Development of a Visual Assay for Detection of Viable Cronobacter sakazakii Using RT-PSR and Hydroxynaphthol Blue Indicator
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Chemicals and Reagents
2.2. RNA Extraction of C. sakazakii
2.3. Primers of RT-PSR Reaction
2.4. Visual Inspection of C. sakazakii
2.5. Optimization of Experimental Conditions
2.6. Specificity of RT-PSR Visual Assay
2.7. Sensitivity of RT-PSR Visual Assay
2.8. Accuracy Evaluation Using Artificially Contaminated Samples
2.9. Experimental Design and Statistical Analysis
3. Results and Discussion
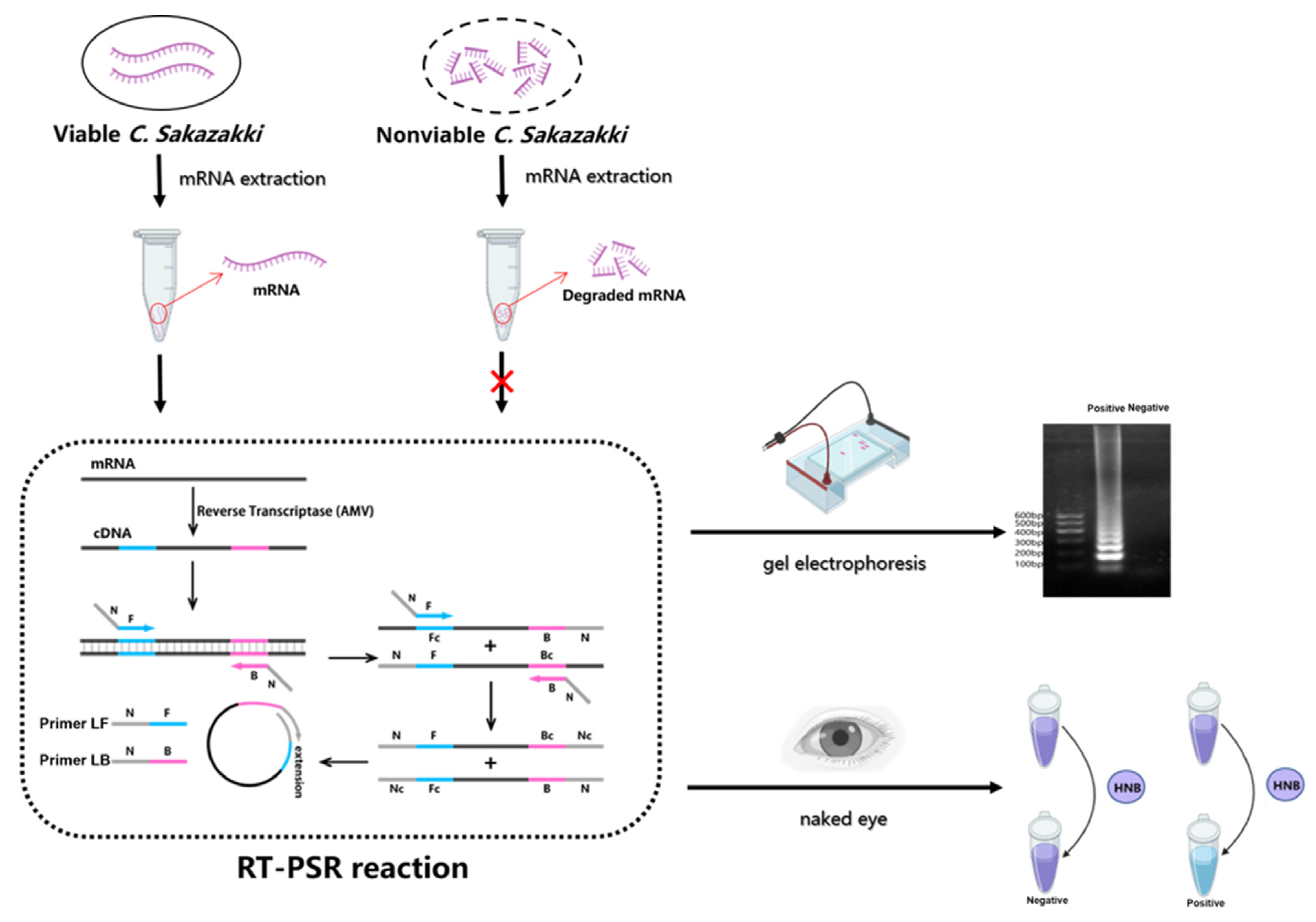
3.1. Principle of RT-PSR-Based Visual Detection
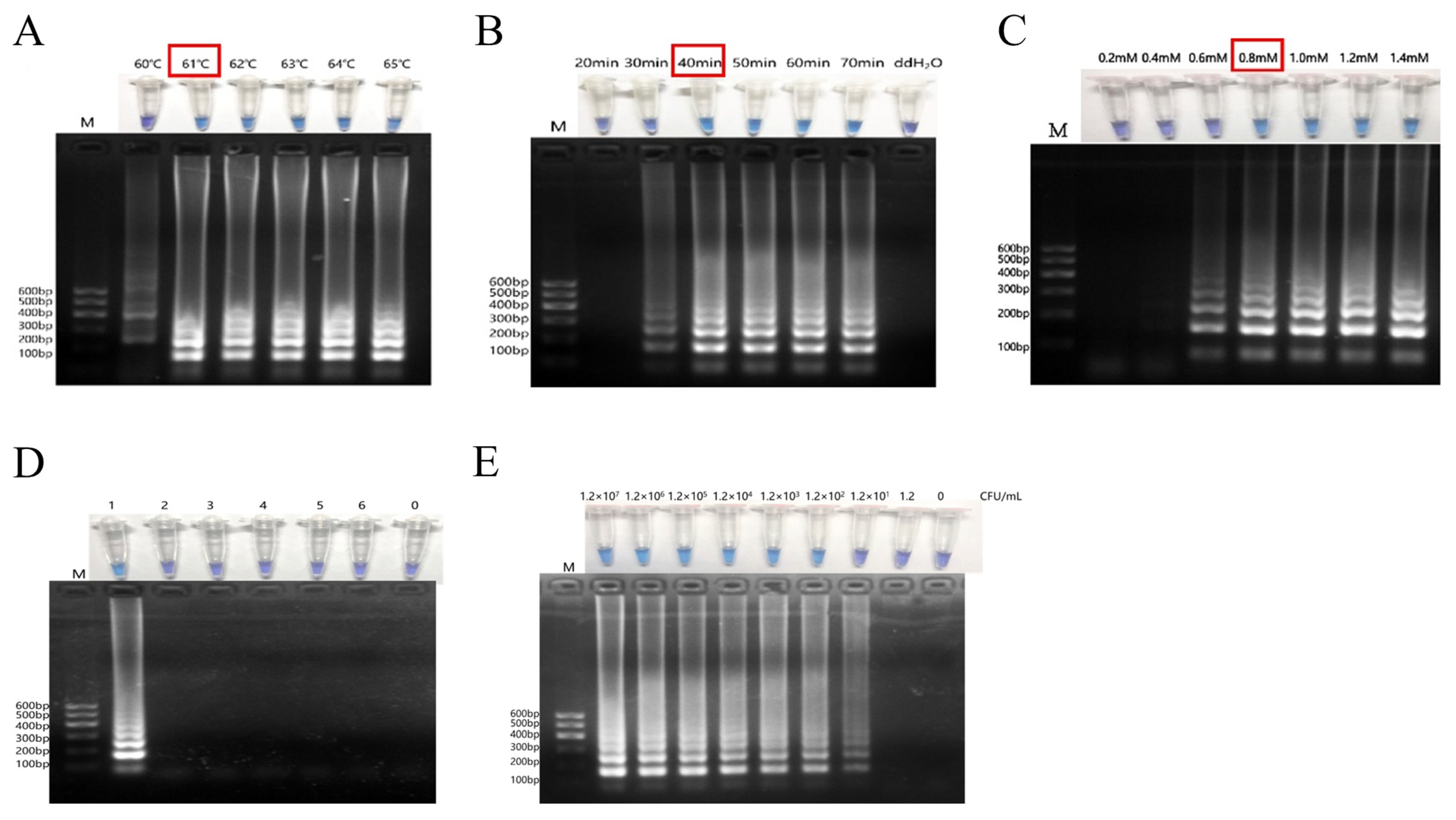
3.2. Optimization of Reaction Temperature, Reaction Time and dNTP Concentration for RT-PSR Visual Assay
3.3. Evaluation of RT-PSR-Based Visual Assay Specificity
3.4. Determination of RT-PSR-Based Visual Assay Sensitivity
3.5. Accuracy Evaluation Using Artificially Contaminated Samples
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Pina-Pérez, M.C.; Rodrigo, D.; Martínez, A. Nonthermal inactivation of Cronobacter sakazakii in infant formula milk: A review. Crit. Rev. Food Sci. Nutr. 2016, 56, 1620–1629. [Google Scholar] [CrossRef]
- Friedemann, M. Enterobacter sakazakii in food and beverages (other than infant formula and milk powder). Int. J. Food Microbiol. 2007, 116, 1–10. [Google Scholar] [CrossRef]
- Beuchat, L.R.; Kim, H.; Gurtler, J.B.; Lin, L.C.; Ryu, J.H.; Richards, G.M. Cronobacter sakazakii in foods and factors affecting its survival, growth, and inactivation. Int. J. Food Microbiol. 2009, 136, 204–213. [Google Scholar] [CrossRef] [PubMed]
- Elgar, D.; Evers, J.M.; Holroyd, S.E.; Johnson, R.; Rowan, A. The measurement of protein in powdered milk products and infant formulas: A review and recent developments. J. AOAC Int. 2016, 99, 26–29. [Google Scholar] [CrossRef]
- Chauhan, R.; Singh, N.; Pal, G.K.; Goel, G. Trending biocontrol strategies against Cronobacter sakazakii: A recent updated review. Food Res. Int. 2020, 137, 109385. [Google Scholar] [CrossRef] [PubMed]
- Giovannini, M.; Verduci, E.; Ghisleni, D.; Salvatici, E.; Riva, E.; Agostoni, C. Enterobacter sakazakii: An emerging problem in paediatric nutrition. J. Int. Med. Res. 2008, 36, 394–399. [Google Scholar] [CrossRef] [PubMed]
- Simmons, B.P.; Gelfand, M.S.; Haas, M.; Metts, L.; Ferguson, J. Enterobacter sakazakii infections in neonates associated with intrinsic contamination of a powdered infant formulaInfection Control. Hosp. Epidemiol. 1989, 10, 398–401. [Google Scholar] [CrossRef]
- van Acker, J.; de Smet, F.; Muyldermans, G.; Bougatef, A.; Naessens, A.; Lauwers, S. Outbreak of necrotizing enterocolitis associated with Enterobacter sakazakii in powdered milk formula. J. Clin. Microbiol. 2001, 39, 293–297. [Google Scholar] [CrossRef]
- Hunter, C.J.; Bean, J.F. Cronobacter: An emerging opportunistic pathogen associated with neonatal meningitis, sepsis and necrotizing enterocolitis. J. Perinatol. 2013, 33, 581–585. [Google Scholar] [CrossRef]
- Lehner, A.; Tall, B.D.; Fanning, S.; Srikumar, S. Cronobacter spp.—Opportunistic foodborne pathogens: An update on evolution, osmotic adaptation and pathogenesis. Curr. Clin. Microbiol. Rep. 2018, 5, 97–105. [Google Scholar] [CrossRef]
- O’Grady, J.; Cronin, U.; Tierney, J.; Piterina, A.V.; O’Meara, E.; Wilkinson, M.G. Gaps in the assortment of rapid assays for microorganisms of interest to the dairy industry. Adv. Appl. Microbiol. 2020, 113, 1–56. [Google Scholar] [CrossRef] [PubMed]
- Keyser, M.; Witthuhn, R.C.; Ronquest, L.C.; Britz, T.J. Treatment of winery effluent with upflow anaerobic sludge blanket (UASB)—Granular sludges enriched with Enterobacter sakazakii. Biotechnol. Lett. 2003, 25, 1893–1898. [Google Scholar] [CrossRef]
- Gičová, A.; Oriešková, M.; Oslanecová, L.; Drahovská, H.; Kaclíková, E. Identification and characterization of Cronobacter strains isolated from powdered infant foods. Lett. Appl. Microbiol. 2014, 58, 242–247. [Google Scholar] [CrossRef] [PubMed]
- Park, S.; Shukla, S.; Kim, Y.; Oh, S.; Hun Kim, S.; Kim, M. Development of sandwich enzyme-linked immunosorbent assay for the detection of Cronobacter muytjensii (formerly called Enterobacter sakazakii). Microbiol. Immunol. 2012, 56, 472–479. [Google Scholar] [CrossRef]
- Shukla, S.; Haldorai, Y.; Bajpai, V.K.; Rengaraj, A.; Hwang, S.K.; Song, X.; Kim, M.; Huh, Y.S.; Han, Y.-K. Electrochemical coupled immunosensing platform based on graphene oxide/gold nanocomposite for sensitive detection of Cronobacter sakazakii in powdered infant formula. Biosens.Bioelectron. 2018, 109, 139–149. [Google Scholar] [CrossRef]
- Muthubharathi, B.C.; Ravichandiran, V.; Balamurugan, K. Cronobacter sakazakii cue for the attraction and its impact on the immunity of Caenorhabditis elegans. Infect. Immun. 2022, 90, e0028122. [Google Scholar] [CrossRef] [PubMed]
- Ye, Y.; Wu, Q.; Zhang, J.; Jiang, H.; Hu, W. Detection of viable Cronobacter spp. (Enterobacter sakazakii) by one-step RT-PCR in dry aquatic product. J. Food Sci. 2012, 77, M616–M619. [Google Scholar] [CrossRef]
- Gao, J.X.; Li, P.; Du, X.J.; Han, Z.H.; Xue, R.; Liang, B.; Wang, S. A negative regulator of cellulose biosynthesis, bcsR, affects biofilm formation, and adhesion/invasion ability of Cronobacter sakazakii. Front. Microbiol. 2017, 8, 1839. [Google Scholar] [CrossRef]
- Minami, J.; Soejima, T.; Yaeshima, T.; Iwatsuki, K. Direct real-time PCR with ethidium monoazid.e: A method for the rapid detection of viable Cronobacter sakazakii in powdered infant formula. J. Food Prot. 2012, 75, 1572–1579. [Google Scholar] [CrossRef]
- Yuan, Y.; Wu, X.; Liu, Z.; Ning, Q.; Fu, L.; Wu, S. A signal cascade amplification strategy based on RT-PCR triggering of a G-quadruplex DNAzyme for a novel electrochemical detection of viable Cronobacter sakazakii. Analyst 2020, 145, 4477–4483. [Google Scholar] [CrossRef]
- Liang, T.; Zhou, P.; Zhou, B.; Xu, Q.; Zhou, Z.; Wu, X.; Aguilar, Z.P.; Xu, H. Simultaneous quantitative detection of viable Escherichia coli O157:H7, Cronobacter spp., and Salmonella spp. using sodium deoxycholate-propidium monoazide with multiplex real-time PCR. J. Dairy Sci. 2019, 102, 2954–2965. [Google Scholar] [CrossRef] [PubMed]
- Lv, X.; Wang, L.; Zhang, J.; He, X.; Shi, L.; Zhao, L. Quantitative detection of trace VBNC Cronobacter sakazakii by immunomagnetic separation in combination with PMAxx-ddPCR in dairy products. Food Microbiol. 2021, 99, 103831. [Google Scholar] [CrossRef]
- Yu, S.; Yan, L.; Wu, X.; Li, F.; Wang, D.; Xu, H. Multiplex PCR coupled with propidium monoazide for the detection of viable Cronobacter sakazakii, Bacillus cereus, and Salmonella spp. in milk and milk products. J. Dairy Sci. 2017, 100, 7874–7882. [Google Scholar] [CrossRef] [PubMed]
- Hu, L.; Zhang, S.; Xue, Y.; Zhang, Y.; Zhang, W.; Wang, S. Quantitative detection of viable but nonculturable Cronobacter sakazakii using photosensitive nucleic acid dye PMA combined with isothermal amplification LAMP in raw milk. Foods 2022, 11, 11172653. [Google Scholar] [CrossRef]
- Liu, J.; Xie, G.; Xiong, Q.; Mu, D.; Xu, H. A simple and sensitive aptasensor with rolling circle amplification for viable Cronobacter sakazakii detection in powdered infant formula. J. Dairy Sci. 2021, 104, 12365–12374. [Google Scholar] [CrossRef] [PubMed]
- Gupta, V.; Chakravarti, S.; Chander, V.; Majumder, S.; Bhat, S.A.; Gupta, V.K.; Nandi, S. Polymerase spiral reaction (PSR): A novel, visual isothermal amplification method for detection of canine parvovirus 2 genomic DNA. Arch. Virol. 2017, 162, 1995–2001. [Google Scholar] [CrossRef]
- Somboonna, N.; Choopara, I. Point-of-Care Chlamydia trachomatis detection using loop-mediated isothermal amplification and hydroxynaphthol blue. Methods Mol. Biol. 2019, 2042, 11–17. [Google Scholar] [CrossRef]
- Dong, D.; Zou, D.; Liu, H.; Yang, Z.; Huang, S.; Liu, N.; He, X.; Liu, W.; Huang, L. Rapid detection of Pseudomonas aeruginosa targeting the toxA gene in intensive care unit patients from Beijing, China. Front. Microbiol. 2015, 6, 1100. [Google Scholar] [CrossRef]
- Aliotta, J.M.; Pelletier, J.J.; Ware, J.L.; Moran, L.S.; Benner, J.S.; Kong, H. Thermostable Bst DNA polymerase I lacks a 3′→5′ proofreading exonuclease activity. Genet. Anal. Biomol. Eng. 1996, 12, 185–195. [Google Scholar] [CrossRef]
- ISO 22964:2017; The International Organization for Standardization. Microbiology of the Food Chain—HORIZONTAL Method for the Detection of Cronobacter spp. ISO: Geneva, Switzerland, 2017.
- Wu, X.; Chen, Q.; Yang, C.; Ning, Q.; Liu, Z. An enhanced visual detection assay for Listeria monocytogenes in food based on isothermal amplified peroxidase-mimicking catalytic beacon. Food Control 2022, 134, 108721. [Google Scholar] [CrossRef]
| Primer | Sequence (5′–3′) |
|---|---|
| PSR-LF | CCCAACCCGCCCTACCCAAAGAGGAGATCCACGCGATGAT |
| PSR-LB | CCCAACCCGCCCTACCCAAATCGTTCATCTGGCGTAGCA |
| PSR-IF | AAACCGCCGATGCCTTTATC |
| PSR-IB | GATCATGGCGAACGGCAAG |
| Assay | RT Time | Total Time | LOD | Reference |
|---|---|---|---|---|
| Aptamer combined rolling circle amplification | N/A | 180 min | 2.7 × 102 CFU/mL | [25] |
| gEMA-DqPCR | N/A | 540 min | 101 CFU/mL | [19] |
| PMA-qLAMP assay | N/A | 40 min | 4.3 × 102 CFU/mL | [24] |
| Multiplex PCR coupled with propidium monoazide | N/A | 80 min | 9.5 × 104 CFU/mL | [23] |
| SD-PMA-mRT-PCR | 75 min | 120 min | 102 CFU/mL | [21] |
| RT-PCR triggering of a G-quadruplex DNAzyme | 90.8 min | 120 min | 5.01 × 102 CFU/mL | [20] |
| IMS-PMAxx-ddPCR | N/A | 110 min | 23 CFU/mL | [22] |
| One-Step RT-PCR | 132.5 min | 150 min | 104 CFU/mL | [17] |
| RT-PSR-based visual assay | 75 min | 130 min | 1.2 × 101 CFU/mL (gel electrophoresis), 1.2 × 102 CFU/mL (naked eye) | This work |
| Samples a | Visual Inspection (Positive/Total) | Culture-Based Method b Detection (Positive/Total) | Accuracy (%) c |
|---|---|---|---|
| Viable C. sakazakii | 10/10 | 10/10 | 100 |
| Nonviable C. sakazakii | 0/10 | 0/10 | 100 |
| Blank control | 0/10 | 0/10 | 100 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, P.; Chen, Q.; Wang, Y.; Sun, X.; Liu, Z. Development of a Visual Assay for Detection of Viable Cronobacter sakazakii Using RT-PSR and Hydroxynaphthol Blue Indicator. Biology 2025, 14, 383. https://doi.org/10.3390/biology14040383
Wang P, Chen Q, Wang Y, Sun X, Liu Z. Development of a Visual Assay for Detection of Viable Cronobacter sakazakii Using RT-PSR and Hydroxynaphthol Blue Indicator. Biology. 2025; 14(4):383. https://doi.org/10.3390/biology14040383
Chicago/Turabian StyleWang, Peng, Qiming Chen, Yikai Wang, Xueting Sun, and Zhanmin Liu. 2025. "Development of a Visual Assay for Detection of Viable Cronobacter sakazakii Using RT-PSR and Hydroxynaphthol Blue Indicator" Biology 14, no. 4: 383. https://doi.org/10.3390/biology14040383
APA StyleWang, P., Chen, Q., Wang, Y., Sun, X., & Liu, Z. (2025). Development of a Visual Assay for Detection of Viable Cronobacter sakazakii Using RT-PSR and Hydroxynaphthol Blue Indicator. Biology, 14(4), 383. https://doi.org/10.3390/biology14040383






