dehydrogenase, PaaZ homolog), the latter containing only an aldehyde dehydrogenase domain, are involved in epoxide rearrangement and ring opening and oxidation. BoxC contains two homologous crotonase fold domains, interacting with each other in a symmetrical manner, although only the N-terminal domain contains an active center [40]. Its active site, hydrophobic in nature, contains two Glu residues positioned in the same locations as in PaaF. This is in line with the proposed BoxC function for conducting two consecutive hydrolysis steps and a variety of reactions, catalyzed by the crotonase protein family [10]. The structure of BoxD [41] revealed a tightly bound homodimer with each monomer made of three domains: Rossmann-fold cofactor binding domain, the catalytic domain and an oligomerization domain. Both PaaZ and BoxD are NADP+-specific enzymes.
The lower part of the Paa pathway is very similar to the fatty acid β-oxidation pathway. In the case of a fatty acid with a double bond, the enoyl-CoA is first isomerized by Δ3-Δ2-enoyl-CoA isomerase (ECI) and Δ3,5-dienoyl-CoA isomerase (DCI), which rearrange the position of one or two conjugated double bonds, correspondingly [42,43,44,45]. The other steps, which involve 2-enoyl-CoA hydratase (ECH), 3-hydroxyacyl-CoA dehydrogenase (HACD) and 3-ketoacyl-CoA thiolase (KACT) activities, may be performed by separate enzymes, multifunctional enzymes or enzyme complexes [32,46,47]. The β-oxidation in bacteria is performed by the αββα heterotetrameric complex (called FOM), where the α-subunit contains domains with 2-enoyl-CoA hydratase (ECH) and 3-hydroxyacyl-CoA dehydrogenase activities (HACD) activities, while the β-subunit is a 3-ketoacyl-CoA thiolase (KACT) [48].
The isomerase PaaG is similar to ECI. The structure of PaaG shows that it possesses only one catalytic glutamate, which is in line with the structure of an active center of isomerases. The shape and properties of the substrate-binding pocket reveal how the enzyme accommodates two structurally diverse substrates to perform two different chemical reactions, the unusual rearrangement of the epoxide to an oxepin, and the rearrangement of the double bond from the Δ3 to the Δ2 position.
The reaction catalyzed by PaaF, hydration of 2,3-dehydroadipyl-CoA, is similar to that performed by ECH and the comparison of their structures shows that the positioning of the two catalytic glutamates is the same in both enzymes. The shape and charge distribution in PaaF show adaptation for binding of the substrate with a negatively-charged carboxyl group as opposed to a fully hydrophobic alkyl moiety for ECH.
Although the architecture of the PaaFG complex bears no similarity to structures of the fatty acid β-oxidation complex FOM [48] and multifunctional enzyme MFE-1 [32], it proves that monofunctional enzymes can also assemble into highly-oligomeric structures with potential functional benefits.
PaaH appears to be a fusion of two hydroxyacyl-CoA dehydrogenase genes and its structure mimics a dimer of the human heart short chain L-3-hydroxyacyl-CoA dehydrogenase [49]. The two fully functional Rossmann-fold domains are connected together via a dimerization domain. However, in the structure of PaaH, only the N-terminal Rossmann-fold domain appears to be fully-functional, while the second Rossmann-fold domain (residues 305–383) is missing the catalytic residues and an NAD(H) binding cleft. By analogy with the human enzyme, the active site is likely to be placed in the cavity between the N-terminal Rossmann-fold domain and a “dimerization” domain. The catalytically important residues His142 and Glu154 are conserved.
Finally, the structure of PaaJ from Thermus Thermophilus (PDB code 1ULQ) is very similar to human mitochondrial acetoacetyl-CoA thiolase (2IB7) ([50] with RMSD of 1.0 Å for common 350 Cα atoms of the two monomers.
5. Conclusions
The phenylacetate (PA) utilization pathway represents the central route for degradation of a variety of organic compounds, whose degradation reactions converge at this pathway. The pathway combines the features of aerobic and anaerobic oxidative metabolic pathways using oxygen to open the aromatic ring and coenzyme A derivatization of phenylacetate. During the last few years the pathway have been characterized and the reactions catalyzed by the Paa enzymes as well as their substrates and products were identified. For many Paa enzymes the structures are known while for the others the structural information can be inferred from the structures of their homologs. Only two protein-protein complexes have been identified among Paa enzymes, that of PaaABCD and PaaFG. The first complex serves an indispensable monooxygenase which inserts one oxygen atom into an aromatic ring to form an epoxide. The same complex has an ability to deoxygenate the epoxide in order to control its intracellular concentration. PaaFG, although being dissimilar to protein complexes and multifunctional enzymes known for a fatty acid β-oxidation pathway, shows a unique assembly of the two enzymes of the crotonase fold with a potential benefit for the speed of the catabolic conversion.
Acknowledgments
This research was supported by the Canadian Institutes for Health Research grant MOP-48370 to Miroslaw Cygler.
Author Contributions
Andrey M. Grishin and Miroslaw Cygler wrote the manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Bugg, T.D.; Ahmad, M.; Hardiman, E.M.; Rahmanpour, R. Pathways for degradation of lignin in bacteria and fungi. Nat. Prod. Rep. 2011, 28, 1883–1896. [Google Scholar] [CrossRef] [PubMed]
- Cao, B.; Nagarajan, K.; Loh, K.C. Biodegradation of aromatic compounds: Current status and opportunities for biomolecular approaches. Appl. Microbiol. Biotechnol. 2009, 85, 207–228. [Google Scholar] [CrossRef] [PubMed]
- Metzler, D. Biochemistry, 2nd ed.; Academic Press: San Diego, CA, USA, 2003. [Google Scholar]
- Fuchs, G.; Boll, M.; Heider, J. Microbial degradation of aromatic compounds—From one strategy to four. Nat. Rev. Microbiol. 2011, 9, 803–816. [Google Scholar] [CrossRef] [PubMed]
- Carmona, M.; Zamarro, M.T.; Blazquez, B.; Durante-Rodriguez, G.; Juarez, J.F.; Valderrama, J.A.; Barragan, M.J.; Garcia, J.L.; Diaz, E. Anaerobic catabolism of aromatic compounds: A genetic and genomic view. Microbiol. Mol. Biol. Rev. 2009, 73, 71–133. [Google Scholar] [CrossRef] [PubMed]
- Fuchs, G. Anaerobic metabolism of aromatic compounds. Ann. N.Y. Acad. Sci. 2008, 1125, 82–99. [Google Scholar] [CrossRef] [PubMed]
- Ferrandez, A.; Minambres, B.; Garcia, B.; Olivera, E.R.; Luengo, J.M.; Garcia, J.L.; Diaz, E. Catabolism of phenylacetic acid in Escherichia coli. Characterization of a new aerobic hybrid pathway. J. Biol. Chem. 1998, 273, 25974–25986. [Google Scholar] [CrossRef] [PubMed]
- Ismail, W.; El-Said Mohamed, M.; Wanner, B.L.; Datsenko, K.A.; Eisenreich, W.; Rohdich, F.; Bacher, A.; Fuchs, G. Functional genomics by NMR spectroscopy. Phenylacetate catabolism in Escherichia coli. Eur. J. Biochem. 2003, 270, 3047–3054. [Google Scholar] [CrossRef] [PubMed]
- Teufel, R.; Mascaraque, V.; Ismail, W.; Voss, M.; Perera, J.; Eisenreich, W.; Haehnel, W.; Fuchs, G. Bacterial phenylalanine and phenylacetate catabolic pathway revealed. Proc. Natl. Acad. Sci. USA 2010, 107, 14390–14395. [Google Scholar] [CrossRef] [PubMed]
- Rather, L.J.; Knapp, B.; Haehnel, W.; Fuchs, G. Coenzyme A-dependent aerobic metabolism of benzoate via epoxide formation. J. Biol. Chem. 2010, 285, 20615–20624. [Google Scholar] [CrossRef] [PubMed]
- Boll, M.; Fuchs, G. Benzoyl-coenzyme A reductase (dearomatizing), a key enzyme of anaerobic aromatic metabolism. ATP dependence of the reaction, purification and some properties of the enzyme from Thauera aromatica strain K172. Eur. J. Biochem. 1995, 234, 921–933. [Google Scholar] [CrossRef] [PubMed]
- Kung, J.W.; Loffler, C.; Dorner, K.; Heintz, D.; Gallien, S.; van Dorsselaer, A.; Friedrich, T.; Boll, M. Identification and characterization of the tungsten-containing class of benzoyl-coenzyme A reductases. Proc. Natl. Acad. Sci. USA 2009, 106, 17687–17692. [Google Scholar] [CrossRef] [PubMed]
- Teufel, R.; Gantert, C.; Voss, M.; Eisenreich, W.; Haehnel, W.; Fuchs, G. Studies on the mechanism of ring hydrolysis in phenylacetate degradation: A metabolic branching point. J. Biol. Chem. 2011, 286, 11021–11034. [Google Scholar] [CrossRef] [PubMed]
- Teufel, R.; Friedrich, T.; Fuchs, G. An oxygenase that forms and deoxygenates toxic epoxide. Nature 2012, 483, 359–362. [Google Scholar] [CrossRef] [PubMed]
- El-Said Mohamed, M. Biochemical and molecular characterization of phenylacetate-coenzyme A ligase, an enzyme catalyzing the first step in aerobic metabolism of phenylacetic acid in Azoarcus evansii. J. Bacteriol. 2000, 182, 286–294. [Google Scholar] [CrossRef] [PubMed]
- Grishin, A.M.; Ajamian, E.; Tao, L.; Zhang, L.; Menard, R.; Cygler, M. Structural and functional studies of the Escherichia coli phenylacetyl-CoA monooxygenase complex. J. Biol. Chem. 2011, 286, 10735–10743. [Google Scholar] [CrossRef] [PubMed]
- Nogales, J.; Macchi, R.; Franchi, F.; Barzaghi, D.; Fernandez, C.; Garcia, J.L.; Bertoni, G.; Diaz, E. Characterization of the last step of the aerobic phenylacetic acid degradation pathway. Microbiology 2007, 153, 357–365. [Google Scholar] [CrossRef] [PubMed]
- Gulick, A.M. Conformational dynamics in the Acyl-CoA synthetases, adenylation domains of non-ribosomal peptide synthetases, and firefly luciferase. ACS chemical biology 2009, 4, 811–827. [Google Scholar] [CrossRef] [PubMed]
- Erb, T.J.; Ismail, W.; Fuchs, G. Phenylacetate metabolism in thermophiles: Characterization of phenylacetate-CoA ligase, the initial enzyme of the hybrid pathway in Thermus thermophilus. Curr. Microbiol. 2008, 57, 27–32. [Google Scholar] [CrossRef] [PubMed]
- Law, A.; Boulanger, M.J. Defining a structural and kinetic rationale for paralogous copies of phenylacetate-CoA ligases from the cystic fibrosis pathogen Burkholderia cenocepacia J2315. J. Biol. Chem. 2011, 286, 15577–15585. [Google Scholar] [CrossRef] [PubMed]
- Imolorhe, I.A.; Cardona, S.T. 3-Hydroxyphenylacetic acid induces the Burkholderia cenocepacia phenylacetic acid degradation pathway—Toward understanding the contribution of aromatic catabolism to pathogenesis. Front. Cell. Infect. Microbio. 2011. [Google Scholar] [CrossRef] [PubMed]
- Fernandez, C.; Ferrandez, A.; Minambres, B.; Diaz, E.; Garcia, J.L. Genetic characterization of the phenylacetyl-coenzyme A oxygenase from the aerobic phenylacetic acid degradation pathway of Escherichia coli. Appl. Environ. Microbiol. 2006, 72, 7422–7426. [Google Scholar] [CrossRef] [PubMed]
- Grishin, A.M.; Ajamian, E.; Zhang, L.; Cygler, M. Crystallization and preliminary X-ray analysis of PaaAC, the main component of the hydroxylase of the Escherichia coli phenylacetyl-coenzyme A oxygenase complex. Acta Crystallogr. Sect. F Struct. Biolo. Cryst. Commun. 2010, 66, 1045–1049. [Google Scholar] [CrossRef] [PubMed]
- Rosenzweig, A.C.; Frederick, C.A.; Lippard, S.J.; Nordlund, P. Crystal structure of a bacterial non-haem iron hydroxylase that catalyses the biological oxidation of methane. Nature 1993, 366, 537–543. [Google Scholar] [CrossRef] [PubMed]
- Grishin, A.M.; Ajamian, E.; Tao, L.; Bostina, M.; Zhang, L.; Trempe, J.F.; Menard, R.; Rouiller, I.; Cygler, M. Family of phenylacetyl-CoA monooxygenases differs in subunit organization from other monooxygenases. J. Struct. Biol. 2013, 184, 147–154. [Google Scholar] [CrossRef] [PubMed]
- Correll, C.C.; Batie, C.J.; Ballou, D.P.; Ludwig, M.L. Phthalate dioxygenase reductase: A modular structure for electron transfer from pyridine nucleotides to [2Fe-2S]. Science 1992, 258, 1604–1610. [Google Scholar] [CrossRef] [PubMed]
- Phillips, D.H.; Farmer, P.B. Evidence for DNA and protein binding by styrene and styrene oxide. Crit. Rev. Toxicol. 1994, 24, S35–S46. [Google Scholar] [CrossRef] [PubMed]
- Park, J.B.; Buhler, B.; Habicher, T.; Hauer, B.; Panke, S.; Witholt, B.; Schmid, A. The efficiency of recombinant Escherichia coli as biocatalyst for stereospecific epoxidation. Biotechnol. Bioeng. 2006, 95, 501–512. [Google Scholar] [CrossRef] [PubMed]
- Law, R.J.; Hamlin, J.N.; Sivro, A.; McCorrister, S.J.; Cardama, G.A.; Cardona, S.T. A functional phenylacetic acid catabolic pathway is required for full pathogenicity of Burkholderia cenocepacia in the Caenorhabditis elegans host model. J. Bacteriol. 2008, 190, 7209–7218. [Google Scholar] [CrossRef] [PubMed]
- Song, F.; Zhuang, Z.; Finci, L.; Dunaway-Mariano, D.; Kniewel, R.; Buglino, J.A.; Solorzano, V.; Wu, J.; Lima, C.D. Structure, function, and mechanism of the phenylacetate pathway hot dog-fold thioesterase PaaI. J. Biol. Chem. 2006, 281, 11028–11038. [Google Scholar] [CrossRef] [PubMed]
- Fernandez, C.; Diaz, E.; Garcia, J.L. Insights on the regulation of the phenylacetate degradation pathway from Escherichia coli. Environ. Microbiol. Rep. 2014, 6, 239–250. [Google Scholar] [CrossRef] [PubMed]
- Kasaragod, P.; Venkatesan, R.; Kiema, T.R.; Hiltunen, J.K.; Wierenga, R.K. Crystal structure of liganded rat peroxisomal multifunctional enzyme type 1: A flexible molecule with two interconnected active sites. J. Biol. Chem. 2010, 285, 24089–24098. [Google Scholar] [CrossRef] [PubMed]
- Grishin, A.M.; Ajamian, E.; Zhang, L.; Rouiller, I.; Bostina, M.; Cygler, M. Protein-protein interactions in the beta-oxidation part of the phenylacetate utilization pathway: Crystal structure of the PaaF–PaaG hydratase-isomerase complex. J. Biol. Chem. 2012, 287, 37986–37996. [Google Scholar] [CrossRef] [PubMed]
- Hamed, R.B.; Batchelar, E.T.; Clifton, I.J.; Schofield, C.J. Mechanisms and structures of crotonase superfamily enzymes—How nature controls enolate and oxyanion reactivity. Cell. Mol. Life Sci. 2008, 65, 2507–2527. [Google Scholar] [CrossRef] [PubMed]
- Finn, R.D.; Bateman, A.; Clements, J.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R.; Heger, A.; Hetherington, K.; Holm, L.; Mistry, J.; et al. Pfam: The protein families database. Nucleic Acids Res. 2014, 42, D222–D230. [Google Scholar] [CrossRef] [PubMed]
- Ferrandez, A.; Garcia, J.L.; Diaz, E. Transcriptional regulation of the divergent paa catabolic operons for phenylacetic acid degradation in Escherichia coli. J. Biol. Chem. 2000, 275, 12214–12222. [Google Scholar] [CrossRef] [PubMed]
- Rojas-Altuve, A.; Carrasco-Lopez, C.; Hernandez-Rocamora, V.M.; Sanz, J.M.; Hermoso, J.A. Crystallization and preliminary X-ray diffraction studies of the transcriptional repressor PaaX, the main regulator of the phenylacetic acid degradation pathway in Escherichia coli W. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2011, 67, 1278–1280. [Google Scholar] [CrossRef] [PubMed]
- Rather, L.J.; Weinert, T.; Demmer, U.; Bill, E.; Ismail, W.; Fuchs, G.; Ermler, U. Structure and mechanism of the diiron benzoyl-coenzyme A epoxidase BoxB. J. Biol. Chem. 2011, 286, 29241–29248. [Google Scholar] [CrossRef] [PubMed]
- Zaar, A.; Gescher, J.; Eisenreich, W.; Bacher, A.; Fuchs, G. New enzymes involved in aerobic benzoate metabolism in Azoarcus evansii. Mol. Microbiol. 2004, 54, 223–238. [Google Scholar] [CrossRef] [PubMed]
- Bains, J.; Leon, R.; Boulanger, M.J. Structural and biophysical characterization of BoxC from Burkholderia xenovorans LB400: A novel ring-cleaving enzyme in the crotonase superfamily. J. Biol. Chem. 2009, 284, 16377–16385. [Google Scholar] [CrossRef] [PubMed]
- Bains, J.; Boulanger, M.J. Structural and biochemical characterization of a novel aldehyde dehydrogenase encoded by the benzoate oxidation pathway in Burkholderia xenovorans LB400. J. Mol. Biol. 2008, 379, 597–608. [Google Scholar] [CrossRef] [PubMed]
- Mursula, A.M.; Hiltunen, J.K.; Wierenga, R.K. Structural studies on delta3-delta2-enoyl-CoA isomerase: The variable mode of assembly of the trimeric disks of the crotonase superfamily. FEBS Lett. 2004, 557, 81–87. [Google Scholar] [CrossRef]
- Partanen, S.T.; Novikov, D.K.; Popov, A.N.; Mursula, A.M.; Hiltunen, J.K.; Wierenga, R.K. The 1.3 Å crystal structure of human mitochondrial Delta3-Delta2-enoyl-CoA isomerase shows a novel mode of binding for the fatty acyl group. J. Mol. Biol. 2004, 342, 1197–1208. [Google Scholar] [CrossRef] [PubMed]
- Mursula, A.M.; van Aalten, D.M.; Hiltunen, J.K.; Wierenga, R.K. The crystal structure of delta3-delta2-enoyl-CoA isomerase. J. Mol. Biol. 2001, 309, 845–853. [Google Scholar] [CrossRef] [PubMed]
- Modis, Y.; Filppula, S.A.; Novikov, D.K.; Norledge, B.; Hiltunen, J.K.; Wierenga, R.K. The crystal structure of dienoyl-CoA isomerase at 1.5 Å resolution reveals the importance of aspartate and glutamate sidechains for catalysis. Structure 1998, 6, 957–970. [Google Scholar] [CrossRef]
- Engel, C.K.; Kiema, T.R.; Hiltunen, J.K.; Wierenga, R.K. The crystal structure of enoyl-CoA hydratase complexed with octanoyl-CoA reveals the structural adaptations required for binding of a long chain fatty acid-CoA molecule. J. Mol. Biol. 1998, 275, 847–859. [Google Scholar] [CrossRef] [PubMed]
- Engel, C.K.; Mathieu, M.; Zeelen, J.P.; Hiltunen, J.K.; Wierenga, R.K. Crystal structure of enoyl-coenzyme A (CoA) hydratase at 2.5 angstroms resolution: A spiral fold defines the CoA-binding pocket. EMBO J. 1996, 15, 5135–5145. [Google Scholar] [PubMed]
- Ishikawa, M.; Tsuchiya, D.; Oyama, T.; Tsunaka, Y.; Morikawa, K. Structural basis for channelling mechanism of a fatty acid beta-oxidation multienzyme complex. EMBO J. 2004, 23, 2745–2754. [Google Scholar] [CrossRef] [PubMed]
- Barycki, J.J.; O’Brien, L.K.; Bratt, J.M.; Zhang, R.; Sanishvili, R.; Strauss, A.W.; Banaszak, L.J. Biochemical characterization and crystal structure determination of human heart short chain L-3-hydroxyacyl-CoA dehydrogenase provide insights into catalytic mechanism. Biochemistry 1999, 38, 5786–5798. [Google Scholar] [CrossRef] [PubMed]
- Haapalainen, A.M.; Merilainen, G.; Pirila, P.L.; Kondo, N.; Fukao, T.; Wierenga, R.K. Crystallographic and kinetic studies of human mitochondrial acetoacetyl-CoA thiolase: The importance of potassium and chloride ions for its structure and function. Biochemistry 2007, 46, 4305–4321. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).