Development of a Functionally Minimized Mutant of the R3C Ligase Ribozyme Offers Insight into the Plausibility of the RNA World Hypothesis
Abstract
:1. Introduction
2. Experimental
2.1. Plasmid Construction and in Vitro Transcription
2.2. Analysis of Ligation
2.3. CD Spectroscopy
2.4. Prediction of RNA Secondary Structures
3. Results
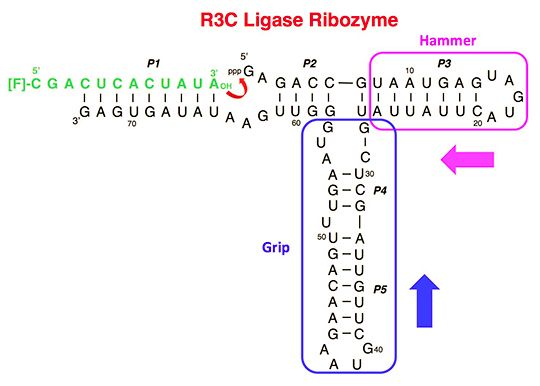
3.1. Ligation Activities of R3C Ribozymes and of “Hammer”- or “Grip”-Completely Deleted Mutant
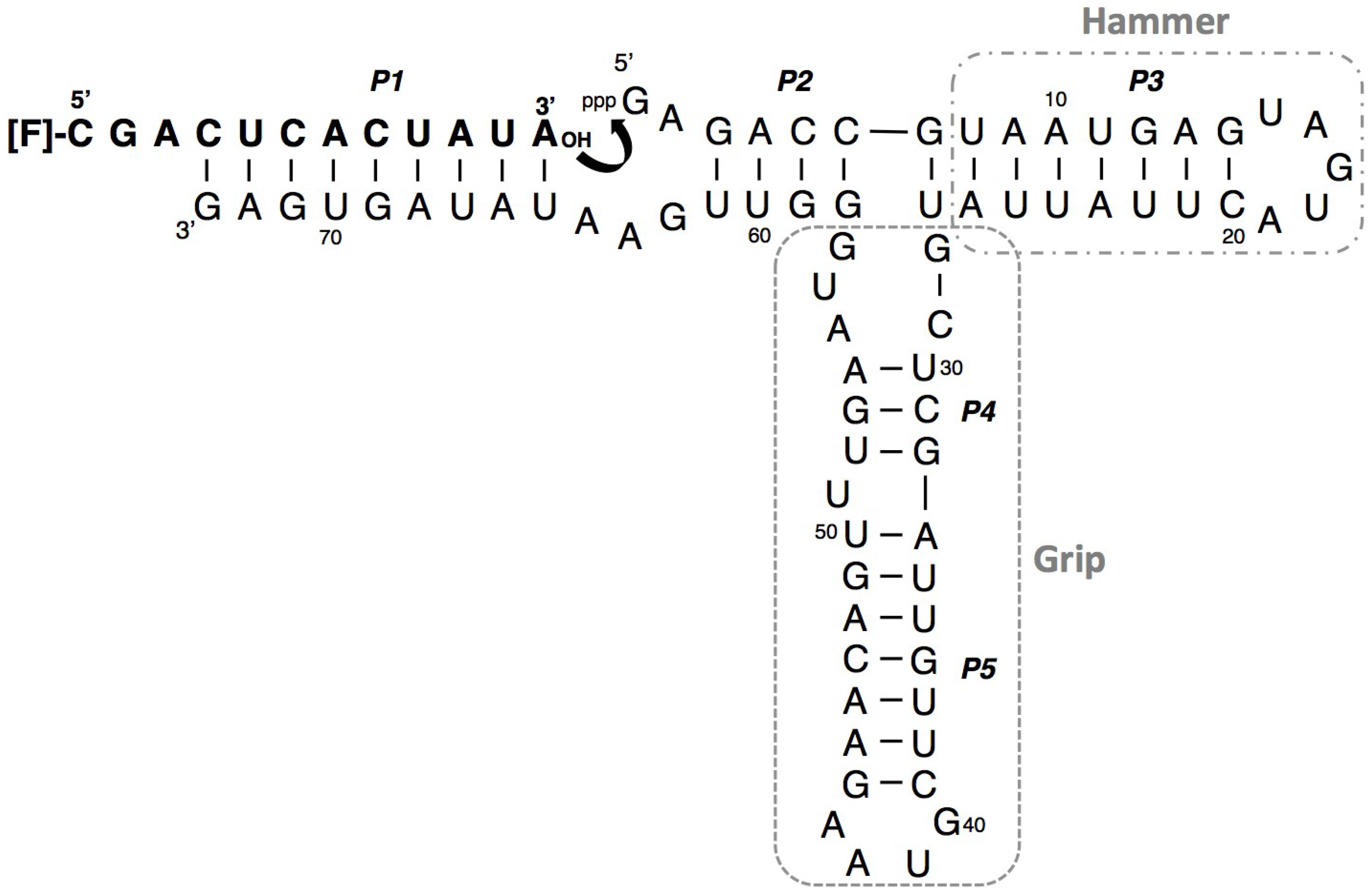
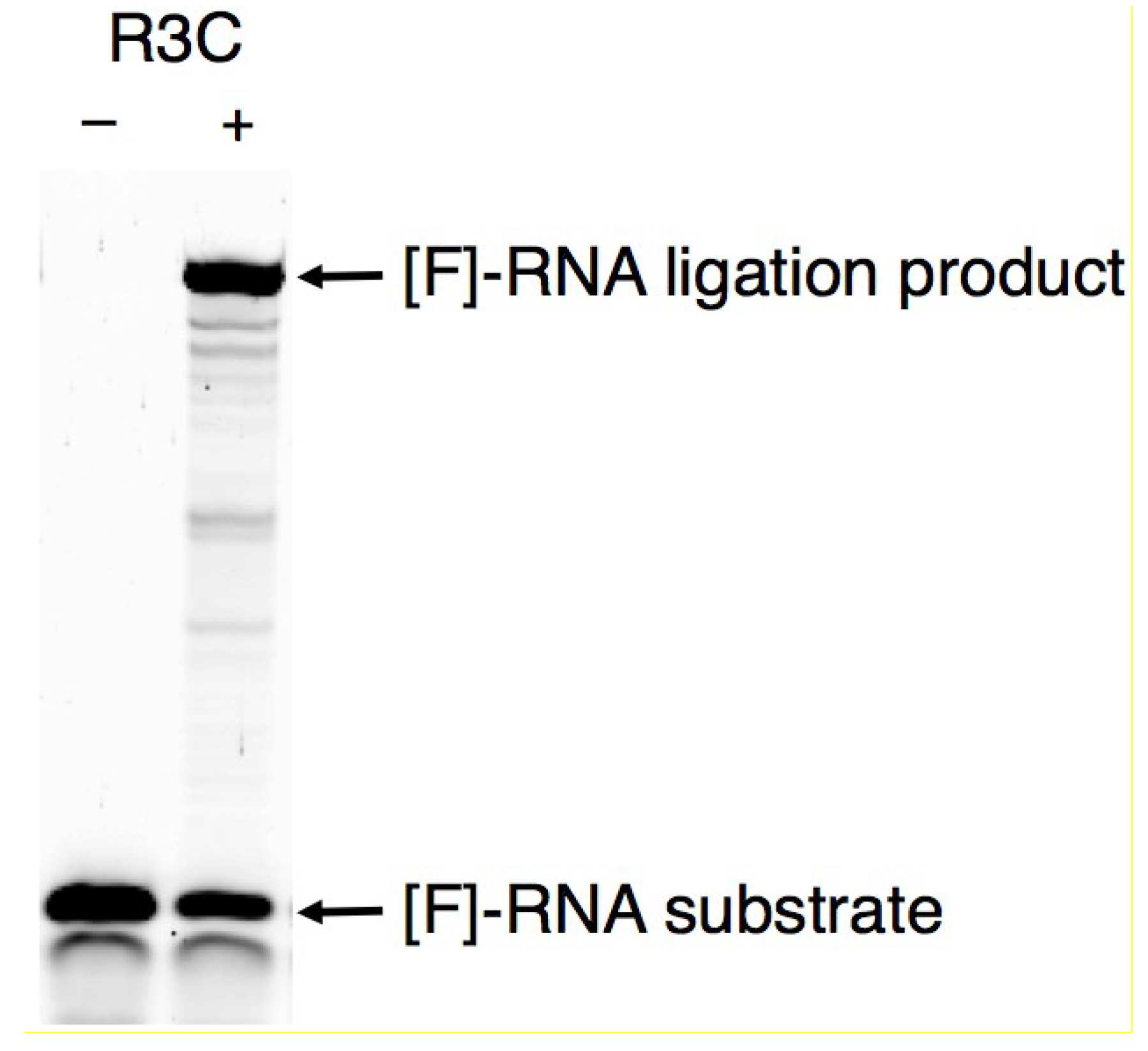
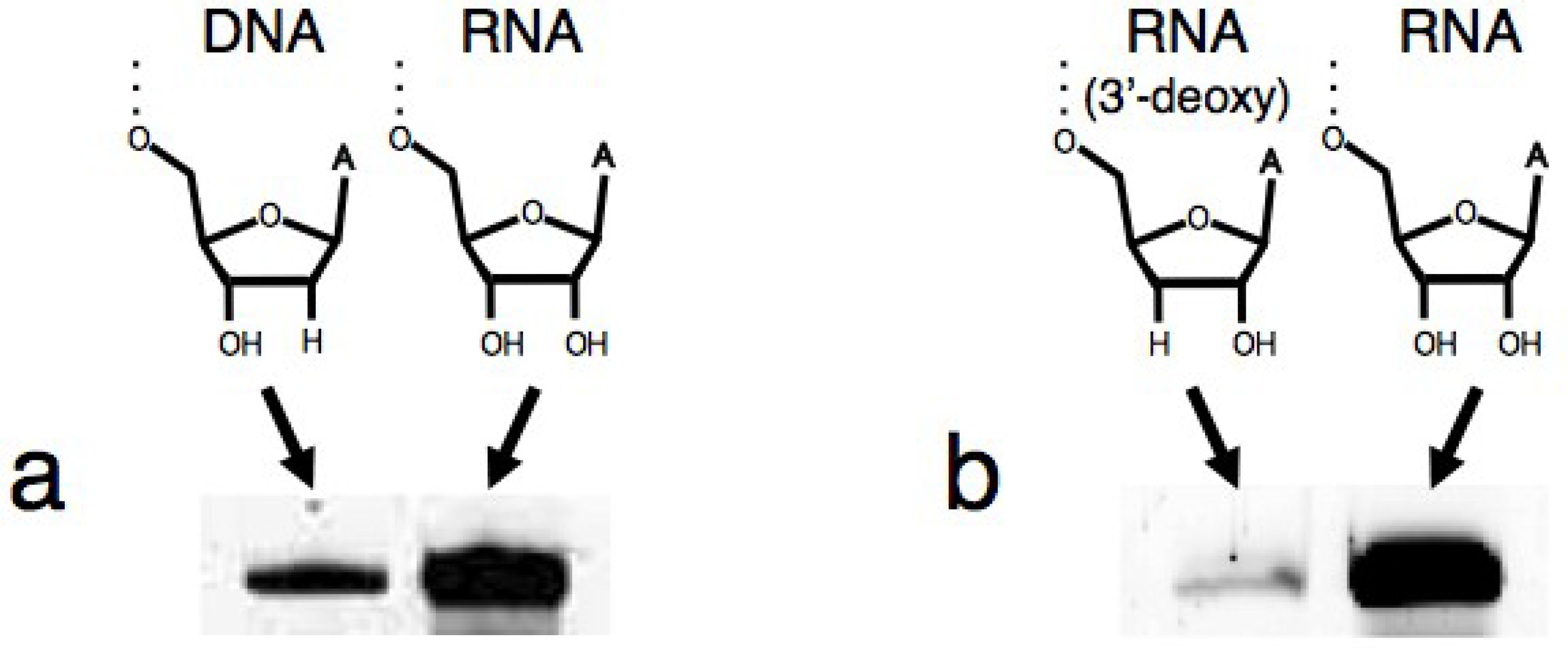
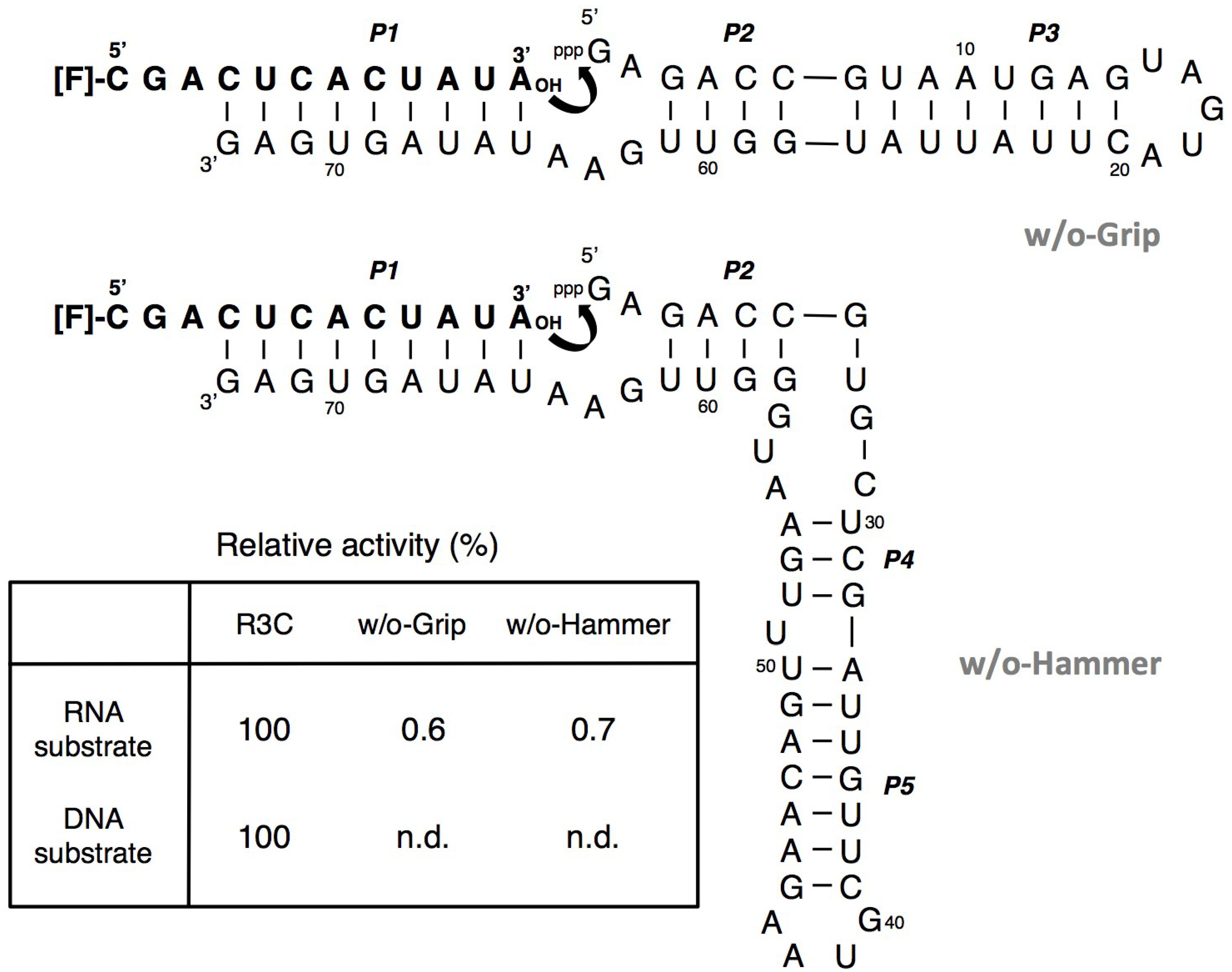
3.2. Ligation Activities of R3C “Grip”-Partially Deleted Mutants

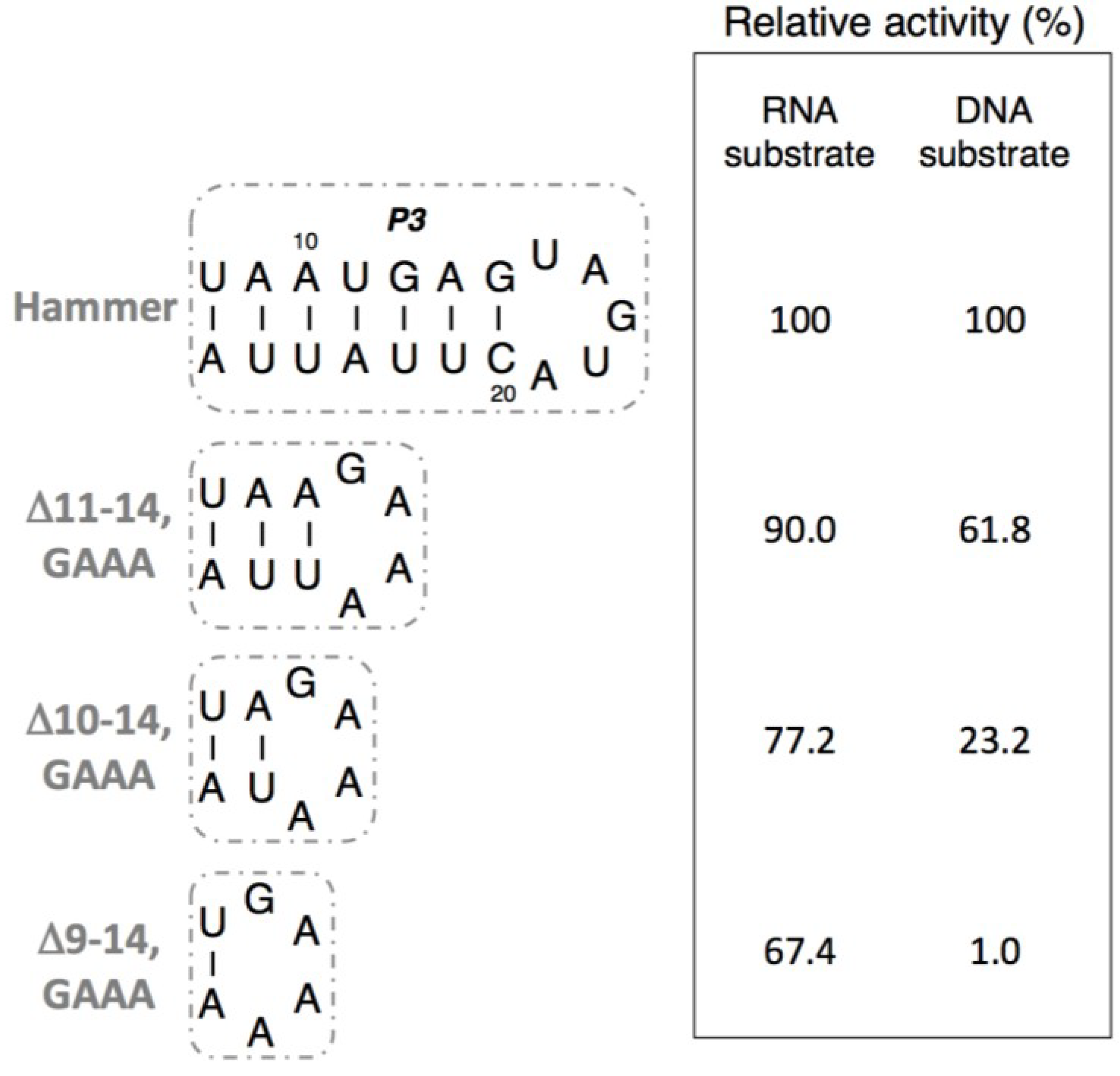
3.3. Ligation Activities of R3C “Hammer”-Partially Deleted Mutants
3.4. Ligation Activities of R3C “Grip” and “Hammer”-Double Partially Deleted Mutants

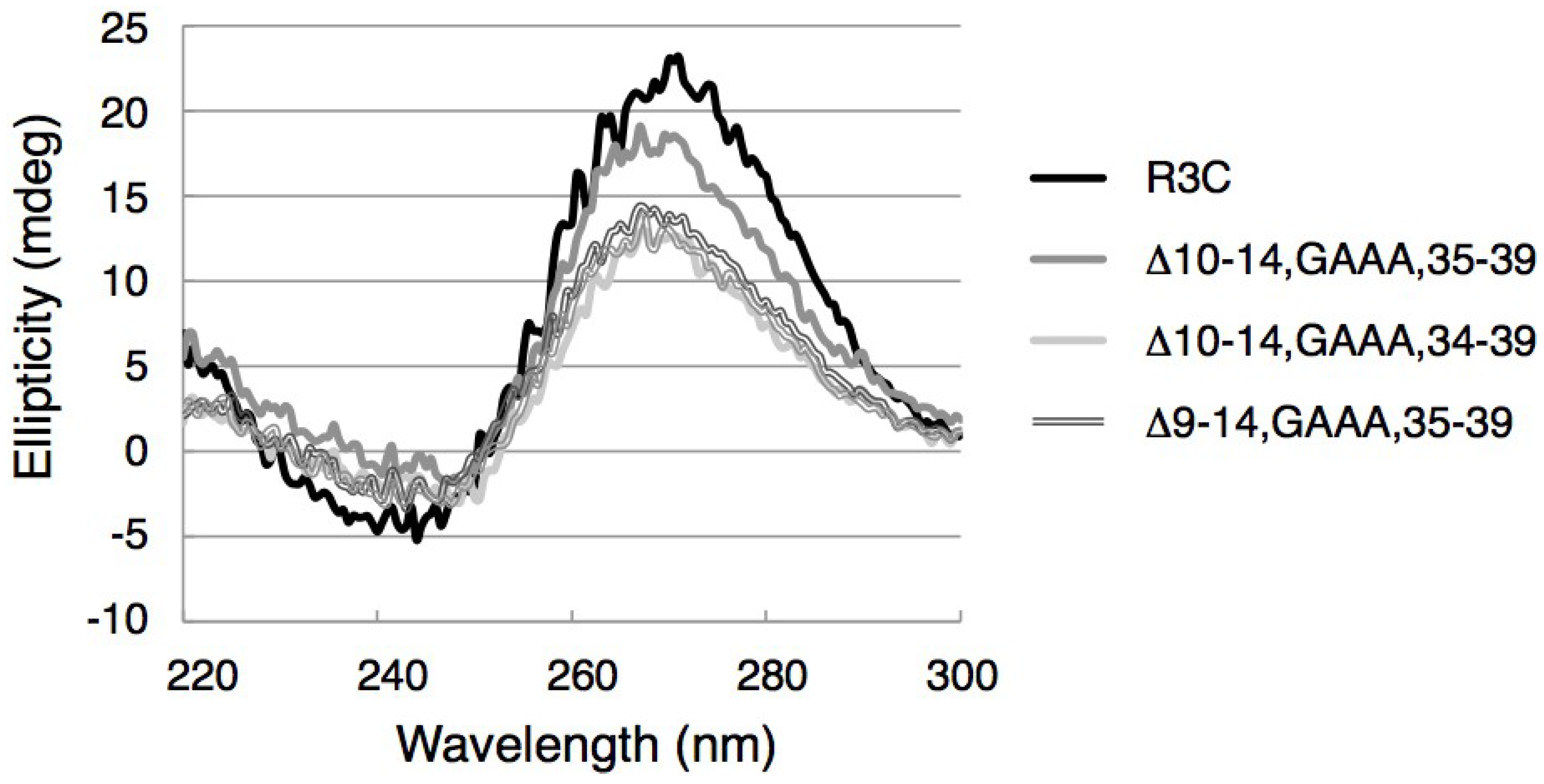
3.5. CD Spectra of the Ligase Ribozymes
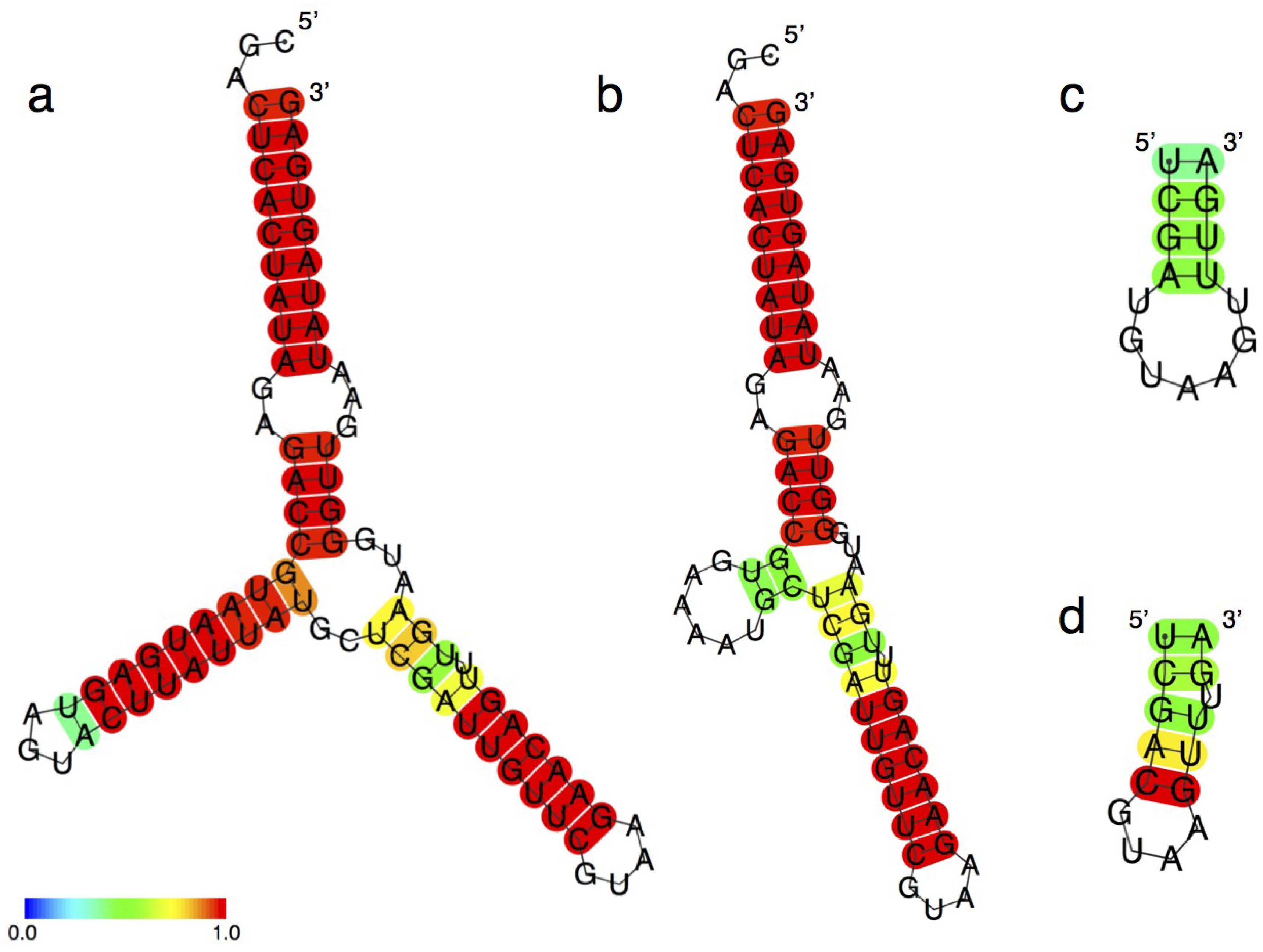
3.6. Secondary Structure Prediction of the Ligase Ribozymes
4. Discussion
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Crick, F.H.C. On protein synthesis. Symp. Soc. Exp. Biol. 1958, 12, 139–163. [Google Scholar]
- Kruger, K.; Grabowski, P.J.; Zaug, A.J.; Sands, J.; Gottschling, D.E.; Cech, T.R. Self-splicing RNA: Autoexcision and autocyclization of the ribosomal RNA intervening sequence of Tetrahymena. Cell 1982, 31, 147–157. [Google Scholar] [CrossRef] [PubMed]
- Guerrier-Takada, C.; Gardiner, K.; Marsh, T.; Pace, N.; Altman, S. The RNA moiety of ribonuclease P is the catalytic subunit of the enzyme. Cell 1983, 35, 849–857. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, W. Origin of life: The RNA world. Nature 1986, 319, 618. [Google Scholar] [CrossRef]
- Joyce, G.F.; Visser, G.M.; van Boeckel, C.A.A.; van Boom, J.H.; Orgel, L.E.; van Westrenen, J. Chiral selection in poly(C)-directed synthesis of oligo(G). Nature 1984, 310, 602–604. [Google Scholar] [CrossRef] [PubMed]
- Bolli, M.; Micura, R.; Eschenmoser, A. Pyranosyl-RNA: Chiroselective self-assembly of base sequences by ligative oligomerization of tetranucleotide-2',3'-cyclophosphates (with a commentary concerning the origin of biomolecular homochirality). Chem. Biol. 1997, 4, 309–320. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Schimmel, P. Chiral-selective aminoacylation of an RNA minihelix. Science 2004, 305, 1253. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Schimmel, P.R. Chiral-selective aminoacylation of an RNA minihelix: Mechanistic features and chiral suppression. Proc. Natl. Acad. Sci. USA 2006, 103, 13750–13752. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K. Origin of amino acid homochirality: Relationship with the RNA world and origin of tRNA aminoacylation. BioSystems 2008, 92, 91–98. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K. Molecular handedness of life: Significance of RNA aminoacylation. J. Biosci. 2009, 34, 991–994. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K. Amino acid homochirality and the RNA world: Necessities for life on Earth. J. Cosmol. 2010, 5, 883–889. [Google Scholar]
- Tamura, K. Molecular basis for chiral selection in RNA aminoacylation. Int. J. Mol. Sci. 2011, 12, 4745–4757. [Google Scholar] [CrossRef] [PubMed]
- Joyce, G.F. Forty years of in vitro evolution. Angew. Chem. Int. Ed. Engl. 2007, 46, 6420–6436. [Google Scholar] [CrossRef] [PubMed]
- Ekland, E.H.; Szostak, J.W.; Bartel, D.P. Structurally complex and highly active RNA ligases derived from random RNA sequences. Science 1995, 269, 364–370. [Google Scholar] [CrossRef] [PubMed]
- Robertson, M.P.; Ellington, A.D. In vitro selection of an allosteric ribozyme that transduces analytes to amplicons. Nat. Biotechnol. 1999, 17, 62–66. [Google Scholar] [CrossRef] [PubMed]
- Rogers, J.; Joyce, G.F. The effect of cytidine on the structure and function of an RNA ligase ribozyme. RNA 2001, 7, 395–404. [Google Scholar] [CrossRef] [PubMed]
- Shechner, D.M.; Grant, R.A.; Bagby, S.C.; Koldobskaya, Y.; Piccirilli, J.A.; Bartel, D.P. Crystal structure of the catalytic core of an RNA-polymerase ribozyme. Science 2009, 326, 1271–1275. [Google Scholar] [CrossRef] [PubMed]
- Shechner, D.M.; Bartel, D.P. The structural basis of RNA-catalyzed RNA polymerization. Nat. Struct. Mol. Biol. 2011, 18, 1036–1042. [Google Scholar] [CrossRef] [PubMed]
- Ekland, E.H.; Bartel, D.P. The secondary structure and sequence optimization of an RNA ligase ribozyme. Nucl. Acids Res. 1995, 23, 3231–3238. [Google Scholar] [CrossRef] [PubMed]
- Rogers, J.; Joyce, G.F. A ribozyme that lacks cytidine. Nature 1999, 402, 323–325. [Google Scholar] [CrossRef] [PubMed]
- Jaeger, L.; Wright, M.C.; Joyce, G.F. A complex ligase ribozyme evolved in vitro from a group I ribozyme domain. Proc. Natl. Acad. Sci. USA 1999, 96, 14712–14717. [Google Scholar] [CrossRef] [PubMed]
- Ikawa, Y.; Tsuda, K.; Matsumura, S.; Inoue, T. De novo synthesis and development of an RNA enzyme. Proc. Natl. Acad. Sci. USA 2004, 101, 13750–13755. [Google Scholar] [CrossRef] [PubMed]
- Robertson, M.P.; Hesselberth, J.R.; Ellington, A.D. Optimization and optimality of a short ribozyme ligase that joins non-Watson-Crick base pairings. RNA 2001, 7, 513–523. [Google Scholar]
- Robertson, M.P.; Scott, W.G. The structural basis of ribozyme-catalyzed RNA assembly. Science 2007, 315, 1549–1553. [Google Scholar] [CrossRef] [PubMed]
- Eigen, M.; Schuster, P. Hypercycle. A principle of natural self-organization. Part A: Emergence of the hypercycle. Naturwissenschaften 1977, 64, 541–565. [Google Scholar]
- Ferris, J.P. Montmorillonite-catalysed formation of RNA oligomers: The possible role of catalysis in the origins of life. Phil. Trans. R. Soc. Lond. B Biol. Sci. 2006, 361, 1777–1786. [Google Scholar] [CrossRef]
- Paul, N.; Joyce, G.F. A self-replicating ligase ribozyme. Proc. Natl. Acad. Sci. USA 2002, 99, 12733–12740. [Google Scholar] [CrossRef] [PubMed]
- Lincoln, T.A.; Joyce, G.F. Self-sustained replication of an RNA enzyme. Science 2009, 323, 1229–1232. [Google Scholar] [CrossRef] [PubMed]
- Hamachi, K.; Hayashi, H.; Shimamura, M.; Yamaji, Y.; Kaneko, A.; Fujisawa, A.; Umehara, T.; Tamura, K. Glycols modulate terminator stem stability and ligand-dependency of a glycine riboswitch. BioSystems 2013, 113, 59–65. [Google Scholar] [CrossRef] [PubMed]
- Umehara, T.; Kitagawa, T.; Nakazawa, Y.; Yoshino, H.; Nemoto, R.; Tamura, K. RNA tetraplex as a primordial peptide synthesis scaffold. BioSystems 2012, 109, 145–150. [Google Scholar] [CrossRef] [PubMed]
- Hamada, M.; Kiryu, H.; Sato, K.; Mituyama, T.; Asai, K. Prediction of RNA secondary structure using generalized centroid estimators. Bioinformatics 2009, 25, 465–473. [Google Scholar] [CrossRef] [PubMed]
- Sato, K.; Hamada, M.; Asai, K.; Mituyama, T. CentroidFold: A web server for RNA secondary structure prediction. Nucleic Acids Res. 2009, 37, W277–W280. [Google Scholar] [CrossRef] [PubMed]
- CentroidFold-ncRNA.org. Available online: http://www.ncrna.org/centroidfold/ (accessed on 27 May 2014).
- Arnott, S.; Chandrasekaran, R.; Millane, R.P.; Park, H.-S. DNA-RNA hybrid secondary structures. J. Mol. Biol. 1986, 188, 631–640. [Google Scholar] [CrossRef] [PubMed]
- Borer, P.M.; Dengler, B.; Tinoco, I., Jr.; Uhlenbeck, O.C. Stability of ribonucleic acid double-stranded helices. J. Mol. Biol. 1974, 86, 843–853. [Google Scholar] [CrossRef] [PubMed]
- Breslauer, K.J.; Frank, R.; Blocker, H.; Marky, L.A. Predicting DNA duplex stability from the base sequence. Proc. Natl. Acad. Sci. USA 1986, 83, 3746–3750. [Google Scholar] [CrossRef] [PubMed]
- Freier, A.M.; Keirzek, R.; Jaeger, J.A.; Sugimoto, N.; Caruthers, M.H.; Neilson, T.; Turner, D.H. Improved free-energy parameters for predictions of RNA duplex stability. Proc. Natl. Acad. Sci. USA 1986, 83, 9373–9377. [Google Scholar] [CrossRef] [PubMed]
- Sugimoto, N.; Nakano, S.; Katoh, M.; Matsumura, A.; Nakamuta, H.; Ohmichi, T.; Yoneyama, M.; Sasaki, M. Thermodynamic parameters to predict stability of RNA/DNA hybrid duplexes. Biochemistry 1995, 34, 11211–11216. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.T.; Samejima, T. Optical rotatory dispersion and circular dichroism of nucleic acids. In Progress in Nucleic Acid Research and Molecular Biology; Davidson, J.N., Cohn, W.E., Eds.; Academic Press: New York, NY, USA, 1969; Volume 9, pp. 224–300. [Google Scholar]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Kurihara, E.; Uchida, S.; Umehara, T.; Tamura, K. Development of a Functionally Minimized Mutant of the R3C Ligase Ribozyme Offers Insight into the Plausibility of the RNA World Hypothesis. Biology 2014, 3, 452-465. https://doi.org/10.3390/biology3030452
Kurihara E, Uchida S, Umehara T, Tamura K. Development of a Functionally Minimized Mutant of the R3C Ligase Ribozyme Offers Insight into the Plausibility of the RNA World Hypothesis. Biology. 2014; 3(3):452-465. https://doi.org/10.3390/biology3030452
Chicago/Turabian StyleKurihara, Eri, Sayuri Uchida, Takuya Umehara, and Koji Tamura. 2014. "Development of a Functionally Minimized Mutant of the R3C Ligase Ribozyme Offers Insight into the Plausibility of the RNA World Hypothesis" Biology 3, no. 3: 452-465. https://doi.org/10.3390/biology3030452
APA StyleKurihara, E., Uchida, S., Umehara, T., & Tamura, K. (2014). Development of a Functionally Minimized Mutant of the R3C Ligase Ribozyme Offers Insight into the Plausibility of the RNA World Hypothesis. Biology, 3(3), 452-465. https://doi.org/10.3390/biology3030452