Identification of the Dof Gene Family in Quinoa and Its Potential Role in Regulating Flavonoid Synthesis Under Different Stress Conditions
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material and Abiotic Stress Treatments
2.2. Identification and Basic Information Analysis of CqDofs
2.3. Multiple Sequence Alignment, Phylogenetic Relationship, and Conserved Domain Analysis of CqDofs
2.4. Chromosomal Localization, Gene Duplication Events, and Collinearity Analysis of CqDofs
2.5. Gene Structure, cis-Acting Element, and Gene Expression Analysis of CqDofs
2.6. RT-PCR Analysis of CqDofs Under Abiotic Stress
2.7. Sample Preparation and LC–MS Analysis
2.8. Statistical Analysis
3. Results
3.1. Identification, Physicochemical Properties, and Chromosomal Locations Analysis of CqDof Genes in Quinoa
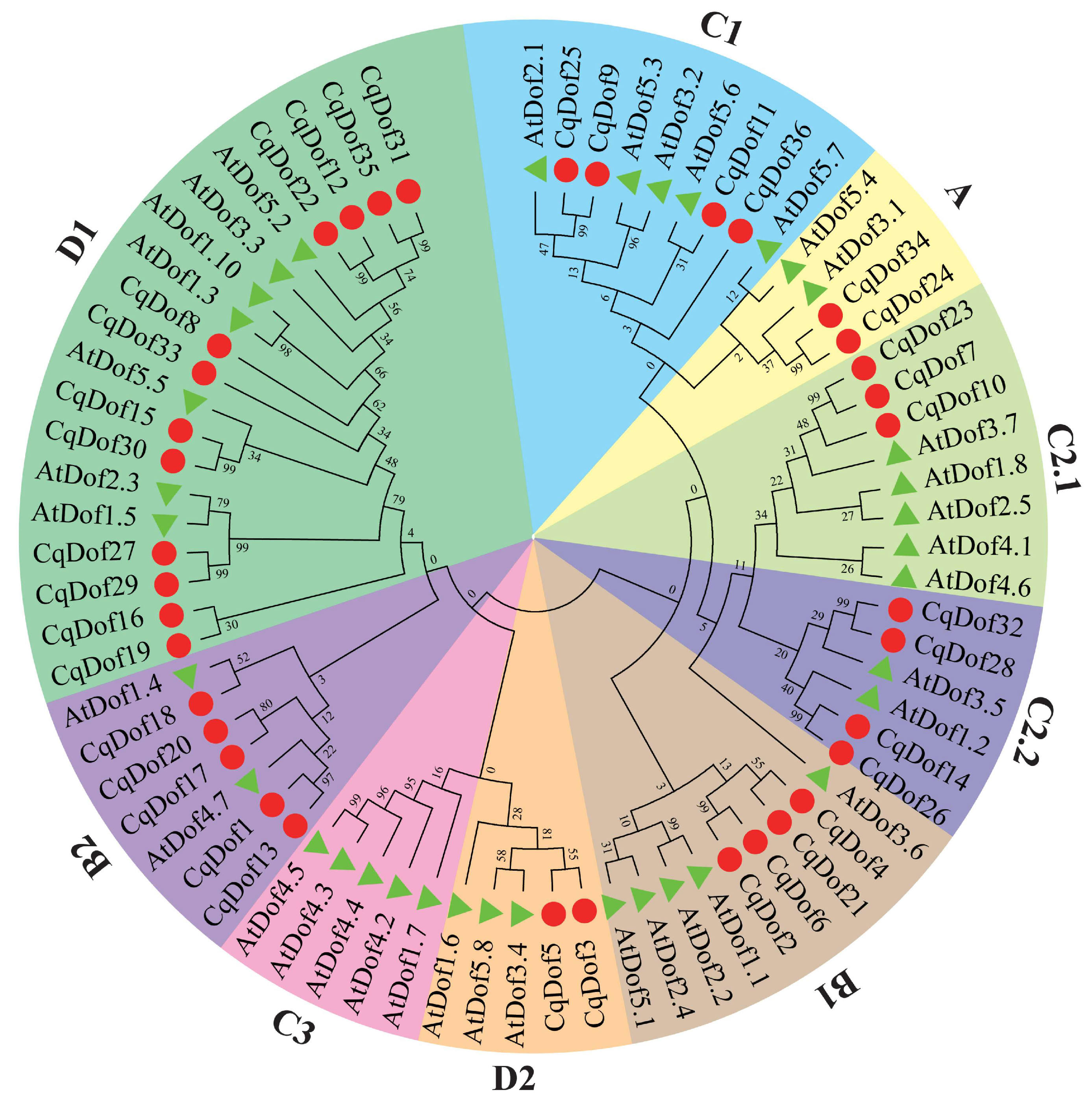
3.2. Phylogenetic Relationship and Classification of CqDof Genes in Quinoa
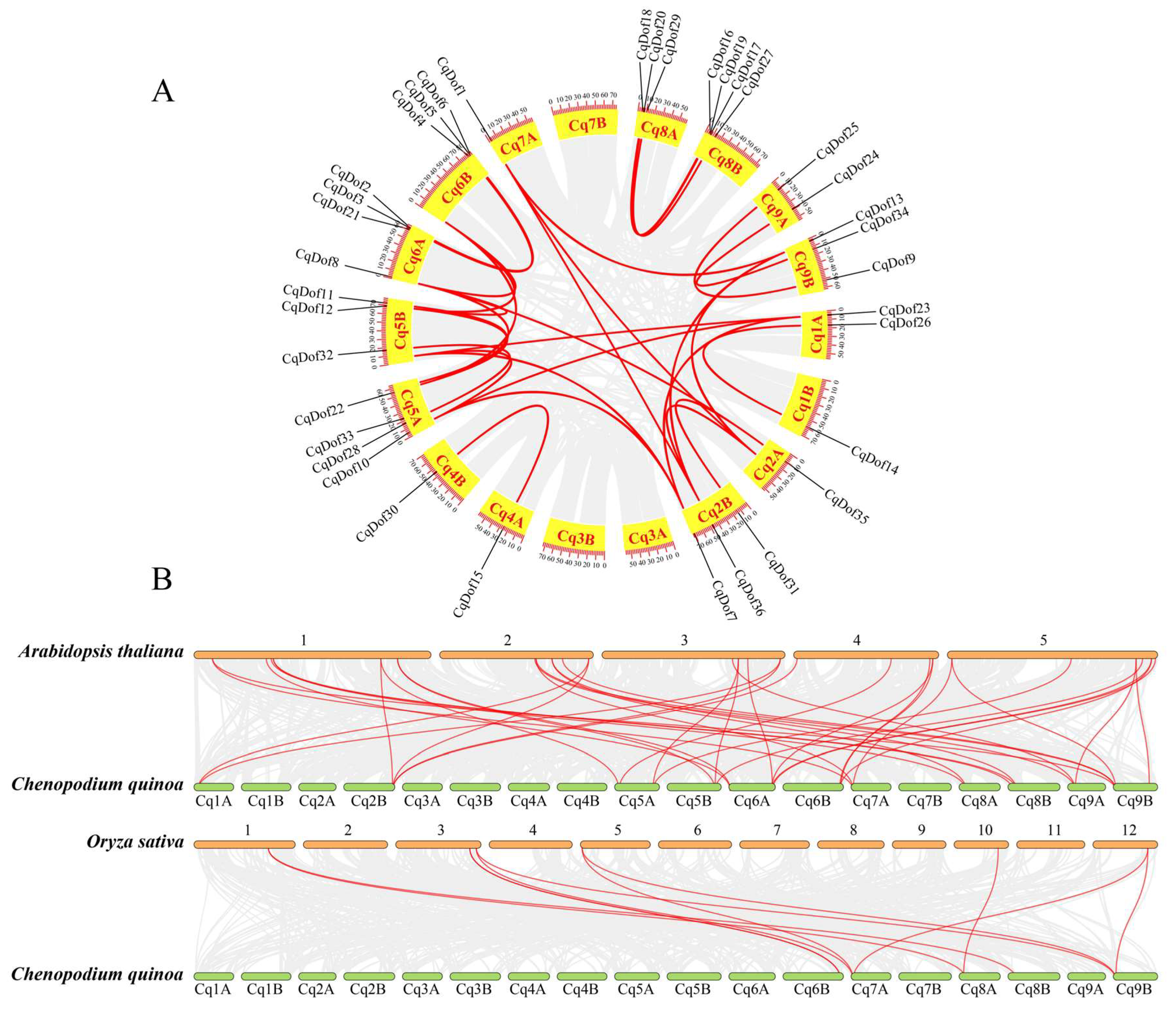
3.3. Gene Duplication and Syntenic Analysis of CqDof Genes in Quinoa
3.4. Gene Structure, Conserved Domain, and Motif Analysis of the CqDof Genes
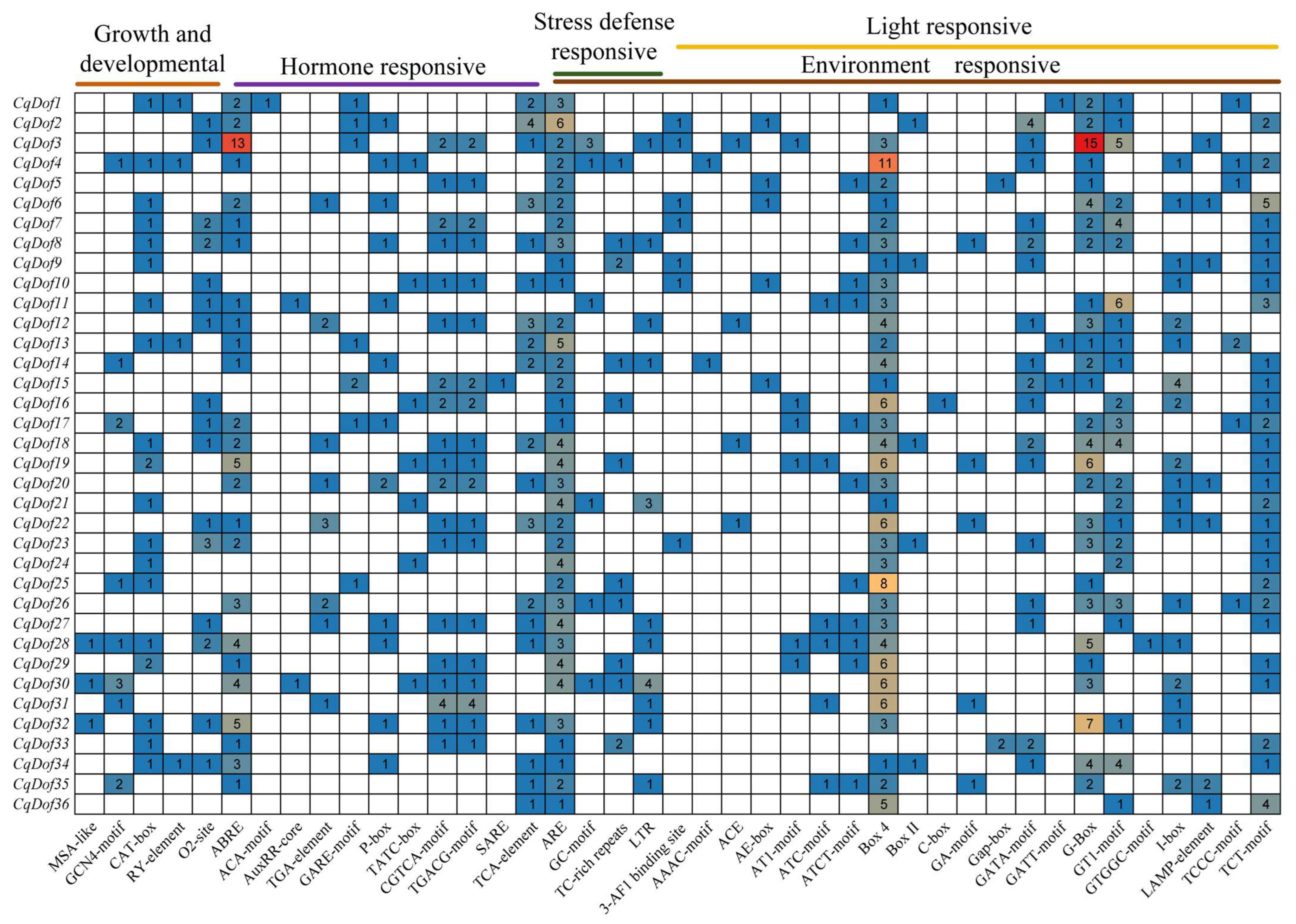
3.5. Promoter cis-Acting Element Analysis of the CqDof Genes
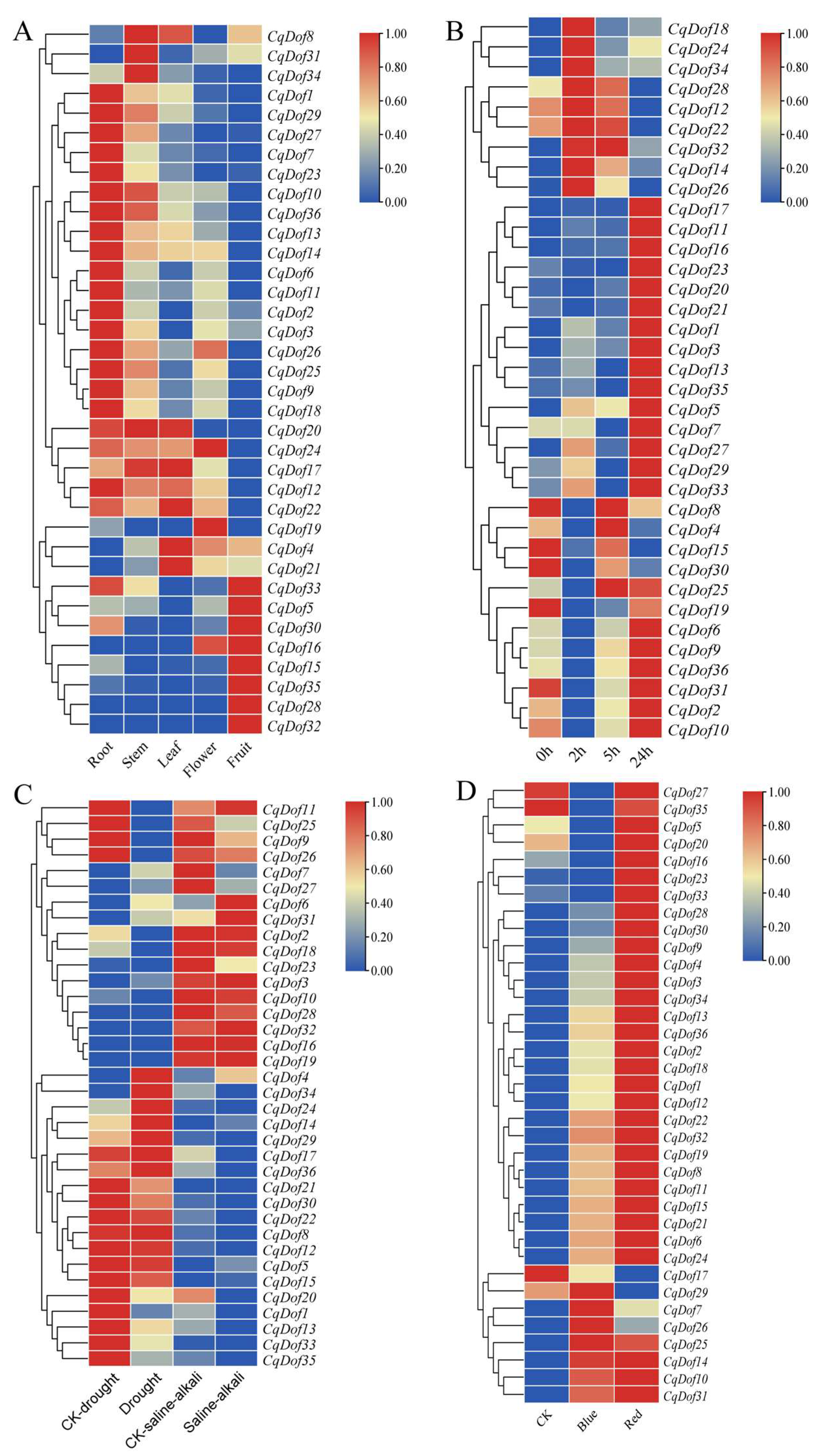
3.6. Expression Pattern of the CqDof Genes in Various Tissues Under Abiotic Stresses
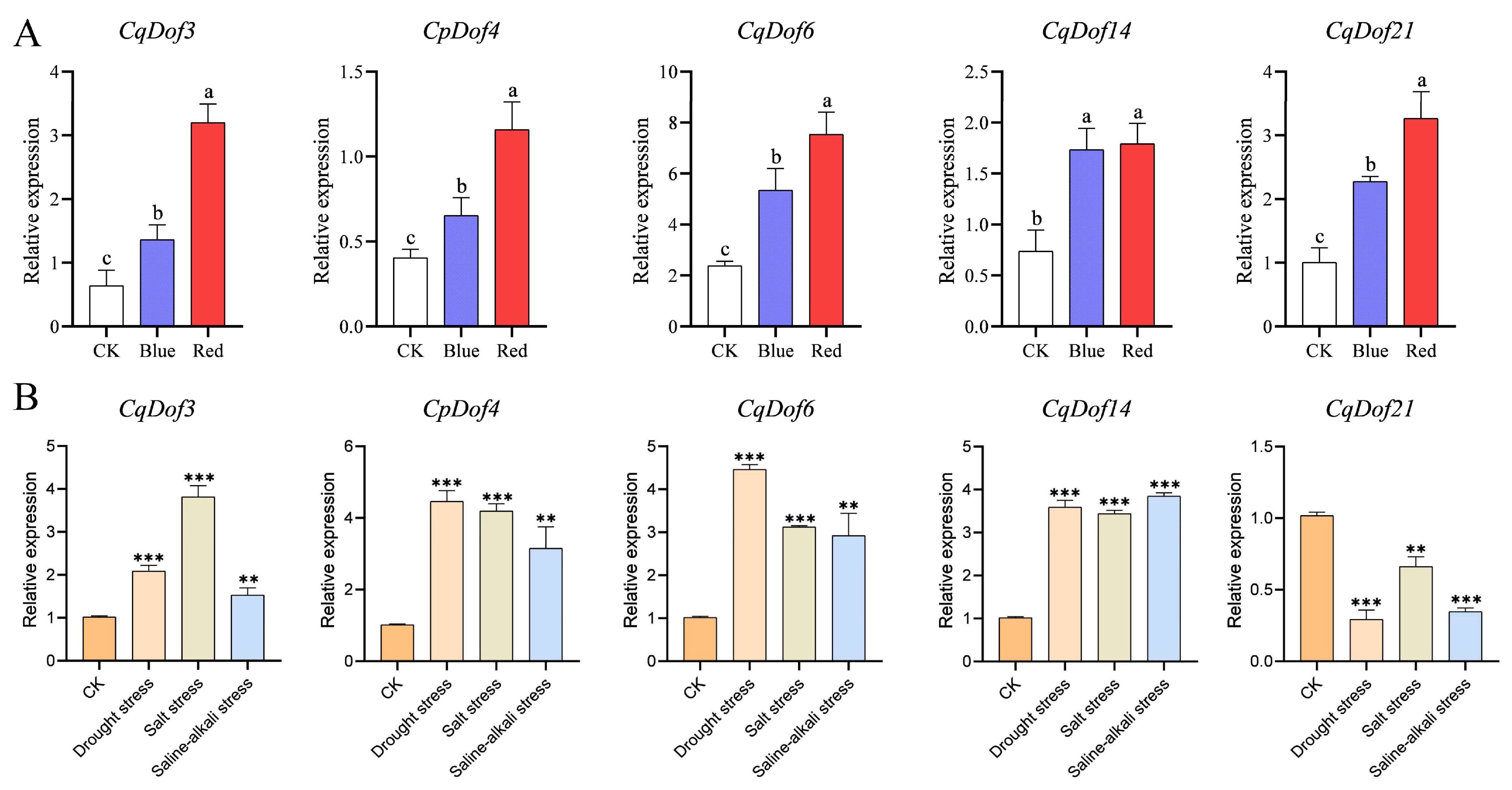
3.7. Expression Analysis of the CqDof Genes Under Abiotic Stress and Light Treatment
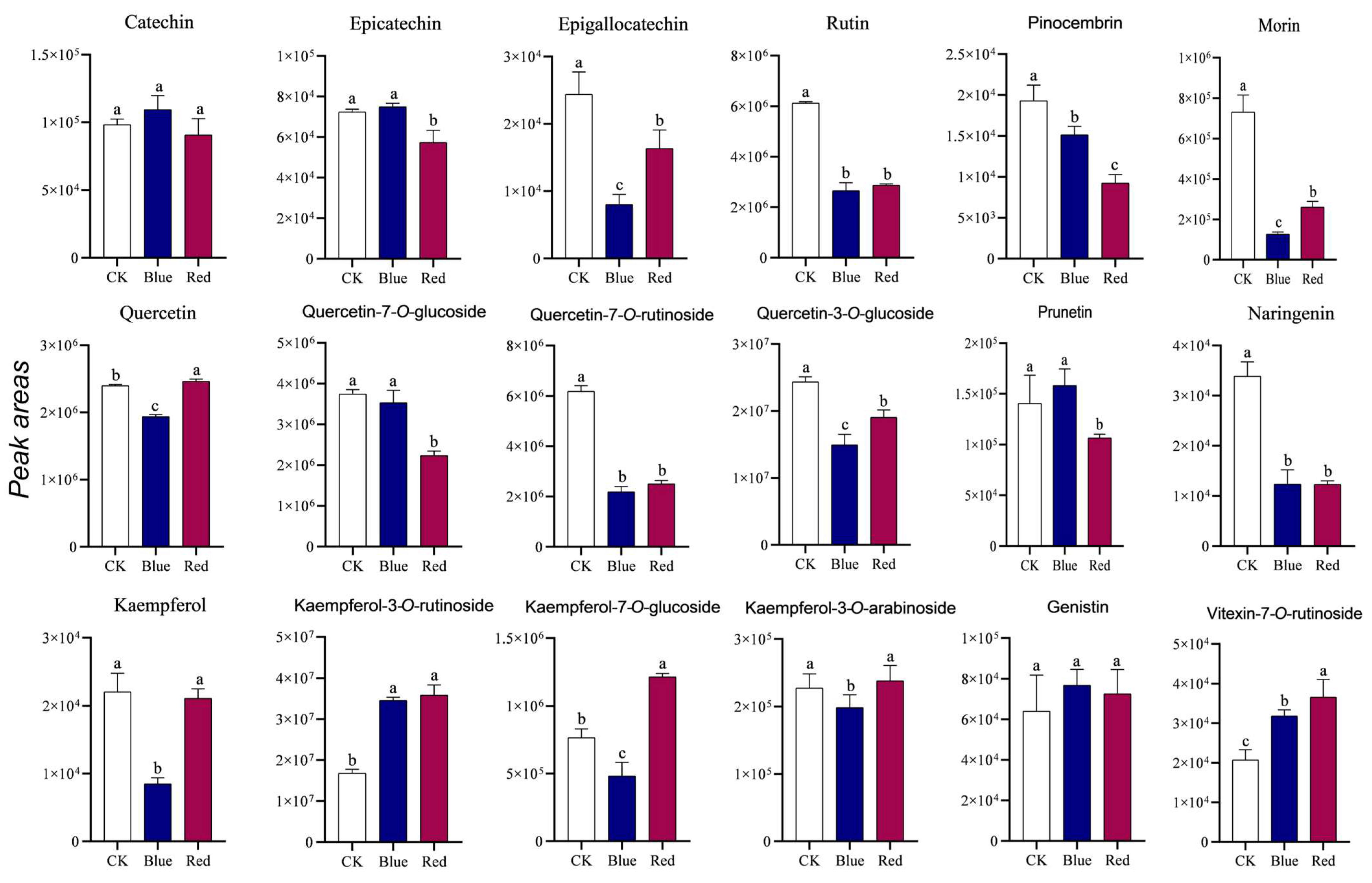
3.8. Analysis of the Flavonoid Content After Light Treatment
3.9. Correlation and Interaction Analysis Between CqDofs and Flavonoids
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Qian, G.; Li, X.; Zhang, H.; Zhang, H.; Zhou, J.; Ma, X.; Sun, W.; Yang, W.; He, R.; Wahab, A.T.; et al. Metabolomics analysis reveals the accumulation patterns of flavonoids and phenolic acids in quinoa (Chenopodium quinoa Willd.) grains of different colors. Food Chem. X 2023, 17, 100594. [Google Scholar] [CrossRef] [PubMed]
- Sampaio, S.L.; Fernandes, A.; Pereira, C.; Calhelha, R.C.; Sokovic, M.; Santos-Buelga, C.; Barros, L.; Ferreira, I. Nutritional value, physicochemical characterization and bioactive properties of the Brazilian quinoa brs piabiru. Food Funct. 2020, 11, 2969–2977. [Google Scholar] [CrossRef]
- Melini, V.; Melini, F. Functional components and anti-nutritional factors in gluten-free grains: A focus on quinoa seeds. Foods 2021, 10, 351. [Google Scholar] [CrossRef] [PubMed]
- Pereira, E.; Cadavez, V.; Barros, L.; Encina-Zelada, C.; Stojkovic, D.; Sokovic, M.; Calhelha, R.C.; Gonzales-Barron, U.; Ferreira, I. Chenopodium quinoa Willd. (quinoa) grains: A good source of phenolic compounds. Food Res. Int. 2020, 137, 109574. [Google Scholar] [CrossRef]
- Zhang, J.; Liu, B.; Li, J.; Zhang, L.; Wang, Y.; Zheng, H.; Lu, M.; Chen, J. Hsf and Hsp gene families in populus: Genome-wide identification, organization and correlated expression during development and in stress responses. BMC Genom. 2015, 16, 181. [Google Scholar] [CrossRef]
- Zhang, C.; Dong, T.; Yu, J.; Hong, H.; Liu, S.; Guo, F.; Ma, H.; Zhang, J.; Zhu, M.; Meng, X. Genome-wide survey and expression analysis of Dof transcription factor family in sweetpotato shed light on their promising functions in stress tolerance. Front. Plant Sci. 2023, 14, 1140727. [Google Scholar] [CrossRef] [PubMed]
- Sellami, M.H.; Pulvento, C.; Lavini, A. Agronomic practices and performances of quinoa under field conditions: A systematic review. Plants 2020, 10, 72. [Google Scholar] [CrossRef]
- Zhu, X.; Wang, B.; Liu, W.; Wei, X.; Wang, X.; Du, X.; Liu, H. Genome-wide analysis of AP2/ERF gene and functional analysis of CqERF24 gene in drought stress in quinoa. Int. J. Biol. Macromol 2023, 253, 127582. [Google Scholar] [CrossRef] [PubMed]
- Sun, W.; Wei, J.; Wu, G.; Xu, H.; Chen, Y.; Yao, M.; Zhan, J.; Yan, J.; Wu, N.; Chen, H.; et al. CqZF-HD14 enhances drought tolerance in quinoa seedlings through interaction with CqHIPP34 and CqNAC79. Plant Sci. 2022, 323, 111406. [Google Scholar] [CrossRef]
- Tovar, J.C.; Berry, J.C.; Quillatupa, C.; Castillo, S.E.; Acosta-Gamboa, L.; Fahlgren, N.; Gehan, M.A. Heat stress changes mineral nutrient concentrations in Chenopodium quinoa seed. Plant Direct 2022, 6, e384. [Google Scholar] [CrossRef]
- Tashi, G.; Zhan, H.; Xing, G.; Chang, X.; Zhang, H.; Nie, X.; Ji, W. Genome-wide identification and expression analysis of heat shock transcription factor family in Chenopodium quinoa Willd. Agronomy 2018, 8, 103. [Google Scholar] [CrossRef]
- Hussin, S.A.; Ali, S.H.; Lotfy, M.E.; El-Samad, E.; Eid, M.A.; Abd-Elkader, A.M.; Eisa, S.S. Morpho-physiological mechanisms of two different quinoa ecotypes to resist salt stress. BMC Plant Biol. 2023, 23, 374. [Google Scholar] [CrossRef]
- Yang, F.; Fan, Y.; Wu, X.; Cheng, Y.; Liu, Q.; Feng, L.; Chen, J.; Wang, Z.; Wang, X.; Yong, T.; et al. Auxin-to-gibberellin ratio as a signal for light intensity and quality in regulating soybean growth and matter partitioning. Front. Plant Sci. 2018, 9, 56. [Google Scholar] [CrossRef]
- Zheng, C.; Ma, J.Q.; Ma, C.L.; Shen, S.Y.; Liu, Y.F.; Chen, L. Regulation of growth and flavonoid formation of tea plants (Camellia sinensis) by blue and green light. J. Agric. Food. Chem. 2019, 67, 2408–2419. [Google Scholar] [CrossRef] [PubMed]
- Manivannan, A.; Soundararajan, P.; Park, Y.G.; Jeong, B.R. Physiological and proteomic insights into red and blue light-mediated enhancement of in vitro growth in scrophularia kakudensis—A potential medicinal plant. Front. Plant Sci. 2020, 11, 607007. [Google Scholar] [CrossRef]
- Huarancca Reyes, T.; Scartazza, A.; Castagna, A.; Cosio, E.G.; Ranieri, A.; Guglielminetti, L. Physiological effects of short acute UVB treatments in Chenopodium quinoa Willd. Sci. Rep. 2018, 8, 371. [Google Scholar] [CrossRef]
- Prado, F.E.; Perez, M.L.; González, Y.J.A. Efectos de la radiación ultravioleta B (UV-B) sobre diferentes variedades de quinoa: II.- efectos sobre la síntesis de pigmentos fotosintéticos, protectores y azúcares solubles en condiciones controladas. Boletín La Soc. Argent. Botánica 2016, 51, 665–673. [Google Scholar] [CrossRef]
- Gupta, S.; Malviya, N.; Kushwaha, H.; Nasim, J.; Bisht, N.C.; Singh, V.K.; Yadav, D. Insights into structural and functional diversity of Dof (DNA binding with one finger) transcription factor. Planta 2015, 241, 549–562. [Google Scholar] [CrossRef]
- Yanagisawa, S.; Schmidt, R.J. Diversity and similarity among recognition sequences of Dof transcription factors. Plant J. 1999, 17, 209–214. [Google Scholar] [CrossRef]
- Wei, Z.; Zhang, H.; Fang, M.; Lin, S.; Zhu, M.; Li, Y.; Jiang, L.; Cui, T.; Cui, Y.; Kui, H.; et al. The Dof transcription factor COG1 acts as a key regulator of plant biomass by promoting photosynthesis and starch accumulation. Mol. Plant 2023, 16, 1759–1772. [Google Scholar] [CrossRef]
- Ramachandran, V.; Tobimatsu, Y.; Masaomi, Y.; Sano, R.; Umezawa, T.; Demura, T.; Ohtani, M. Plant-specific dof transcription factors vascular-related dof1 and vascular-related dof2 regulate vascular cell differentiation and lignin biosynthesis in Arabidopsis. Plant Mol. Biol. 2020, 104, 263–281. [Google Scholar] [CrossRef]
- Li, Y.; Tian, M.; Feng, Z.; Zhang, J.; Lu, J.; Fu, X.; Ma, L.; Wei, H.; Wang, H. GhDof1.7, a dof transcription factor, plays positive regulatory role under salinity stress in upland cotton. Plants 2023, 12, 3740. [Google Scholar] [CrossRef] [PubMed]
- Jarvis, D.E.; Ho, Y.S.; Lightfoot, D.J.; Schmockel, S.M.; Li, B.; Borm, T.J.; Ohyanagi, H.; Mineta, K.; Michell, C.T.; Saber, N.; et al. The genome of Chenopodium quinoa. Nature 2017, 542, 307–312. [Google Scholar] [CrossRef]
- Rey, E.; Maughan, P.J.; Maumus, F.; Lewis, D.; Wilson, L.; Fuller, J.; Schmockel, S.M.; Jellen, E.N.; Tester, M.; Jarvis, D.E. A chromosome-scale assembly of the quinoa genome provides insights into the structure and dynamics of its subgenomes. Commun. Biol. 2023, 6, 1263. [Google Scholar] [CrossRef]
- Qian, G.; Wang, M.; Zhou, J.; Wang, X.; Zhang, Y.; Liu, Y.; Zhu, P.; Han, L.; Li, X.; Liu, C.; et al. Analysis of widely targeted metabolites of quinoa sprouts (Chenopodium quinoa Willd.) under saline-alkali stress provides new insights into nutritional value. Food Chem. 2024, 448, 138575. [Google Scholar] [CrossRef] [PubMed]
- Cai, Z.Q.; Gao, Q. Comparative physiological and biochemical mechanisms of salt tolerance in five contrasting highland quinoa cultivars. BMC Plant Biol. 2020, 20, 70. [Google Scholar] [CrossRef]
- Wang, Y.; Wu, Y.; Bao, Q.; Shi, H.; Zhang, Y. Integrating physiology, transcriptome, and metabolome analyses reveals the drought response in two quinoa cultivars with contrasting drought tolerance. Int. J. Mol. Sci. 2024, 25, 12188. [Google Scholar] [CrossRef] [PubMed]
- Qian, G.; Meng, X.; Wang, S.; Mi, Y.; Qin, Z.; Liu, T.; Zhang, Y.; Wan, H.; Chen, W.; Sun, W.; et al. Genome-wide identification of HSF gene family and their expression analysis in vegetative tissue of young seedlings of hemp under different light treatments. Ind. Crop Prod. 2023, 204, 117375. [Google Scholar] [CrossRef]
- Xiaolin, Z.; Baoqiang, W.; Xian, W.; Xiaohong, W. Identification of the CIPK-CBL family gene and functional characterization of CqCIPK14 gene under drought stress in quinoa. BMC Genom. 2022, 23, 447. [Google Scholar] [CrossRef]
- Yang, W.; Qian, G.; Chen, Y.; Liu, T.; Wan, H.; Wang, S.; Meng, X.; Chen, W.; Su, Y.; Zhang, Y.; et al. Profiling of polyphenols for in-depth understanding of Tartary buckwheat sprouts: Correlation between cultivars and active components, dynamic changes and the effects of ultraviolet B stress. Food Chem. X 2022, 14, 100295. [Google Scholar] [CrossRef]
- Ren, G.; Teng, C.; Fan, X.; Guo, S.; Zhao, G.; Zhang, L.; Liang, Z.; Qin, P. Nutrient composition, functional activity and industrial applications of quinoa (Chenopodium quinoa Willd.). Food Chem. 2023, 410, 135290. [Google Scholar] [CrossRef] [PubMed]
- Fu, B.; Ji, X.; Zhao, M.; He, F.; Wang, X.; Wang, Y.; Liu, P.; Niu, L. The influence of light quality on the accumulation of flavonoids in tobacco (Nicotiana tabacum L.) leaves. J. Photochem. Photobiol. B 2016, 162, 544–549. [Google Scholar] [CrossRef] [PubMed]
- Zou, X.; Sun, H. DOF transcription factors: Specific regulators of plant biological processes. Front. Plant Sci. 2023, 14, 1044918. [Google Scholar] [CrossRef]
- Yang, L.; Min, X.; Wei, Z.; Liu, N.; Li, J.; Zhang, Y.; Yang, Y. Genome-wide identification and expression analysis of the dof transcription factor in Annual Alfalfa Medicago polymorpha. Plants 2023, 12, 1831. [Google Scholar] [CrossRef] [PubMed]
- Luo, T.; Song, Y.; Gao, H.; Wang, M.; Cui, H.; Ji, C.; Wang, J.; Yuan, L.; Li, R. Genome-wide identification and functional analysis of Dof transcription factor family in Camelina sativa. BMC Genom. 2022, 23, 812. [Google Scholar] [CrossRef]
- Lohani, N.; Babaei, S.; Singh, M.B.; Bhalla, P.L. Genome-wide in silico identification and comparative analysis of dof gene family in Brassica napus. Plants 2021, 10, 709. [Google Scholar] [CrossRef]
- Zhou, Y.; Cheng, Y.; Wan, C.; Li, J.; Yang, Y.; Chen, J. Genome-wide characterization and expression analysis of the Dof gene family related to abiotic stress in watermelon. PeerJ 2020, 8, e8358. [Google Scholar] [CrossRef]
- Yue, Y.; Du, J.; Li, Y.; Thomas, H.R.; Frank, M.H.; Wang, L.; Hu, H. Insight into the petunia Dof transcription factor family reveals a new regulator of male-sterility. Ind. Crop Prod 2021, 161, 113196. [Google Scholar] [CrossRef]
- Li, T.; Wang, X.; Elango, D.; Zhang, W.; Li, M.; Zhang, F.; Pan, Q.; Wu, Y. Genome-wide identification, phylogenetic and expression pattern analysis of Dof transcription factors in blueberry (Vaccinium corymbosum L.). PeerJ 2022, 10, e14087. [Google Scholar] [CrossRef]
- Liu, H.; Lyu, H.M.; Zhu, K.; Van de Peer, Y.; Max, C.Z. The emergence and evolution of intron-poor and intronless genes in intron-rich plant gene families. Plant J. 2021, 105, 1072–1082. [Google Scholar] [CrossRef]
- Tabassum, J.; Raza, Q.; Riaz, A.; Ahmad, S.; Rashid, M.; Javed, M.A.; Ali, Z.; Kang, F.; Khan, I.A.; Atif, R.M.; et al. Exploration of the genomic atlas of Dof transcription factor family across genus Oryza provides novel insights on rice breeding in changing climate. Front. Plant Sci. 2022, 13, 1004359. [Google Scholar] [CrossRef] [PubMed]
- Nan, H.; Ludlow, R.A.; Lu, M.; An, H. Genome-wide analysis of Dof genes and their response to abiotic stress in rose (Rosa chinensis). Front. Genet. 2021, 12, 538733. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Zhang, Y.; Xu, L.; Wang, C.; Luo, Y.; Feng, S.; Yuan, Y.; Yang, Q.; Feng, B. Genome-wide identification of dna binding with one finger (Dof) gene family in Tartary Buckwheat (Fagopyrum tataricum) and analysis of its expression pattern after exogenous hormone stimulation. Biology 2022, 11, 173. [Google Scholar] [CrossRef]
- Meng, X.; Liu, S.; Zhang, C.; He, J.; Ma, D.; Wang, X.; Dong, T.; Guo, F.; Cai, J.; Long, T.; et al. The unique sweet potato NAC transcription factor IbNAC3 modulates combined salt and drought stresses. Plant Physiol. 2023, 191, 747–771. [Google Scholar] [CrossRef]
- Wu, Z.; Cheng, J.; Cui, J.; Xu, X.; Liang, G.; Luo, X.; Chen, X.; Tang, X.; Hu, K.; Qin, C. Genome-wide identification and expression profile of dof transcription factor gene family in pepper (Capsicum annuum L.). Front. Plant Sci. 2016, 7, 574. [Google Scholar] [CrossRef]
- Wen, C.L.; Cheng, Q.; Zhao, L.; Mao, A.; Yang, J.; Yu, S.; Weng, Y.; Xu, Y. Identification and characterisation of Dof transcription factors in the cucumber genome. Sci. Rep. 2016, 6, 23072. [Google Scholar] [CrossRef]
- Gao, H.; Song, W.; Severing, E.; Vayssieres, A.; Huettel, B.; Franzen, R.; Richter, R.; Chai, J.; Coupland, G. PIF4 enhances DNA binding of CDF2 to co-regulate target gene expression and promote Arabidopsis hypocotyl cell elongation. Nat. Plants 2022, 8, 1082–1093. [Google Scholar] [CrossRef]
- Santopolo, S.; Boccaccini, A.; Lorrai, R.; Ruta, V.; Capauto, D.; Minutello, E.; Serino, G.; Costantino, P.; Vittorioso, P. Dof affecting germination 2 is a positive regulator of light-mediated seed germination and is repressed by dof affecting germination 1. BMC Plant Biol. 2015, 15, 72. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Hu, L.; Kong, W.; Yang, C.; Xi, W. Red light-transmittance bagging promotes carotenoid accumulation of grapefruit during ripening. Commun. Biol. 2022, 5, 303. [Google Scholar] [CrossRef]
- Yang, L.; Zhou, S.; Hou, Y.; Ji, B.; Pei, L.; Su, X.; Zhong, H.; Dong, C. Blue light induces biosynthesis of flavonoids in Epimedium sagittatum (Sieb.et Zucc.) Maxim. leaves, a study on a light-demanding medicinal shade herb. Ind. Crop. Prod. 2022, 187, 115512. [Google Scholar] [CrossRef]
- Nam, T.G.; Kim, D.O.; Eom, S.H. Effects of light sources on major flavonoids and antioxidant activity in common buckwheat sprouts. Food Sci. Biotechnol. 2018, 27, 169–176. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Ma, J.; Zou, H.; Zhang, L.; Li, S.; Wang, Y. The combination of blue and red LED light improves growth and phenolic acid contents in Salvia miltiorrhiza Bunge. Ind. Crop. Prod. 2020, 158, 112959. [Google Scholar] [CrossRef]



| Gene ID | Gene Name | Length (Aa) | MV (kDa) | PI | Instability Index | GRAVY | Aliphatic Index | Subcellular Location |
|---|---|---|---|---|---|---|---|---|
| AUR62001807 | CqDof1 | 305 | 32.51 | 9.06 | 44.57 | −0.742 | 49.61 | Nucleus |
| AUR62001970 | CqDof2 | 196 | 21.93 | 9.38 | 43.76 | −0.886 | 52.76 | Nucleus |
| AUR62001976 | CqDof3 | 186 | 19.17 | 8.72 | 50.76 | −0.356 | 55.97 | Nucleus |
| AUR62003691 | CqDof4 | 291 | 31.13 | 9.40 | 51.63 | −0.604 | 61.07 | Nucleus |
| AUR62003828 | CqDof5 | 558 | 59.10 | 9.09 | 56.97 | −0.253 | 66.49 | Nucleus |
| AUR62003833 | CqDof6 | 337 | 35.84 | 9.18 | 49.13 | −0.620 | 53.23 | Nucleus |
| AUR62004520 | CqDof7 | 196 | 21.93 | 9.38 | 43.76 | −0.886 | 52.76 | Nucleus |
| AUR62005809 | CqDof8 | 457 | 50.13 | 5.60 | 50.34 | −0.765 | 55.30 | Nucleus |
| AUR62006501 | CqDof9 | 304 | 33.90 | 8.07 | 52.68 | −0.838 | 55.49 | Nucleus |
| AUR62007029 | CqDof10 | 223 | 25.32 | 9.44 | 51.32 | −0.904 | 55.96 | Nucleus |
| AUR62008205 | CqDof11 | 346 | 36.87 | 8.03 | 50.31 | −0.640 | 48.50 | Nucleus |
| AUR62008425 | CqDof12 | 450 | 49.65 | 5.13 | 55.16 | −0.901 | 48.56 | Nucleus |
| AUR62009593 | CqDof13 | 398 | 42.41 | 6.65 | 51.28 | −0.816 | 49.30 | Nucleus |
| AUR62013510 | CqDof14 | 255 | 28.01 | 4.52 | 57.45 | −0.656 | 51.29 | Nucleus |
| AUR62014301 | CqDof15 | 304 | 33.44 | 6.03 | 45.24 | −0.560 | 62.96 | Nucleus |
| AUR62014841 | CqDof16 | 313 | 34.96 | 11.46 | 85.15 | −0.708 | 67.48 | Nucleus |
| AUR62014843 | CqDof17 | 169 | 18.78 | 8.31 | 46.39 | −0.805 | 47.46 | Nucleus |
| AUR62016967 | CqDof18 | 267 | 29.85 | 6.37 | 57.96 | −0.910 | 47.12 | Nucleus |
| AUR62017038 | CqDof19 | 276 | 30.08 | 8.22 | 56.50 | −0.691 | 62.10 | Nucleus |
| AUR62017040 | CqDof20 | 143 | 16.08 | 8.24 | 32.55 | −0.859 | 42.45 | Nucleus |
| AUR62018002 | CqDof21 | 332 | 36.19 | 8.07 | 45.31 | −0.772 | 55.84 | Nucleus |
| AUR62021670 | CqDof22 | 449 | 49.55 | 5.13 | 54.30 | −0.902 | 48.66 | Nucleus |
| AUR62022735 | CqDof23 | 204 | 22.64 | 9.60 | 48.23 | −0.948 | 49.75 | Nucleus |
| AUR62023916 | CqDof24 | 223 | 23.49 | 9.32 | 56.20 | −0.535 | 59.15 | Nucleus |
| AUR62025032 | CqDof25 | 301 | 33.49 | 8.07 | 54.93 | −0.829 | 56.05 | Nucleus |
| AUR62026763 | CqDof26 | 274 | 30.31 | 4.61 | 58.22 | −0.626 | 51.64 | Nucleus |
| AUR62027677 | CqDof27 | 113 | 12.69 | 9.26 | 50.58 | −0.536 | 56.99 | Nucleus |
| AUR62030727 | CqDof28 | 210 | 23.52 | 9.11 | 47.08 | −0.592 | 60.76 | Nucleus |
| AUR62031206 | CqDof29 | 110 | 12.22 | 8.69 | 45.05 | −0.592 | 45.27 | Nucleus |
| AUR62034094 | CqDof30 | 335 | 37.29 | 6.55 | 51.77 | −0.626 | 65.25 | Nucleus |
| AUR62034427 | CqDof31 | 512 | 56.46 | 5.47 | 45.21 | −0.684 | 64.20 | Nucleus |
| AUR62036367 | CqDof32 | 202 | 22.65 | 8.96 | 47.13 | −0.760 | 51.14 | Nucleus |
| AUR62036527 | CqDof33 | 353 | 38.87 | 7.14 | 61.18 | −0.620 | 61.81 | Nucleus |
| AUR62038328 | CqDof34 | 224 | 23.79 | 9.41 | 53.53 | −0.631 | 51.47 | Nucleus |
| AUR62039802 | CqDof35 | 442 | 48.70 | 6.80 | 43.72 | −0.778 | 53.46 | Nucleus |
| AUR62041860 | CqDof36 | 331 | 35.74 | 8.93 | 53.75 | −0.662 | 57.73 | Nucleus |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Qian, G.; Yang, J.; Wang, M.; Li, L. Identification of the Dof Gene Family in Quinoa and Its Potential Role in Regulating Flavonoid Synthesis Under Different Stress Conditions. Biology 2025, 14, 446. https://doi.org/10.3390/biology14040446
Qian G, Yang J, Wang M, Li L. Identification of the Dof Gene Family in Quinoa and Its Potential Role in Regulating Flavonoid Synthesis Under Different Stress Conditions. Biology. 2025; 14(4):446. https://doi.org/10.3390/biology14040446
Chicago/Turabian StyleQian, Guangtao, Jinrong Yang, Mingyu Wang, and Lixin Li. 2025. "Identification of the Dof Gene Family in Quinoa and Its Potential Role in Regulating Flavonoid Synthesis Under Different Stress Conditions" Biology 14, no. 4: 446. https://doi.org/10.3390/biology14040446
APA StyleQian, G., Yang, J., Wang, M., & Li, L. (2025). Identification of the Dof Gene Family in Quinoa and Its Potential Role in Regulating Flavonoid Synthesis Under Different Stress Conditions. Biology, 14(4), 446. https://doi.org/10.3390/biology14040446






