Mitigating Diabetic Cardiomyopathy: The Synergistic Potential of Sea Buckthorn and Metformin Explored via Bioinformatics and Chemoinformatics
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Application of System Biology Analysis
2.2. Molecular Docking Methodology
2.3. Animal Study
2.4. Mice Behavioral Tests
2.4.1. Elevated Plus Maze Assessment of Anxiety-Related Behavior
2.4.2. Open-Field Test for Assessment of Locomotor Activity
2.5. Biochemical Tests
2.6. Quantitative Real-Time PCR (qRT-PCR)
2.7. Protocol for Extracting Sea Buckthorn (Hippophae rhamnoides L.)
2.8. Metformin Administration
2.9. Diagnostic Evaluation by Pathological Assessment
2.10. Statistical Analysis
3. Results
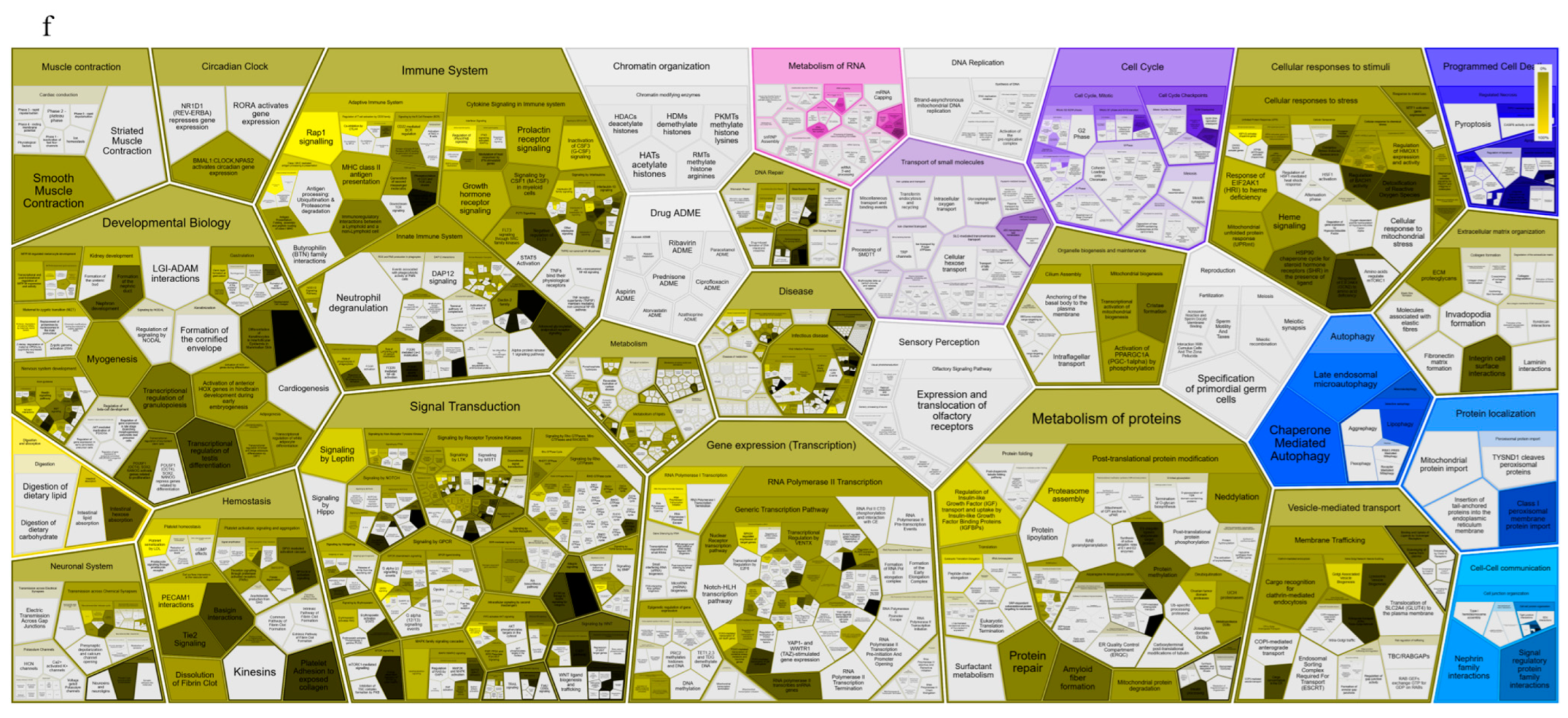
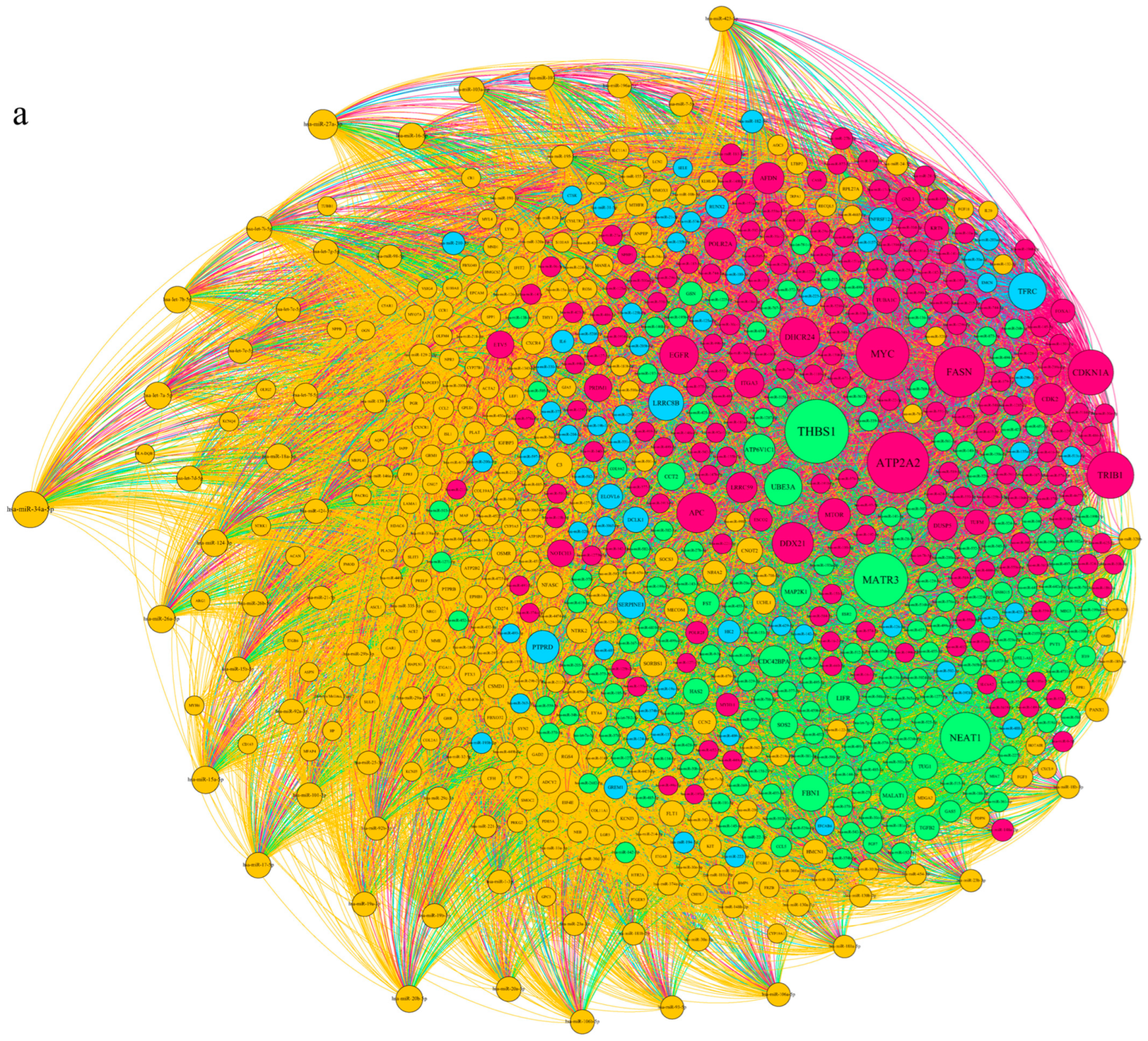
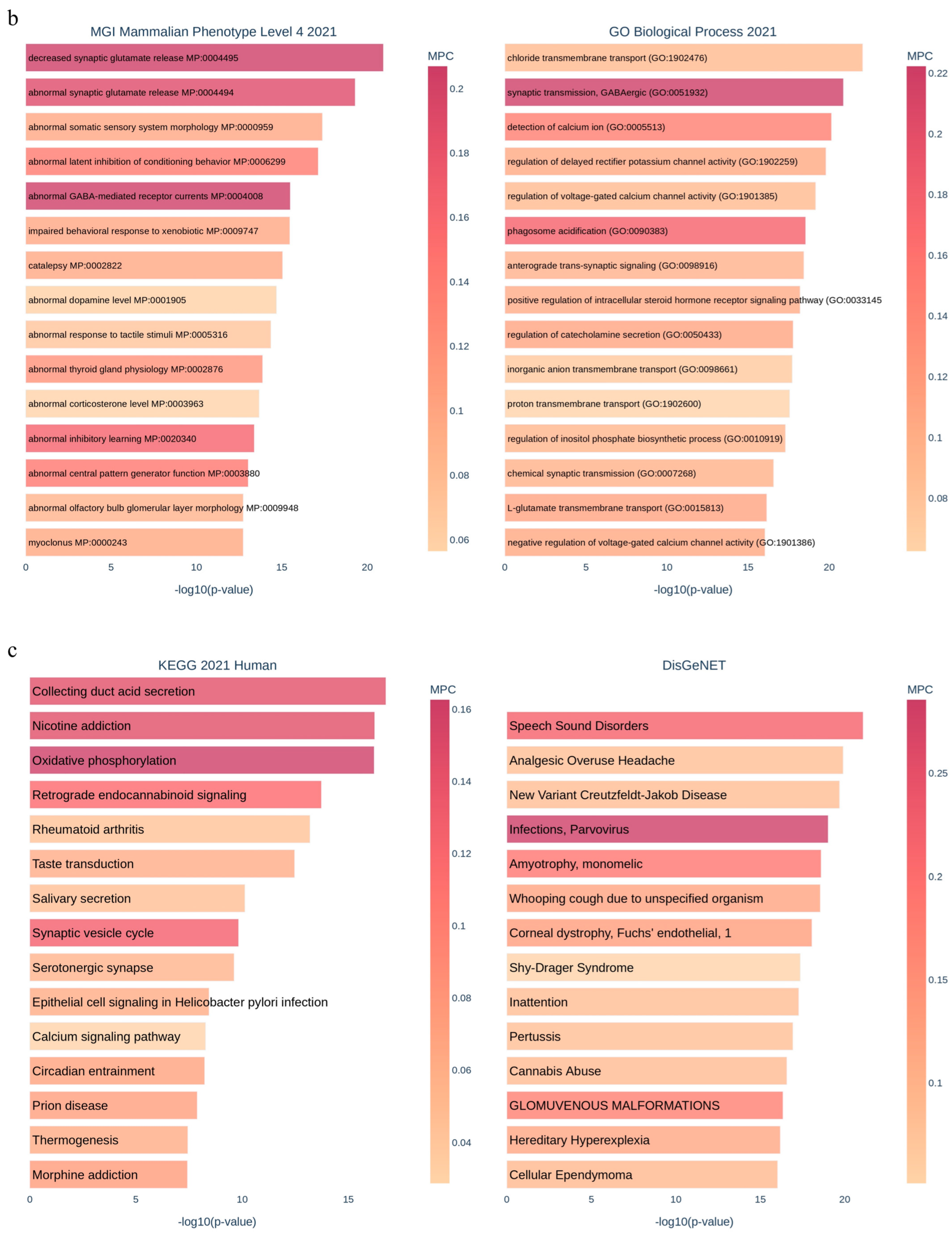
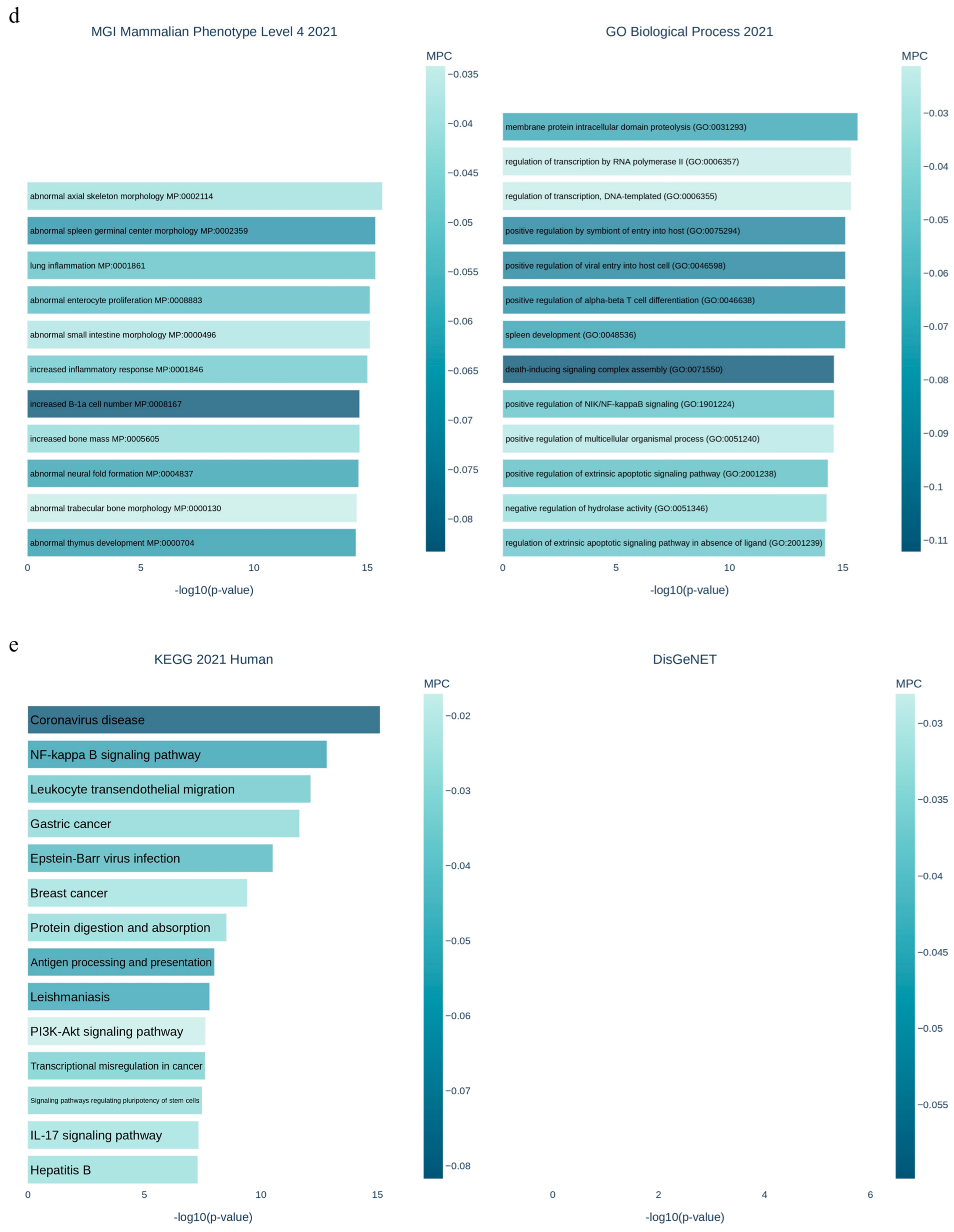
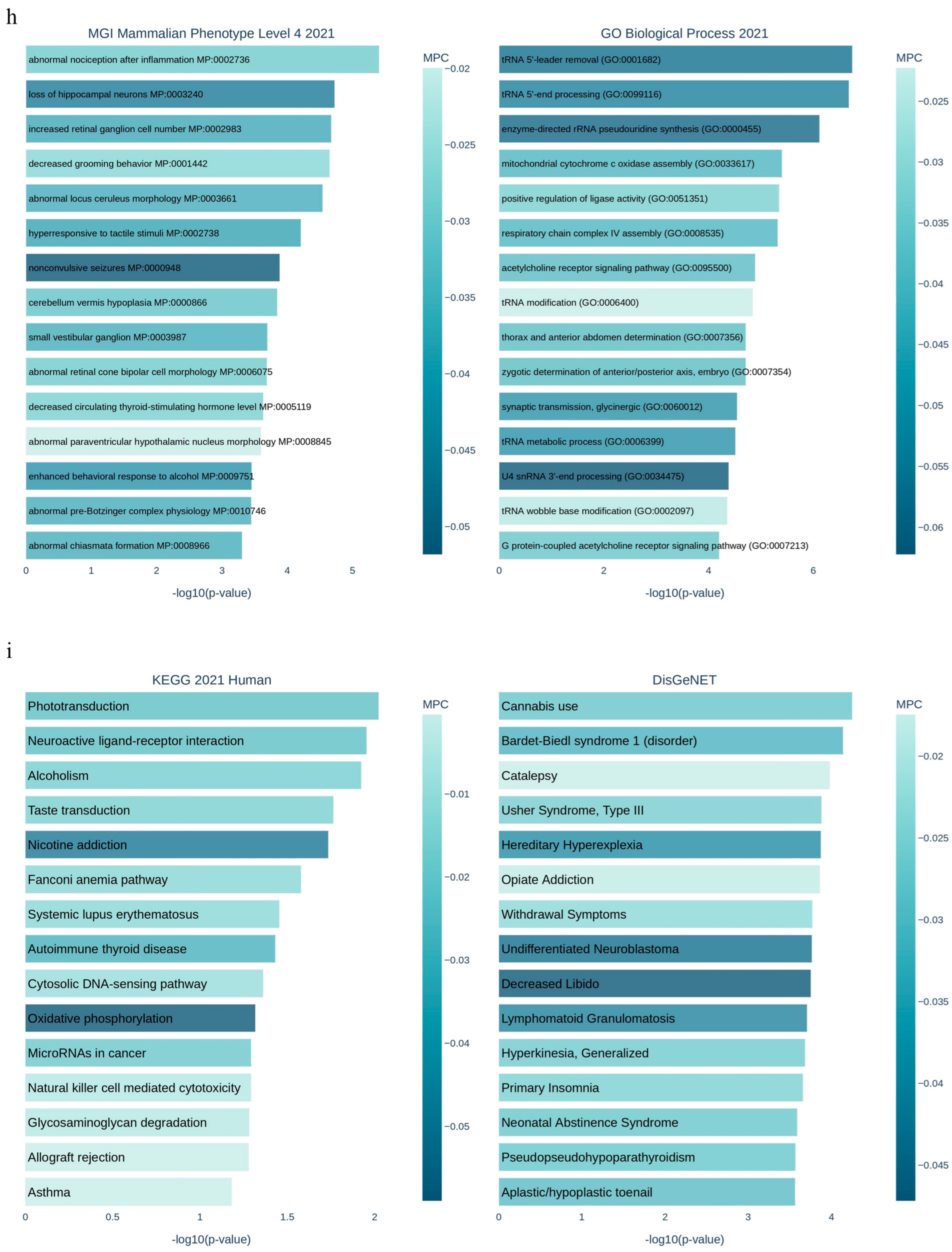
3.1. Applying System Biology Research and Computational Molecular Docking Insights
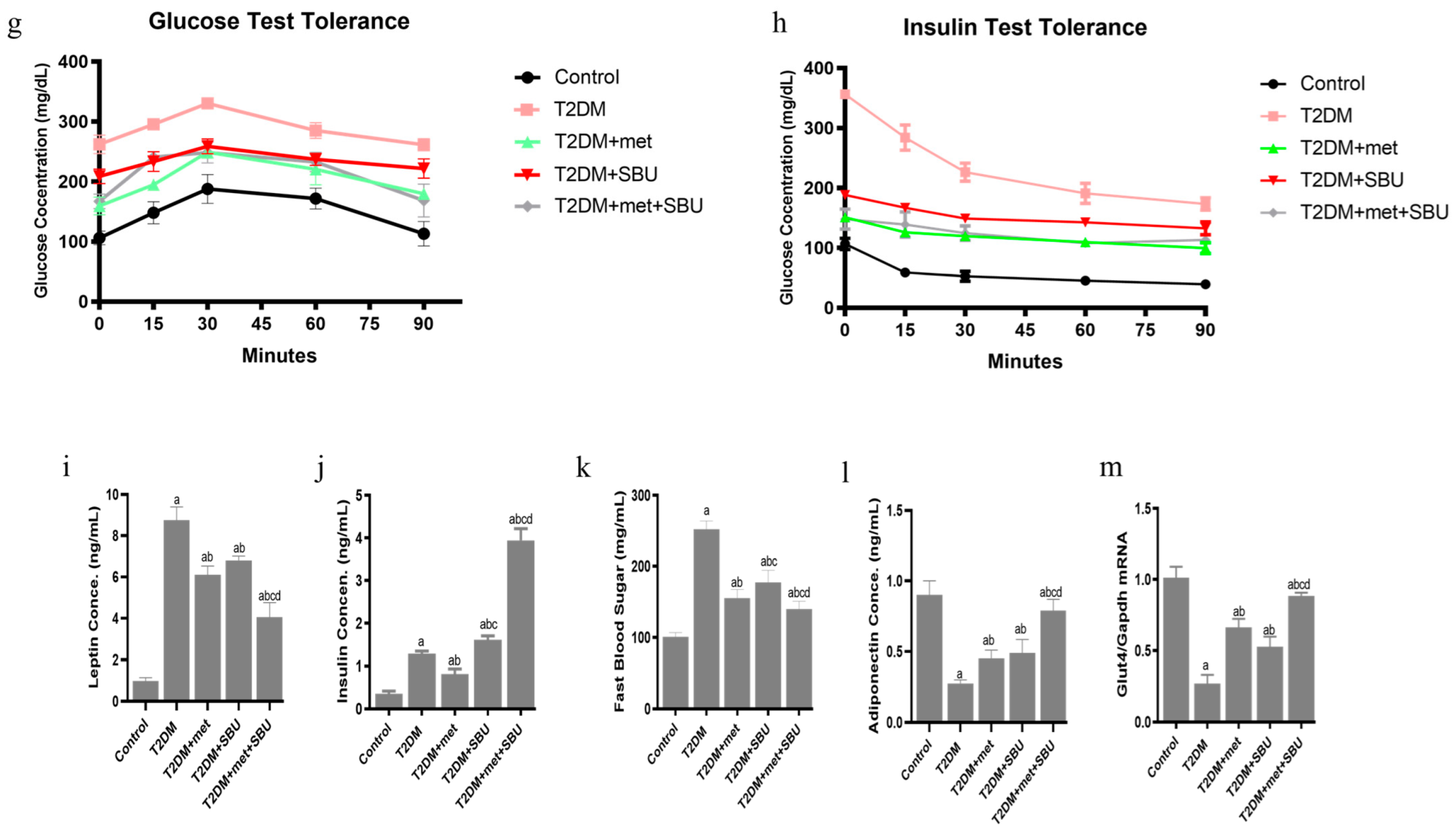
3.2. Metabolic, Physiological, and Behavior Effects of SBU and Metformin in Diabetic Mice
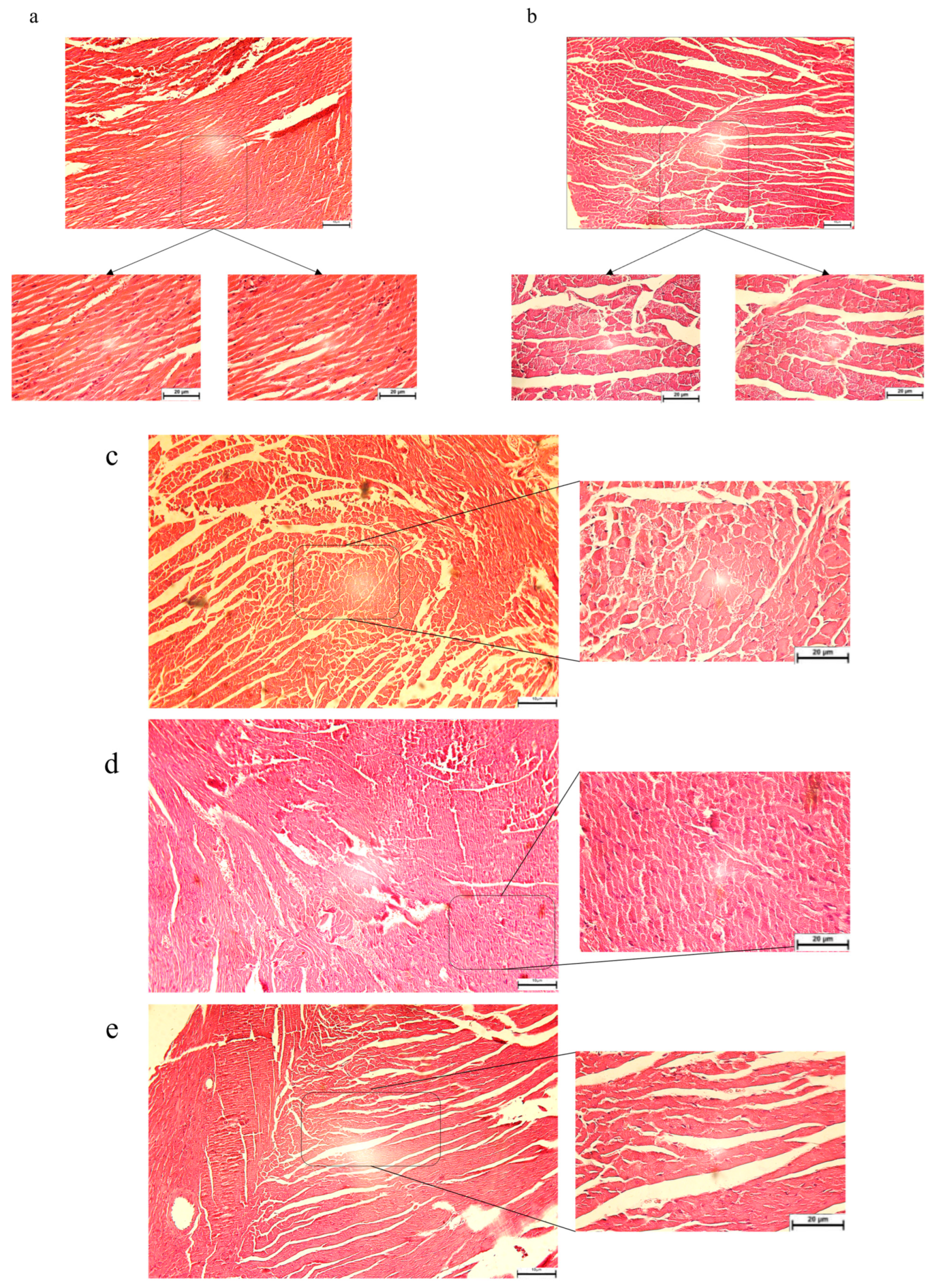
3.3. The Pathological Features Were Evaluated in Diabetic Mice: SBU and Metformin Improved the Pathological Features in Diabetic Mice
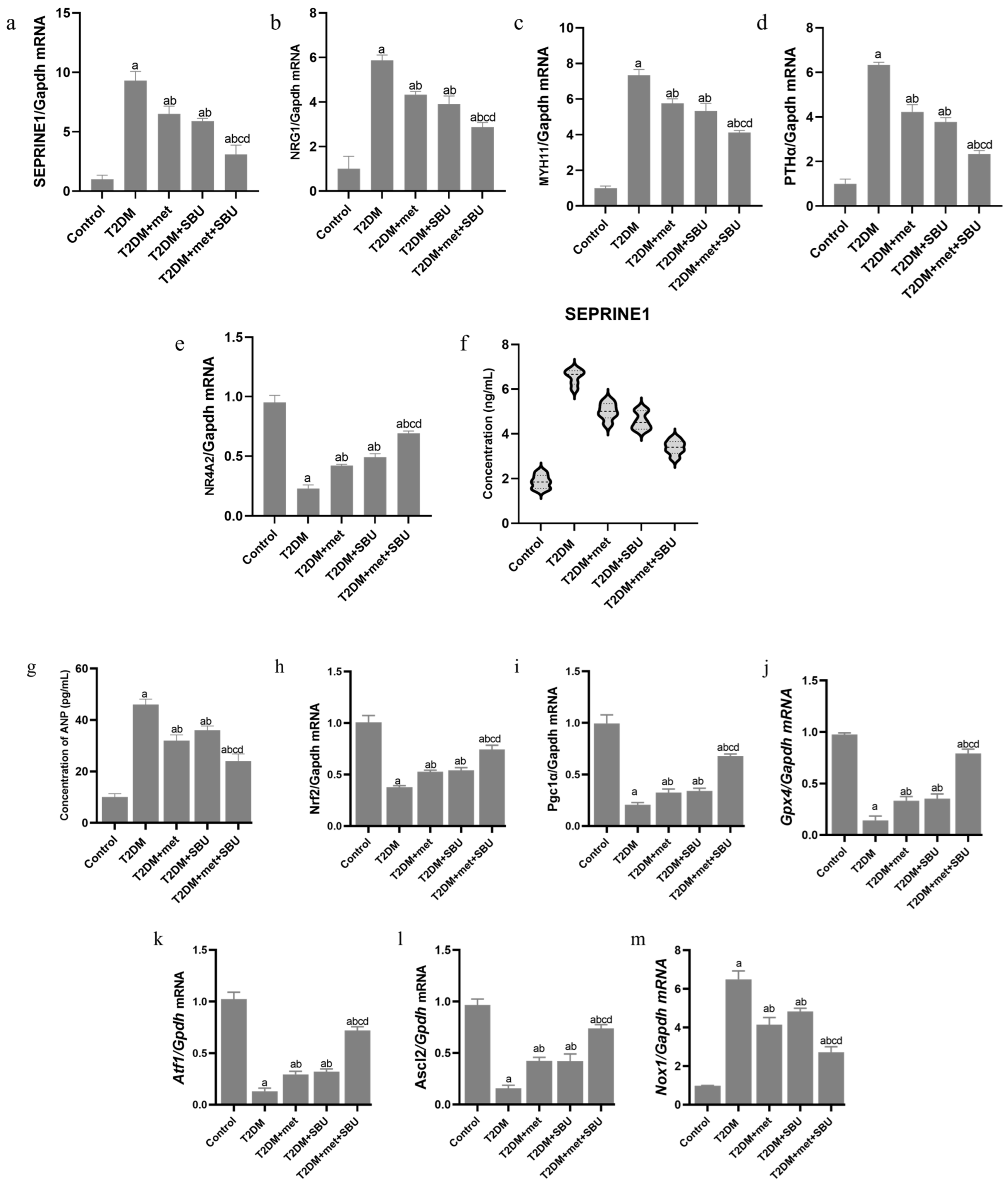
3.4. The Pattern Expression of SEPRINE1/NRG1/MYH11/PTH/NR4A in the Heart of Diabetic Mice: SBU and Metformin Improved the SEPRINE1/NRG1/MYH11/PTH/NR4A Level
3.5. ANP Concentration Was Evaluated in Serum of Diabetic Mice: SBU and Metformin Improved the ANP Concentration in Diabetic Mice
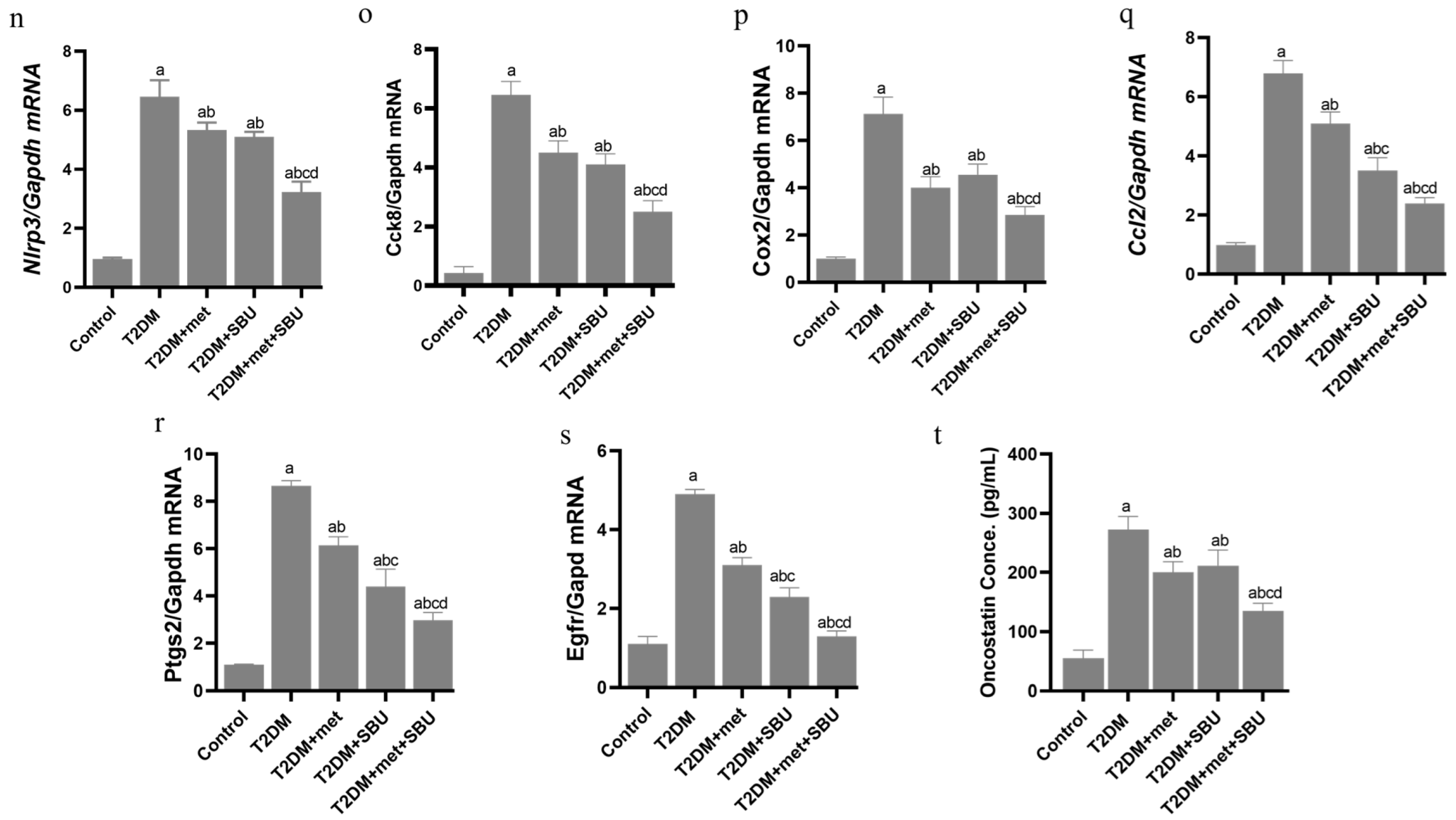
3.6. Dysregulation of the NRF2/PGC1α/ATF1/Ascl2/NOX1/GPX4/NLRP3/CCK8/COX2/CCL2/PTGS2/EGFR Network in the PBMC of Diabetic Mice
3.7. SBU and Metformin Mediated the NRF2/PGC1α/ATF1/ASCL2/NOX1/GPX4/NLRP3/CCK8/COX2/CCL2/PTGS2/EGFR Network in PBMC
3.8. Oncostatin as a Cell Death Biomarker Was Increased in Serum of Diabetic Mice
3.9. SBU and Metformin Regulated Oncostatin in Serum of Diabetic Mice
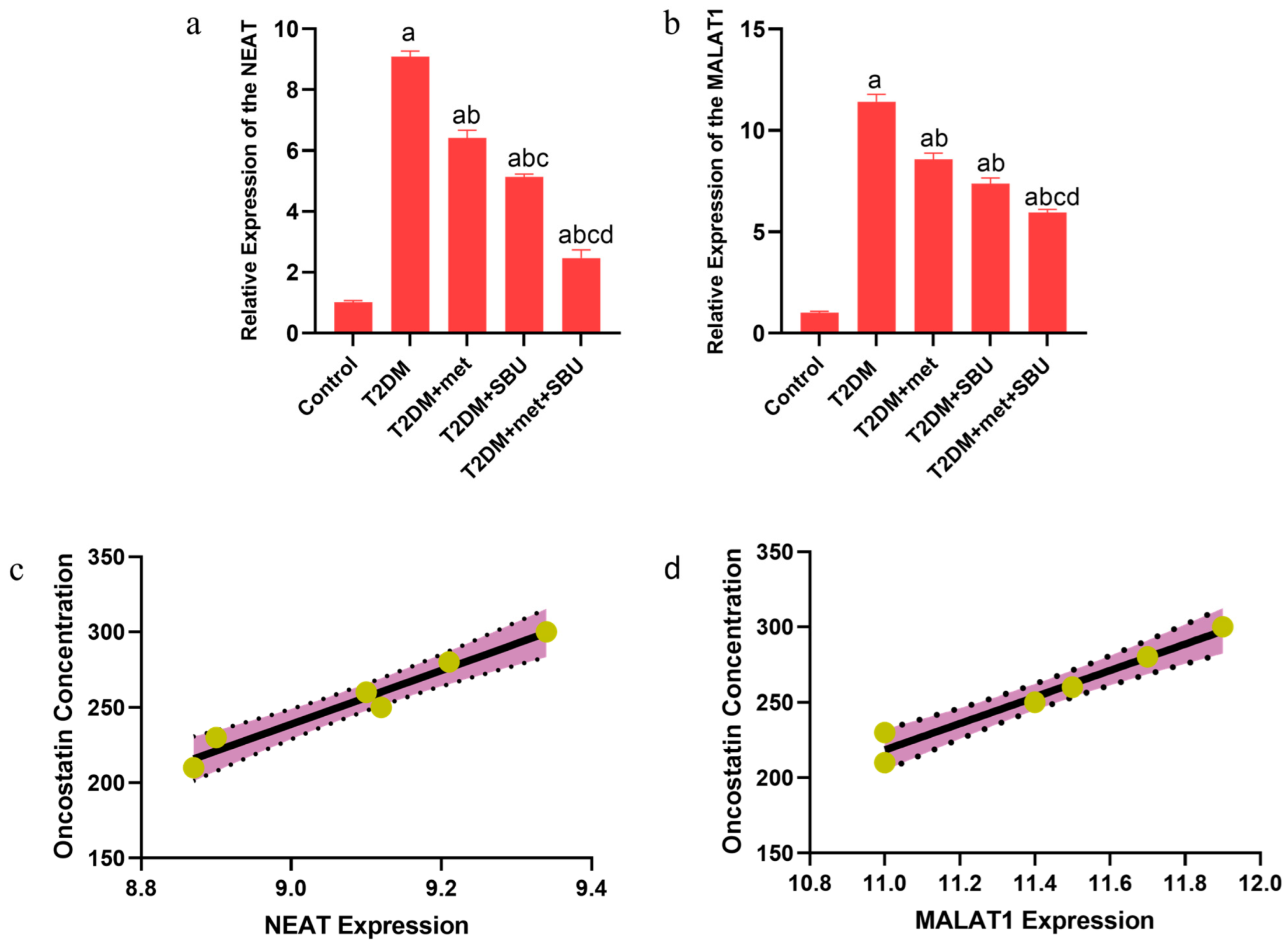
3.10. The lncRNAs (NEAT1 and MALAT1) Are Implicated in the Diabetic Condition: SBU and Metformin Ameliorated the Expression of lncRNAs (NEAT1 and MALAT1) in the Heart Tissue
3.11. The Correlation Between lncRNAs (NEAT1 and MALAT1) and Oncostatin
4. Discussion
Limitations and Future Perspectives
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sugandh, F.; Chandio, M.; Raveena, F.; Kumar, L.; Karishma, F.; Khuwaja, S.; Memon, U.A.; Bai, K.; Kashif, M.; Varrassi, G. Advances in the management of diabetes mellitus: A focus on personalized medicine. Cureus 2023, 15, e43697. [Google Scholar] [CrossRef] [PubMed]
- Antar, S.A.; Ashour, N.A.; Sharaky, M.; Khattab, M.; Ashour, N.A.; Zaid, R.T.; Roh, E.J.; Elkamhawy, A.; Al-Karmalawy, A.A. Diabetes mellitus: Classification, mediators, and complications; A gate to identify potential targets for the development of new effective treatments. Biomed. Pharmacother. 2023, 168, 115734. [Google Scholar]
- Sun, H.; Saeedi, P.; Karuranga, S.; Pinkepank, M.; Ogurtsova, K.; Duncan, B.B.; Stein, C.; Basit, A.; Chan, J.C.; Mbanya, J.C. IDF Diabetes Atlas: Global, regional and country-level diabetes prevalence estimates for 2021 and projections for 2045. Diabetes Res. Clin. Pract. 2022, 183, 109119. [Google Scholar]
- Mendis, S.; Graham, I.; Narula, J. Addressing the global burden of cardiovascular diseases; need for scalable and sustainable frameworks. Glob. Heart 2022, 17, 48. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, S.S.; Epstein, S.; Corkey, B.E.; Grant, S.F.; Gavin, J.R., III; Aguilar, R.B.; Herman, M.E. A unified pathophysiological construct of diabetes and its complications. Trends Endocrinol. Metab. 2017, 28, 645–655. [Google Scholar]
- Akasheva, D.U.; Utina, T.G.; Dzhioeva, O.N.; Drapkina, O.M. Subclinical left ventricular dysfunction over seven-year follow-up in type 2 diabetes patients without cardiovascular diseases. Biomedicines 2024, 12, 2031. [Google Scholar] [CrossRef] [PubMed]
- Bogle, C.; Colan, S.D.; Miyamoto, S.D.; Choudhry, S.; Baez-Hernandez, N.; Brickler, M.M.; Feingold, B.; Lal, A.K.; Lee, T.M.; Canter, C.E. Treatment strategies for cardiomyopathy in children: A scientific statement from the American Heart Association. Circulation 2023, 148, 174–195. [Google Scholar]
- Quaiyoom, A.; Kumar, R. An Overview of Diabetic Cardiomyopathy. Curr. Diabetes Rev. 2024, 20, 97–109. [Google Scholar]
- Malinow, I.; Fong, D.C.; Miyamoto, M.; Badran, S.; Hong, C.C. Pediatric dilated cardiomyopathy: A review of current clinical approaches and pathogenesis. Front. Pediatr. 2024, 12, 1404942. [Google Scholar]
- Prandi, F.R.; Evangelista, I.; Sergi, D.; Palazzuoli, A.; Romeo, F. Mechanisms of cardiac dysfunction in diabetic cardiomyopathy: Molecular abnormalities and phenotypical variants. Heart Fail. Rev. 2023, 28, 597–606. [Google Scholar]
- Fang, Z.; Raza, U.; Song, J.; Lu, J.; Yao, S.; Liu, X.; Zhang, W.; Li, S. Systemic aging fuels heart failure: Molecular mechanisms and therapeutic avenues. ESC Heart Fail. 2024, 12, 1059–1080. [Google Scholar] [CrossRef] [PubMed]
- Mauricio, D.; Alonso, N. Chronic Complications of Diabetes Mellitus: Current Outlook and Novel Pathophysiological Insights; Elsevier: Amsterdam, The Netherlands, 2024. [Google Scholar]
- Makrecka-Kuka, M.; Liepinsh, E.; Murray, A.J.; Lemieux, H.; Dambrova, M.; Tepp, K.; Puurand, M.; Käämbre, T.; Han, W.H.; de Goede, P. Altered mitochondrial metabolism in the insulin-resistant heart. Acta Physiol. 2020, 228, e13430. [Google Scholar] [CrossRef] [PubMed]
- Cai, K.; Jiang, H.; Zou, Y.; Song, C.; Cao, K.; Chen, S.; Wu, Y.; Zhang, Z.; Geng, D.; Zhang, N. Programmed death of cardiomyocytes in cardiovascular disease and new therapeutic approaches. Pharmacol. Res. 2024, 206, 107281. [Google Scholar] [CrossRef]
- Vitale, I.; Pietrocola, F.; Guilbaud, E.; Aaronson, S.A.; Abrams, J.M.; Adam, D.; Agostini, M.; Agostinis, P.; Alnemri, E.S.; Altucci, L. Apoptotic cell death in disease—Current understanding of the NCCD 2023. Cell Death Differ. 2023, 30, 1097–1154. [Google Scholar] [PubMed]
- Chen, Y.; Hua, Y.; Li, X.; Arslan, I.M.; Zhang, W.; Meng, G. Distinct types of cell death and the implication in diabetic cardiomyopathy. Front. Pharmacol. 2020, 11, 42. [Google Scholar] [CrossRef]
- Hajibabaie, F.; Abedpoor, N.; Mohamadynejad, P. Types of cell death from a molecular perspective. Biology 2023, 12, 1426. [Google Scholar] [CrossRef]
- Dixon, S.J.; Lemberg, K.M.; Lamprecht, M.R.; Skouta, R.; Zaitsev, E.M.; Gleason, C.E.; Patel, D.N.; Bauer, A.J.; Cantley, A.M.; Yang, W.S. Ferroptosis: An iron-dependent form of nonapoptotic cell death. Cell 2012, 149, 1060–1072. [Google Scholar] [CrossRef]
- Feng, S.; Tang, D.; Wang, Y.; Li, X.; Bao, H.; Tang, C.; Dong, X.; Li, X.; Yang, Q.; Yan, Y. The mechanism of ferroptosis and its related diseases. Mol. Biomed. 2023, 4, 33. [Google Scholar]
- Kaviani, E.; Hajibabaie, F.; Abedpoor, N.; Safavi, K.; Ahmadi, Z.; Karimy, A. System biology analysis to develop diagnostic biomarkers, monitoring pathological indexes, and novel therapeutic approaches for immune targeting based on maggot bioactive compounds and polyphenolic cocktails in mice with gastric cancer. Environ. Res. 2023, 238, 117168. [Google Scholar] [CrossRef]
- Tay, Y.; Rinn, J.; Pandolfi, P.P. The multilayered complexity of ceRNA crosstalk and competition. Nature 2014, 505, 344–352. [Google Scholar] [CrossRef]
- Jin, W.; Liu, J.; Yang, J.; Feng, Z.; Feng, Z.; Huang, N.; Yang, T.; Yu, L. Identification of a key ceRNA network associated with ferroptosis in gastric cancer. Sci. Rep. 2022, 12, 20088. [Google Scholar]
- Yang, Z.; He, K.; Chen, W.; Chen, Y. A ferroptosis-related ceRNA network in hepatocellular carcinoma for potential clinical applications. Am. J. Transl. Res. 2023, 15, 3912. [Google Scholar]
- Zhu, H.; Tan, J.; Wang, Z.; Wu, Z.; Zhou, W.; Zhang, Z.; Li, M.; Zhao, Y. Bioinformatics analysis constructs potential ferroptosis-related ceRNA network involved in the formation of intracranial aneurysm. Front. Cell. Neurosci. 2022, 16, 1016682. [Google Scholar]
- Bao, T.; Zhang, X.; Xie, W.; Wang, Y.; Li, X.; Tang, C.; Yang, Y.; Sun, J.; Gao, J.; Yu, T. Natural compounds efficacy in complicated diabetes: A new twist impacting ferroptosis. Biomed. Pharmacother. 2023, 168, 115544. [Google Scholar]
- Mu, W.; Jiang, Y.; Liang, G.; Feng, Y.; Qu, F. Metformin: A promising antidiabetic medication for cancer treatment. Curr. Drug Targets 2023, 24, 41–54. [Google Scholar]
- Xu, L.; Li, W.; Chen, Y.; Liu, S.; Liu, G.; Luo, W.; Cao, G.; Wang, S. Metformin regulates cardiac ferroptosis to reduce metabolic syndrome-induced cardiac dysfunction. Appl. Biochem. Biotechnol. 2024, 197, 179–193. [Google Scholar]
- Ma, W.-Q.; Sun, X.-J.; Zhu, Y.; Liu, N.-F. Metformin attenuates hyperlipidaemia-associated vascular calcification through anti-ferroptotic effects. Free. Radic. Biol. Med. 2021, 165, 229–242. [Google Scholar]
- Zhang, T.; Wang, M.-Y.; Wang, G.-D.; Lv, Q.-Y.; Huang, Y.-Q.; Zhang, P.; Wang, W.; Zhang, Y.; Bai, Y.-P.; Guo, L.-Q. Metformin improves nonalcoholic fatty liver disease in db/db mice by inhibiting ferroptosis. Eur. J. Pharmacol. 2024, 966, 176341. [Google Scholar]
- Wang, Z.; Zhao, F.; Wei, P.; Chai, X.; Hou, G.; Meng, Q. Phytochemistry, health benefits, and food applications of sea buckthorn (Hippophae rhamnoides L.): A comprehensive review. Front. Nutr. 2022, 9, 1036295. [Google Scholar]
- Xunlin, C.; Yongshan, L. The geographical distribution patterns and its formative factors on the genus Hippophae L. Acta Bot. Boreali-Occident. Sin. 1994, 14, 105–110. [Google Scholar]
- Stobdan, T.; Targais, K.; Lamo, D.; Srivastava, R.B. Judicious use of natural resources: A case study of traditional uses of seabuckthorn (Hippophae rhamnoides L.) in trans-Himalayan Ladakh, India. Natl. Acad. Sci. Lett. 2013, 36, 609–613. [Google Scholar]
- Gâtlan, A.-M.; Gutt, G. Sea buckthorn in plant based diets. An analytical approach of sea buckthorn fruits composition: Nutritional value, applications, and health benefits. Int. J. Environ. Res. Public Health 2021, 18, 8986. [Google Scholar] [CrossRef] [PubMed]
- Ghendov-Mosanu, A.; Cristea, E.; Patras, A.; Sturza, R.; Padureanu, S.; Deseatnicova, O.; Turculet, N.; Boestean, O.; Niculaua, M. Potential application of Hippophae Rhamnoides in wheat bread production. Molecules 2020, 25, 1272. [Google Scholar] [CrossRef]
- Suryakumar, G.; Gupta, A. Medicinal and therapeutic potential of Sea buckthorn (Hippophae rhamnoides L.). J. Ethnopharmacol. 2011, 138, 268–278. [Google Scholar] [CrossRef]
- Masoodi, K.Z.; Wani, W.; Dar, Z.A.; Mansoor, S.; Anam-ul-Haq, S.; Farooq, I.; Hussain, K.; Wani, S.A.; Nehvi, F.A.; Ahmed, N. Sea buckthorn (Hippophae rhamnoides L.) inhibits cellular proliferation, wound healing and decreases expression of prostate specific antigen in prostate cancer cells in vitro. J. Funct. Foods 2020, 73, 104102. [Google Scholar]
- Rédei, D.; Kúsz, N.; Rafai, T.; Bogdanov, A.; Burián, K.; Csorba, A.; Mándi, A.; Kurtán, T.; Vasas, A.; Hohmann, J. 14-Noreudesmanes and a phenylpropane heterodimer from sea buckthorn berry inhibit Herpes simplex type 2 virus replication. Tetrahedron 2019, 75, 1364–1370. [Google Scholar]
- Tanwar, H.; Shweta; Singh, D.; Singh, S.B.; Ganju, L. Anti-inflammatory activity of the functional groups present in Hippophae rhamnoides (Seabuckthorn) leaf extract. Inflammopharmacology 2018, 26, 291–301. [Google Scholar]
- Yang, F.; Suo, Y.; Chen, D.; Tong, L. Protection against vascular endothelial dysfunction by polyphenols in sea buckthorn berries in rats with hyperlipidemia. Biosci. Trends 2016, 10, 188–196. [Google Scholar] [CrossRef]
- Solà Marsiñach, M.; Cuenca, A.P. The impact of sea buckthorn oil fatty acids on human health. Lipids Health Dis. 2019, 18, 145. [Google Scholar]
- Szklarczyk, D.; Kirsch, R.; Koutrouli, M.; Nastou, K.; Mehryary, F.; Hachilif, R.; Gable, A.L.; Fang, T.; Doncheva, N.T.; Pyysalo, S. The STRING database in 2023: Protein–protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 2023, 51, D638–D646. [Google Scholar] [CrossRef]
- Otasek, D.; Morris, J.H.; Bouças, J.; Pico, A.R.; Demchak, B. Cytoscape automation: Empowering workflow-based network analysis. Genome Biol. 2019, 20, 185. [Google Scholar]
- Bastian, M.; Heymann, S.; Jacomy, M. Gephi: An open source software for exploring and manipulating networks. In Proceedings of the International AAAI Conference on Web and Social Media, San Jose, CA, USA, 17–20 May 2009; Volume 3, pp. 361–362. [Google Scholar]
- Chen, G.; Wang, Z.; Wang, D.; Qiu, C.; Liu, M.; Chen, X.; Zhang, Q.; Yan, G.; Cui, Q. LncRNADisease: A database for long-non-coding RNA-associated diseases. Nucleic Acids Res. 2012, 41, D983–D986. [Google Scholar]
- Zhao, H.; Yin, X.; Xu, H.; Liu, K.; Liu, W.; Wang, L.; Zhang, C.; Bo, L.; Lan, X.; Lin, S. LncTarD 2.0: An updated comprehensive database for experimentally-supported functional lncRNA–target regulations in human diseases. Nucleic Acids Res. 2023, 51, D199–D207. [Google Scholar]
- Marino, G.B.; Wojciechowicz, M.L.; Clarke, D.J.; Kuleshov, M.V.; Xie, Z.; Jeon, M.; Lachmann, A.; Ma’ayan, A. lncHUB2: Aggregated and inferred knowledge about human and mouse lncRNAs. Database 2023, 2023, baad009. [Google Scholar] [PubMed]
- Chang, L.; Xia, J. MicroRNA regulatory network analysis using miRNet 2.0. In Transcription Factor Regulatory Networks; Springer: Berlin/Heidelberg, Germany, 2022; pp. 185–204. [Google Scholar]
- Yang, X.; Wang, Q.; Pang, Z.-R.; Pan, M.-R.; Zhang, W. Flavonoid-enriched extract from Hippophae rhamnoides seed reduces high fat diet induced obesity, hypertriglyceridemia, and hepatic triglyceride accumulation in C57BL/6 mice. Pharm. Biol. 2017, 55, 1207–1214. [Google Scholar]
- de Figueiredo Cerqueira, M.M.; Castro, M.M.L.; Vieira, A.A.; Kurosawa, J.A.A.; do Amaral Junior, F.L.; de Siqueira, F.D.C.C.; Sosthenes, M.C.K. Comparative analysis between Open Field and Elevated Plus Maze tests as a method for evaluating anxiety-like behavior in mice. Heliyon 2023, 9, e14522. [Google Scholar] [PubMed]
- Pang, X.; Zhao, J.; Zhang, W.; Zhuang, X.; Wang, J.; Xu, R.; Xu, Z.; Qu, W. Antihypertensive effect of total flavones extracted from seed residues of Hippophae rhamnoides L. in sucrose-fed rats. J. Ethnopharmacol. 2008, 117, 325–331. [Google Scholar]
- Rahimi, G.; Heydari, S.; Rahimi, B.; Abedpoor, N.; Niktab, I.; Safaeinejad, Z.; Peymani, M.; Seyed Forootan, F.; Derakhshan, Z.; Esfahani, M.H.N. A combination of herbal compound (SPTC) along with exercise or metformin more efficiently alleviated diabetic complications through down-regulation of stress oxidative pathway upon activating Nrf2-Keap1 axis in AGE rich diet-induced type 2 diabetic mice. Nutr. Metab. 2021, 18, 14. [Google Scholar]
- Ngo, V.; Duennwald, M.L. Nrf2 and oxidative stress: A general overview of mechanisms and implications in human disease. Antioxidants 2022, 11, 2345. [Google Scholar] [CrossRef]
- Liu, Y.; Wan, Y.; Jiang, Y.; Zhang, L.; Cheng, W. GPX4: The hub of lipid oxidation, ferroptosis, disease and treatment. Biochim. Et Biophys. Acta (BBA)-Rev. Cancer 2023, 1878, 188890. [Google Scholar]
- Tao, L.; Yang, K.; Wang, K.; Yang, Y. NOX1-mediated oxidative stress induces chondrocyte ferroptosis by inhibiting the Nrf2/HO-1 pathway. Sci. Rep. 2024, 14, 19877. [Google Scholar]
- Rius-Pérez, S.; Torres-Cuevas, I.; Millán, I.; Ortega, Á.L.; Pérez, S. PGC-1α, inflammation, and oxidative stress: An integrative view in metabolism. Oxidative Med. Cell. Longev. 2020, 2020, 1452696. [Google Scholar]
- Yang, W.S.; Stockwell, B.R. Ferroptosis: Death by lipid peroxidation. Trends Cell Biol. 2016, 26, 165–176. [Google Scholar] [PubMed]
- Vainorius, G.; Novatchkova, M.; Michlits, G.; Baar, J.C.; Raupach, C.; Lee, J.; Yelagandula, R.; Wernig, M.; Elling, U. Ascl1 and Ngn2 convert mouse embryonic stem cells to neurons via functionally distinct paths. Nat. Commun. 2023, 14, 5341. [Google Scholar] [PubMed]
- Lin, W.; Wang, X.; Diao, M.; Wang, Y.; Zhao, R.; Chen, J.; Liao, Y.; Long, Q.; Meng, Y. Promoting reactive oxygen species accumulation to overcome tyrosine kinase inhibitor resistance in cancer. Cancer Cell Int. 2024, 24, 239. [Google Scholar]
- Zhu, Q.; Han, Y.; He, Y.; Meng, P.; Fu, Y.; Yang, H.; He, G.; Long, M.; Shi, Y. Quercetin inhibits neuronal Ferroptosis and promotes immune response by targeting lipid metabolism-related gene PTGS2 to alleviate breast cancer-related depression. Phytomedicine 2024, 130, 155560. [Google Scholar] [CrossRef]
- Ma, S.; Sun, L.; Wu, W.; Wu, J.; Sun, Z.; Ren, J. USP22 protects against myocardial ischemia–reperfusion injury via the SIRT1-p53/SLC7A11–dependent inhibition of ferroptosis–induced cardiomyocyte death. Front. Physiol. 2020, 11, 551318. [Google Scholar] [CrossRef]
- Zhao, Y.; Pan, B.; Lv, X.; Chen, C.; Li, K.; Wang, Y.; Liu, J. Ferroptosis: Roles and molecular mechanisms in diabetic cardiomyopathy. Front. Endocrinol. 2023, 14, 1140644. [Google Scholar]
- Zhou, Y.; Suo, W.; Zhang, X.; Liang, J.; Zhao, W.; Wang, Y.; Li, H.; Ni, Q. Targeting mitochondrial quality control for diabetic cardiomyopathy: Therapeutic potential of hypoglycemic drugs. Biomed. Pharmacother. 2023, 168, 115669. [Google Scholar]
- Cai, Y.; Zhou, Y.; Li, Z.; Xia, P.; ChenFu, X.; Shi, A.; Zhang, J.; Yu, P. Non-coding RNAs in necroptosis, pyroptosis, and ferroptosis in cardiovascular diseases. Front. Cardiovasc. Med. 2022, 9, 909716. [Google Scholar]
- Dutta, S.; Shah, R.B.; Singhal, S.; Dutta, S.B.; Bansal, S.; Sinha, S.; Haque, M. Metformin: A review of potential mechanism and therapeutic utility beyond diabetes. Drug Des. Dev. Ther. 2023, 17, 1907–1932. [Google Scholar] [CrossRef] [PubMed]
- Shang, R.; Miao, J. Mechanisms and effects of metformin on skeletal muscle disorders. Front. Neurol. 2023, 14, 1275266. [Google Scholar] [CrossRef] [PubMed]
- Rena, G.; Hardie, D.G.; Pearson, E.R. The mechanisms of action of metformin. Diabetologia 2017, 60, 1577–1585. [Google Scholar] [CrossRef]
- Chen, Q.; Wang, D.; Tan, C.; Hu, Y.; Sundararajan, B.; Zhou, Z. Profiling of flavonoid and antioxidant activity of fruit tissues from 27 Chinese local citrus cultivars. Plants 2020, 9, 196. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Liu, L.Q.; Xu, L.L.; Xing, Y.; Ye, S. Metformin alleviates renal injury in diabetic rats by inducing Sirt1/FoxO1 autophagic signal axis. Clin. Exp. Pharmacol. Physiol. 2020, 47, 599–608. [Google Scholar] [CrossRef]
- Kopčeková, J.; Mrázová, J.; Fatrcová-Šramková, K.; Chlebo, P.; Bihari, M.; Gažarová, M.; Lenártová, P.; Habánová, M.; Navrátilová, A. Modification of lipid profile, ldl subfractions and other cardiovascular risk markers in hypercholesterolemic women through the sea buckthorn juice consumption. arXiv 2023, arXiv:202309.1474.v1. [Google Scholar]
- Zhao, C.; Ye, Z.; Ma, Z.-X.; Wildman, S.A.; Blaszczyk, S.A.; Hu, L.; Guizei, I.A.; Tang, W. A general strategy for diversifying complex natural products to polycyclic scaffolds with medium-sized rings. Nat. Commun. 2019, 10, 4015. [Google Scholar] [CrossRef]
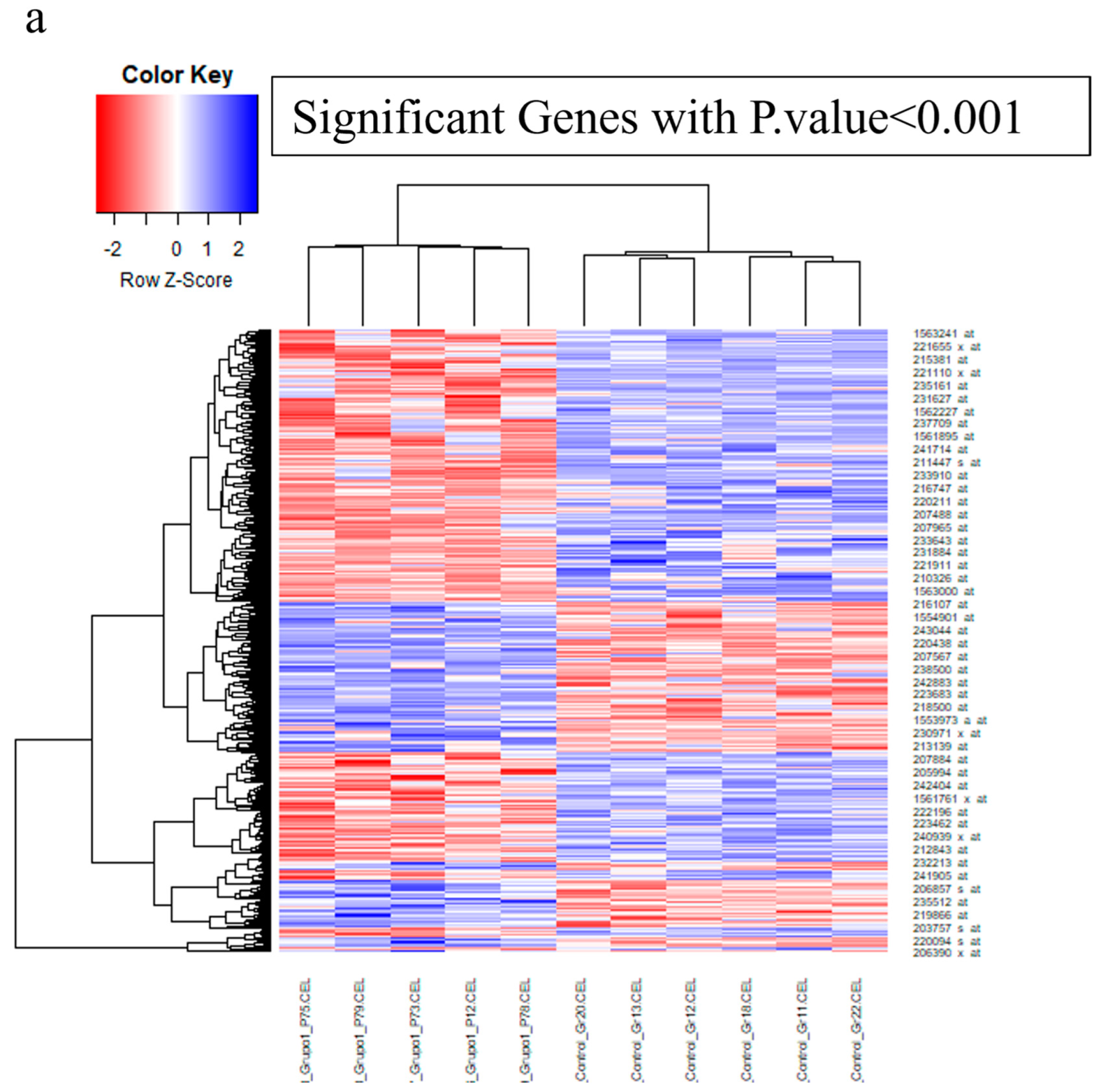
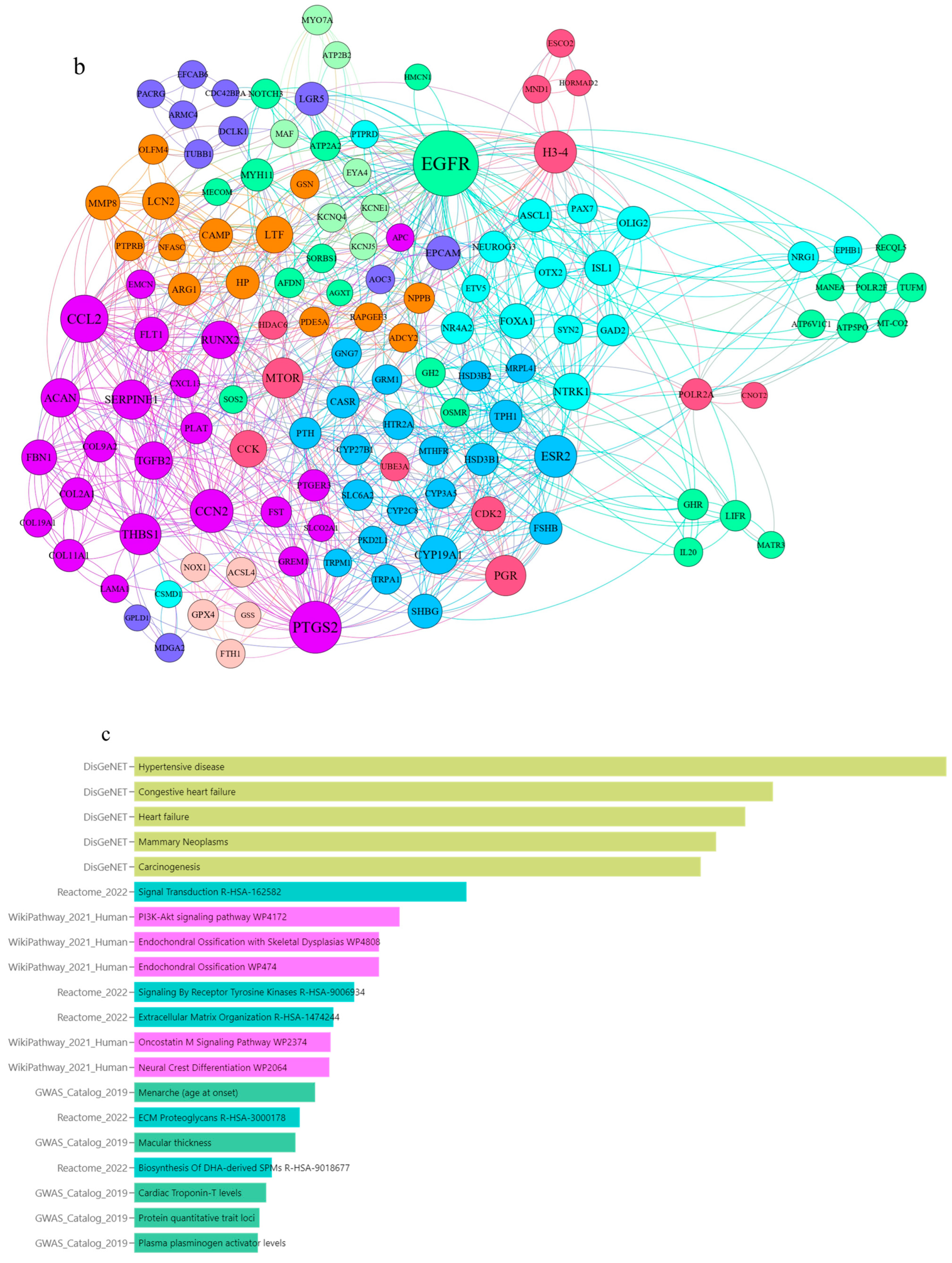
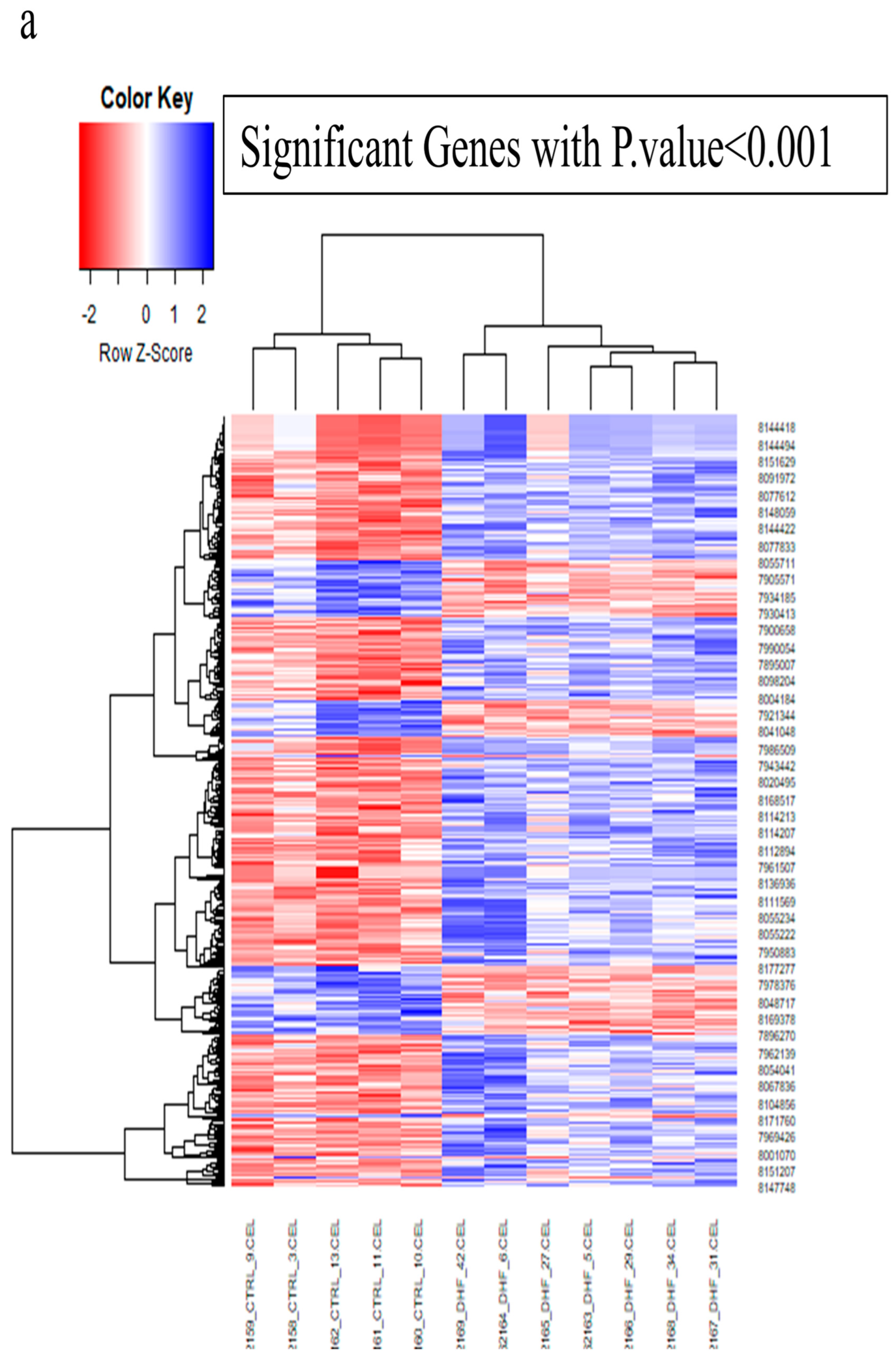
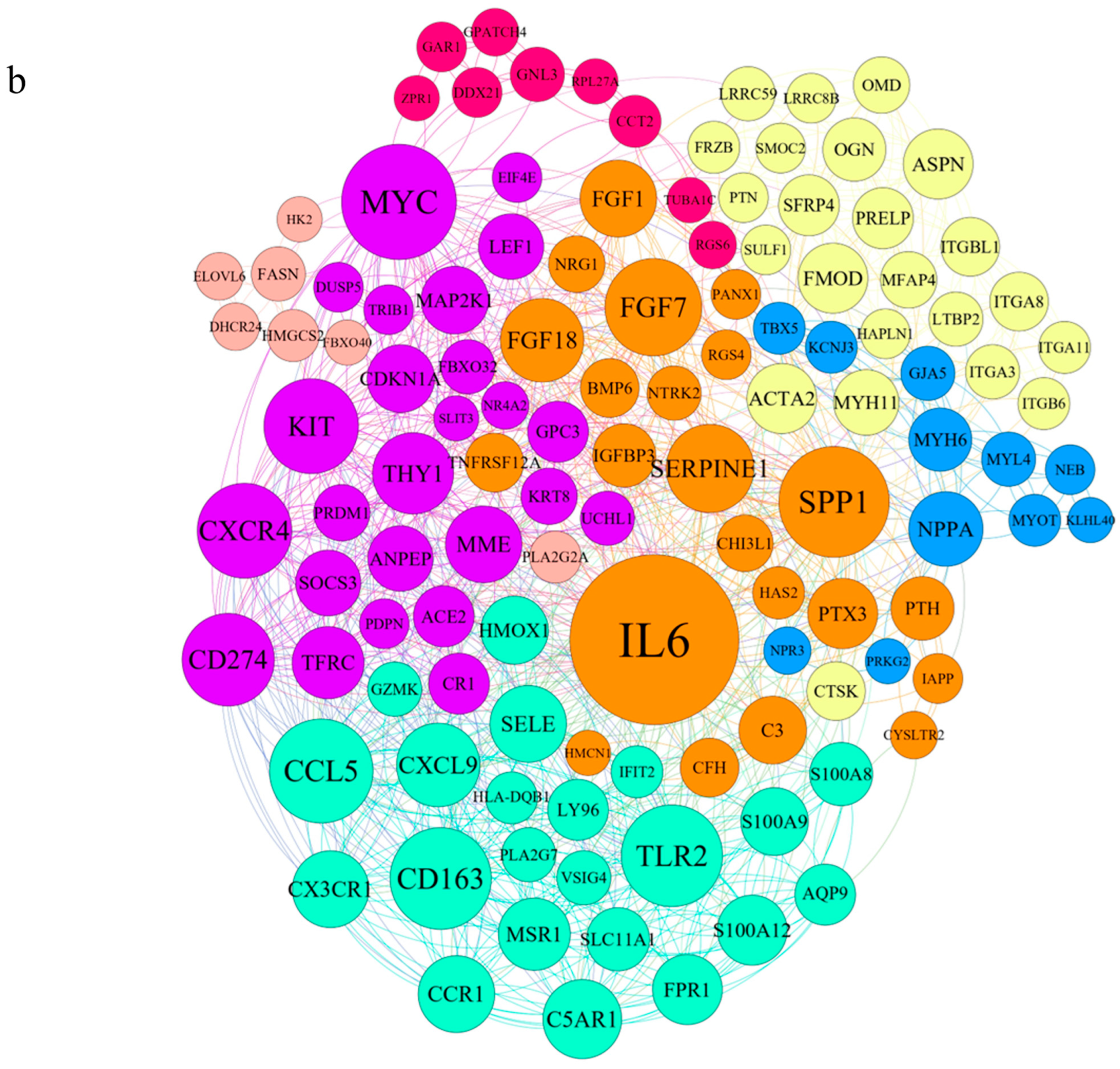




| Gene | Sequence of Primers | Annealing Temperature |
|---|---|---|
| NRF2 | Forward: 5′-CAGCATAGAGCAGGACATGGAG-3′ | 60 °C |
| Reverse: 5′-GAACAGCGGTAGTATCAGCCAG-3′ | ||
| PGC1α | Forward: 5′-GAATCAAGCCACTACAGACACCG-3′ | 60 °C |
| Reverse: 5′-CATCCCTCTTGAGCCTTTCGTG-3′ | ||
| ATF1 | Forward: 5′-ATCAGACGAGCAGCGGACAGTA-3′ | 60 °C |
| Reverse: 5′-GTCAGAGGTCTGTGCATACTGG-3′ | ||
| ASCL2 | Forward: 5′-TTTCCTGTGCCGCACCAGAACT-3′ | 60 °C |
| Reverse: 5′-CAGCGACTCCAGACGAGGTGG-3′ | ||
| NOX1 | Forward: 5′-CTCCAGCCTATCTCATCCTGAG-3′ | 60 °C |
| Reverse: 5′-AGTGGCAATCACTCCAGTAAGGC-3′ | ||
| GPX4 | Forward: 5′-CCTCTGCTGCAAGAGCCTCCC-3′ | 60 °C |
| Reverse: 5′-CTTATCCAGGCAGACCATGTGC-3′ | ||
| NLRP3 | Forward: 5′-TCACAACTCGCCCAAGGAGGAA-3′ | 60 °C |
| Reverse: 5′-AAGAGACCACGGCAGAAGCTAG-3′ | ||
| CCK8 | Forward: 5′-TAGCGCGATACATCCAGCAGGT-3′ | 60 °C |
| Reverse: 5′-GGTATTCGTAGTCCTCGGCACT-3′ | ||
| CCL2 | Forward: 5′- GCTACAAGAGGATCACCAGCAG-3′ | 60 °C |
| Reverse: 5′- GTCTGGACCCATTCCTTCTTGG-3′ | ||
| PTGS2 | Forward: 5′-GCGACATACTCAAGCAGGAGCA-3′ | 60 °C |
| Reverse: 5′-AGTGGTAACCGCTCAGGTGTTG-3′ | ||
| EGFR | Forward: 5′-GGACTGTGTCTCCTGCCAGAAT-3′ | 60 °C |
| Reverse: 5′- GGCAGACATTCTGGATGGCACT-3′ | ||
| GLUT4 | Forward: 5′-GGTGTGGTCAATACGGTCTTCAC-3′ | 60 °C |
| Reverse: 5′-AGCAGAGCCACGGTCATCAAGA-3′ | ||
| SERPINE1 | Forward: 5′-CCTCTTCCACAAGTCTGATGGC-3′ | 60 °C |
| Reverse: 5′-GCAGTTCCACAACGTCATACTCG-3′ | ||
| NRG1 | Forward: 5′-GCTCATCACTCCACGACTGTCA-3′ | 60 °C |
| Reverse: 5′-TGCCTGCTGTTCTCTACCGATG-3′ | ||
| MYH11 | Forward: 5′-GCAACTACAGGCTGAGAGGAAG-3′ | 60 °C |
| Reverse: 5′-TCAGCCGTGACCTTCTCTAGCT-3′ | ||
| PTH-α | Forward: 5′-CAAACGGATGGGAAACCCGTGA-3′ | 60 °C |
| Reverse: 5′-TGGCAGCCATTTGGACTCCAAG-3′ | ||
| NR4A2 | Forward: 5′-CCGCCGAAATCGTTGTCAGTAC-3′ | 60 °C |
| Reverse: 5′-TTCGGCTTCGAGGGTAAACGAC-3′ | ||
| NEAT1 | Forward: 5′-GCTCTGGGACCTTCGTGACTCT-3′ | 60 °C |
| Reverse: 5′-CTGCCTTGGCTTGGAAATGTAA-3′ | ||
| MALAT1 | Forward: 5′-GGCAGAATGCCTTTGAAGAG-3′ | 60 °C |
| Reverse: 5′-GGTCAGCTGCCAATGCTAGT-3′ | ||
| 18S rRNA | Forward: 5′-CGGACACGGACAGGATTG-3′ | 59 °C |
| Reverse: 5′-TCGCTCCACCAACTAAGAAC-3′ | ||
| GAPDH | Forward: 5′-TGCCGCCTGGAGAAACC-3′ | 60 °C |
| Reverse: 5′-TGAAGTCGCAGGAGACAACC-3′ |
| Macromolecule | Phytochemical Compound PubChem ID | RMSD Upper Bound | RMSD Lower Bound | Molecular Docking: Binding Affinity (kcal/mol) |
|---|---|---|---|---|
| PTGS | PTGS2PDB_157822370_uff_E = 101.29 | 0 | 0 | −5.5 |
| PTGS2PDB_54670067_uff_E = 200.65 | 0 | 0 | −5.8 | |
| PTGS2PDB_25244964_uff_E = 572.46 | 0 | 0 | −8.5 | |
| PTGS2PDB_25203368_uff_E = 541.72 | 0 | 0 | −8.9 | |
| PTGS2PDB_12795736_uff_E = 643.77 | 0 | 0 | −8.8 | |
| PTGS2PDB_11063337_uff_E = 244.73 | 0 | 0 | −7.2 | |
| PTGS2PDB_9828626_uff_E = 669.99 | 0 | 0 | −8.8 | |
| PTGS2PDB_9548595_uff_E = 638.65 | 0 | 0 | −9.1 | |
| PTGS2PDB_6419725_uff_E = 609.04 | 0 | 0 | −8.6 | |
| PTGS2PDB_5481663_uff_E = 826.93 | 0 | 0 | −10.5 | |
| PTGS2PDB_5318645_uff_E = 688.07 | 0 | 0 | −9.2 | |
| PTGS2PDB_5282761_uff_E = 74.80 | 0 | 0 | −5.9 | |
| PTGS2PDB_5281654_uff_E = 450.92 | 0 | 0 | −8.9 | |
| PTGS2PDB_5281243_uff_E = 655.10 | 0 | 0 | −8.5 | |
| PTGS2PDB_5281235_uff_E = 709.09 | 0 | 0 | −8.4 | |
| PTGS2PDB_5281230_uff_E = 422.22 | 0 | 0 | −10.8 | |
| PTGS2PDB_5281119_uff_E = 95.80 | 0 | 0 | −6.6 | |
| PTGS2PDB_5280934_uff_E = 142.21 | 0 | 0 | −4.9 | |
| PTGS2PDB_5280933_uff_E = 157.30 | 0 | 0 | −5.7 | |
| PTGS2PDB_5280899_uff_E = 704.15 | 0 | 0 | −8.3 | |
| PTGS2PDB_5280863_uff_E = 362.50 | 0 | 0 | −9.1 | |
| PTGS2PDB_5280805_uff_E = 751.59 | 0 | 0 | −10.8 | |
| PTGS2PDB_5280794_uff_E = 546.19 | 0 | 0 | −8.5 | |
| PTGS2PDB_5280791_uff_E = 556.51 | 0 | 0 | −10.7 | |
| PTGS2PDB_5280489_uff_E = 674.37 | 0 | 0 | −10.8 | |
| PTGS2PDB_5280450_uff_E = 147.99 | 0 | 0 | −5.8 | |
| PTGS2PDB_5280343_uff_E = 380.43 | 0 | 0 | −8.2 | |
| PTGS2PDB_689043_uff_E = 98.60 | 0 | 0 | −6.9 | |
| PTGS2PDB_638072_uff_E = 248.91 | 0 | 0 | −5.9 | |
| PTGS2PDB_637542_uff_E = 90.83 | 0 | 0 | −6.5 | |
| PTGS2PDB_637541_uff_E = 189.36 | 0 | 0 | −6.2 | |
| PTGS2PDB_637540_uff_E = 280.62 | 0 | 0 | −6.6 | |
| PTGS2PDB_493570_uff_E = 317.94 | 0 | 0 | −7.8 | |
| PTGS2PDB_445858_uff_E = 177.42 | 0 | 0 | −7.1 | |
| PTGS2PDB_445639_uff_E = 80.35 | 0 | 0 | −6.8 | |
| PTGS2PDB_445638_uff_E = 85.31 | 0 | 0 | −5.4 | |
| PTGS2PDB_445354_uff_E = 360.80 | 0 | 0 | −6.8 | |
| PTGS2PDB_444539_uff_E = 86.61 | 0 | 0 | −6.5 | |
| PTGS2PDB_444305_uff_E = 68.76 | 0 | 0 | −5.3 | |
| PTGS2PDB_241572_uff_E = 557.95 | 0 | 0 | −8.6 | |
| PTGS2PDB_222284_uff_E = 590.88 | 0 | 0 | −8.8 | |
| PTGS2PDB_173183_uff_E = 573.30 | 0 | 0 | −9.8 | |
| PTGS2PDB_145742_uff_E = 182.22 | 0 | 0 | −5.0 | |
| PTGS2PDB_101761_uff_E = 714.50 | 0 | 0 | −8.9 | |
| PTGS2PDB_101341_uff_E = 993.36 | 0 | 0 | −8.5 | |
| PTGS2PDB_94204_uff_E = 2069.23 | 0 | 0 | −7.7 | |
| PTGS2PDB_92110_uff_E = 2128.40 | 0 | 0 | −8.3 | |
| PTGS2PDB_73170_uff_E = 681.31 | 0 | 0 | −9.4 | |
| PTGS2PDB_73160_uff_E = 208.21 | 0 | 0 | −8.6 | |
| PTGS2PDB_65252_uff_E = 728.67 | 0 | 0 | −8.6 | |
| PTGS2PDB_33032_uff_E = 55.55 | 0 | 0 | −5.1 | |
| PTGS2PDB_14985_uff_E = 288.57 | 0 | 0 | −7.4 | |
| PTGS2PDB_13849_uff_E = 53.38 | 0 | 0 | −5.4 | |
| PTGS2PDB_11005_uff_E = 51.08 | 0 | 0 | −5.2 | |
| PTGS2PDB_10742_uff_E = 110.08 | 0 | 0 | −6.1 | |
| PTGS2PDB_10465_uff_E = 60.66 | 0 | 0 | −6.6 | |
| PTGS2PDB_8468_uff_E = 150.75 | 0 | 0 | −6.4 | |
| PTGS2PDB_7121_uff_E = 233.36 | 0 | 0 | −6.3 | |
| PTGS2PDB_6613_uff_E = 133.81 | 0 | 0 | −6.6 | |
| PTGS2PDB_6508_uff_E = 156.41 | 0 | 0 | −6.5 | |
| PTGS2PDB_6322_uff_E = 70.50 | 0 | 0 | −5.9 | |
| PTGS2PDB_6306_uff_E = 69.94 | 0 | 0 | −4.9 | |
| PTGS2PDB_6288_uff_E = 64.57 | 0 | 0 | −4.6 | |
| PTGS2PDB_6287_uff_E = 55.97 | 0 | 0 | −4.6 | |
| PTGS2PDB_6274_uff_E = 256.87 | 0 | 0 | −5.4 | |
| PTGS2PDB_6140_uff_E = 104.20 | 0 | 0 | −6.2 | |
| PTGS2PDB_6137_uff_E = 94.18 | 0 | 0 | −4.5 | |
| PTGS2PDB_6106_uff_E = 72.66 | 0 | 0 | −5.1 | |
| PTGS2PDB_6057_uff_E = 109.06 | 0 | 0 | −6.7 | |
| PTGS2PDB_5962_uff_E = 63.08 | 0 | 0 | −4.8 | |
| PTGS2PDB_5960_uff_E = 39.14 | 0 | 0 | −4.9 | |
| PTGS2PDB_5951_uff_E = 74.76 | 0 | 0 | −4.5 | |
| PTGS2PDB_5950_uff_E = 33.09 | 0 | 0 | −4.2 | |
| PTGS2PDB_5862_uff_E = 55.51 | 0 | 0 | −4.0 | |
| PTGS2PDB_4091_uff_E = 136.80 | 0 | 0 | −5.3 | |
| PTGS2PDB_3469_uff_E = 78.01 | 0 | 0 | −6.3 | |
| PTGS2PDB_2518_uff_E = 98.61 | 0 | 0 | −7.0 | |
| PTGS2PDB_1198_uff_E = 74.39 | 0 | 0 | −5.8 | |
| PTGS2PDB_1110_uff_E = 26.16 | 0 | 0 | −4.8 | |
| PTGS2PDB_1060_uff_E = 10.03 | 0 | 0 | −4.5 | |
| PTGS2PDB_1054_uff_E = 125.61 | 0 | 0 | −5.5 | |
| PTGS2PDB_971_uff_E = 8.52 | 0 | 0 | −4.4 | |
| PTGS2PDB_938_uff_E = 58.73 | 0 | 0 | −5.4 | |
| PTGS2PDB_750_uff_E = 27.51 | 0 | 0 | −3.9 | |
| PTGS2PDB_525_uff_E = 46.12 | 0 | 0 | −5.2 | |
| PTGS2PDB_370_uff_E = 77.82 | 0 | 0 | −6.3 | |
| PTGS2PDB_338_uff_E = 73.50 | 0 | 0 | −5.9 | |
| PTGS2PDB_311_uff_E = 79.82 | 0 | 0 | −5.6 | |
| PTGS2PDB_284_uff_E = 2.48 | 0 | 0 | −2.8 | |
| PTGS2PDB_176_uff_E = 4.30 | 0 | 0 | −3.6 |
| Macromolecule | Phytochemical Compound PubChem ID | RMSD Upper Bound | RMSD Lower Bound | Molecular Docking: Binding Affinity (kcal/mol) |
|---|---|---|---|---|
| CCL2 | CCL2PDB_157822370_uff_E = 101.29 | 0 | 0 | −4.1 |
| CCL2PDB_54670067_uff_E = 200.65 | 0 | 0 | −4.2 | |
| CCL2PDB_25244964_uff_E = 572.46 | 0 | 0 | −5.7 | |
| CCL2PDB_25203368_uff_E = 541.72 | 0 | 0 | −6.3 | |
| CCL2PDB_12795736_uff_E = 643.77 | 0 | 0 | −6 | |
| CCL2PDB_11063337_uff_E = 244.73 | 0 | 0 | −5 | |
| CCL2PDB_9828626_uff_E = 669.99 | 0 | 0 | −6.8 | |
| CCL2PDB_9548595_uff_E = 638.65 | 0 | 0 | −6.5 | |
| CCL2PDB_6419725_uff_E = 609.04 | 0 | 0 | −7 | |
| CCL2PDB_5481663_uff_E = 826.93 | 0 | 0 | −6.4 | |
| CCL2PDB_5318645_uff_E = 688.07 | 0 | 0 | −5.8 | |
| CCL2PDB_5282761_uff_E = 74.80 | 0 | 0 | −4.4 | |
| CCL2PDB_5281654_uff_E = 450.92 | 0 | 0 | −6 | |
| CCL2PDB_5281243_uff_E = 655.10 | 0 | 0 | −7.1 | |
| CCL2PDB_5281235_uff_E = 709.09 | 0 | 0 | −7.1 | |
| CCL2PDB_5281230_uff_E = 422.22 | 0 | 0 | −7.0 | |
| CCL2PDB_5281119_uff_E = 95.80 | 0 | 0 | −4.3 | |
| CCL2PDB_5280934_uff_E = 142.21 | 0 | 0 | −4.2 | |
| CCL2PDB_5280933_uff_E = 157.30 | 0 | 0 | −4.0 | |
| CCL2PDB_5280899_uff_E = 704.15 | 0 | 0 | −7.1 | |
| CCL2PDB_5280863_uff_E = 362.50 | 0 | 0 | −6.1 | |
| CCL2PDB_5280805_uff_E = 751.59 | 0 | 0 | −6.7 | |
| CCL2PDB_5280794_uff_E = 546.19 | 0 | 0 | −6.3 | |
| CCL2PDB_5280791_uff_E = 556.51 | 0 | 0 | −7.0 | |
| CCL2PDB_5280489_uff_E = 674.37 | 0 | 0 | −7.3 | |
| CCL2PDB_5280450_uff_E = 147.99 | 0 | 0 | −4.3 | |
| CCL2PDB_5280343_uff_E = 380.43 | 0 | 0 | −6.2 | |
| CCL2PDB_689043_uff_E = 98.60 | 0 | 0 | −4.8 | |
| CCL2PDB_638072_uff_E = 248.91 | 0 | 0 | −5.0 | |
| CCL2PDB_637542_uff_E = 90.83 | 0 | 0 | −4.7 | |
| CCL2PDB_637541_uff_E = 189.36 | 0 | 0 | −4.7 | |
| CCL2PDB_637540_uff_E = 280.62 | 0 | 0 | −5.1 | |
| CCL2PDB_493570_uff_E = 317.94 | 0 | 0 | −5.9 | |
| CCL2PDB_445858_uff_E = 177.42 | 0 | 0 | −4.7 | |
| CCL2PDB_445639_uff_E = 80.35 | 0 | 0 | −4.4 | |
| CCL2PDB_445638_uff_E = 85.31 | 0 | 0 | −3.9 | |
| CCL2PDB_445354_uff_E = 360.80 | 0 | 0 | −5.9 | |
| CCL2PDB_444539_uff_E = 86.61 | 0 | 0 | −4.8 | |
| CCL2PDB_444305_uff_E = 68.76 | 0 | 0 | −3.9 | |
| CCL2PDB_241572_uff_E = 557.95 | 0 | 0 | −5.9 | |
| CCL2PDB_222284_uff_E = 590.88 | 0 | 0 | −5.9 | |
| CCL2PDB_173183_uff_E = 573.30 | 0 | 0 | −6.0 | |
| CCL2PDB_145742_uff_E = 182.22 | 0 | 0 | −3.7 | |
| CCL2PDB_101761_uff_E = 714.50 | 0 | 0 | −6.5 | |
| CCL2PDB_101341_uff_E = 993.36 | 0 | 0 | −7.2 | |
| CCL2PDB_94204_uff_E = 2069.23 | 0 | 0 | −6.1 | |
| CCL2PDB_92110_uff_E = 2128.40 | 0 | 0 | −6.4 | |
| CCL2PDB_73170_uff_E = 681.31 | 0 | 0 | −7.1 | |
| CCL2PDB_73160_uff_E = 208.21 | 0 | 0 | −6.1 | |
| CCL2PDB_65252_uff_E = 728.67 | 0 | 0 | −6.6 | |
| CCL2PDB_33032_uff_E = 55.55 | 0 | 0 | −4.1 | |
| CCL2PDB_14985_uff_E = 288.57 | 0 | 0 | −4.7 | |
| CCL2PDB_13849_uff_E = 53.38 | 0 | 0 | −4.0 | |
| CCL2PDB_11005_uff_E = 51.08 | 0 | 0 | −3.7 | |
| CCL2PDB_10742_uff_E = 110.08 | 0 | 0 | −4.4 | |
| CCL2PDB_10465_uff_E = 60.66 | 0 | 0 | −3.7 | |
| CCL2PDB_8468_uff_E = 150.75 | 0 | 0 | −4.3 | |
| CCL2PDB_7121_uff_E = 233.36 | 0 | 0 | −4.5 | |
| CCL2PDB_6613_uff_E = 133.81 | 0 | 0 | −4.4 | |
| CCL2PDB_6508_uff_E = 156.41 | 0 | 0 | −4.5 | |
| CCL2PDB_6322_uff_E = 70.50 | 0 | 0 | −3.9 | |
| CCL2PDB_6306_uff_E = 69.94 | 0 | 0 | −3.6 | |
| CCL2PDB_6288_uff_E = 64.57 | 0 | 0 | −3.6 | |
| CCL2PDB_6287_uff_E = 55.97 | 0 | 0 | −3.5 | |
| CCL2PDB_6274_uff_E = 256.87 | 0 | 0 | −4.0 | |
| CCL2PDB_6140_uff_E = 104.20 | 0 | 0 | −4.6 | |
| CCL2PDB_6137_uff_E = 94.18 | 0 | 0 | −3.3 | |
| CCL2PDB_6106_uff_E = 72.66 | 0 | 0 | −3.8 | |
| CCL2PDB_6057_uff_E = 109.06 | 0 | 0 | −4.7 | |
| CCL2PDB_5962_uff_E = 63.08 | 0 | 0 | −3.5 | |
| CCL2PDB_5960_uff_E = 39.14 | 0 | 0 | −3.7 | |
| CCL2PDB_5951_uff_E = 74.76 | 0 | 0 | −3.2 | |
| CCL2PDB_5950_uff_E = 33.09 | 0 | 0 | −3.1 | |
| CCL2PDB_5862_uff_E = 55.51 | 0 | 0 | −3.1 | |
| CCL2PDB_4091_uff_E = 136.80 | 0 | 0 | −4.3 | |
| CCL2PDB_3469_uff_E = 78.01 | 0 | 0 | −4.8 | |
| CCL2PDB_2518_uff_E = 98.61 | 0 | 0 | −4.8 | |
| CCL2PDB_1198_uff_E = 74.39 | 0 | 0 | −4.2 | |
| CCL2PDB_1110_uff_E = 26.16 | 0 | 0 | −3.7 | |
| CCL2PDB_1060_uff_E = 10.03 | 0 | 0 | −3.3 | |
| CCL2PDB_1054_uff_E = 125.61 | 0 | 0 | −4.3 | |
| CCL2PDB_971_uff_E = 8.52 | 0 | 0 | −3.3 | |
| CCL2PDB_938_uff_E = 58.73 | 0 | 0 | −4.4 | |
| CCL2PDB_750_uff_E = 27.51 | 0 | 0 | −2.9 | |
| CCL2PDB_525_uff_E = 46.12 | 0 | 0 | −3.6 | |
| CCL2PDB_370_uff_E = 77.82 | 0 | 0 | −4.5 | |
| CCL2PDB_338_uff_E = 73.50 | 0 | 0 | −4.6 | |
| CCL2PDB_311_uff_E = 79.82 | 0 | 0 | −4.2 | |
| CCL2PDB_284_uff_E = 2.48 | 0 | 0 | −2.4 | |
| CCL2PDB_176_uff_E = 4.30 | 0 | 0 | −2.7 |
| Macromolecule | Phytochemical Compound PubChem ID | RMSD Upper Bound | RMSD Down Bound | Molecular Docking: Binding Affinity (kcal/mol) |
|---|---|---|---|---|
| EGFR | EGFRPDB_157822370_uff_E = 101.29 | 0 | 0 | −4.9 |
| EGFRPDB_54670067_uff_E = 200.65 | 0 | 0 | −5.7 | |
| EGFRPDB_25244964_uff_E = 572.46 | 0 | 0 | −7.8 | |
| EGFRPDB_25203368_uff_E = 541.72 | 0 | 0 | −7.6 | |
| EGFRPDB_12795736_uff_E = 643.77 | 0 | 0 | −7.1 | |
| EGFRPDB_11063337_uff_E = 244.73 | 0 | 0 | −6 | |
| EGFRPDB_9828626_uff_E = 669.99 | 0 | 0 | −8.1 | |
| EGFRPDB_9548595_uff_E = 638.65 | 0 | 0 | −7.9 | |
| EGFRPDB_6419725_uff_E = 609.04 | 0 | 0 | −7.7 | |
| EGFRPDB_5481663_uff_E = 826.93 | 0 | 0 | −8 | |
| EGFRPDB_5318645_uff_E = 688.07 | 0 | 0 | −7.4 | |
| EGFRPDB_5282761_uff_E = 74.80 | 0 | 0 | −4 | |
| EGFRPDB_5281654_uff_E = 450.92 | 0 | 0 | −7.2 | |
| EGFRPDB_5281243_uff_E = 655.10 | 0 | 0 | −7.4 | |
| EGFRPDB_5281235_uff_E = 709.09 | 0 | 0 | −7.8 | |
| EGFRPDB_5281230_uff_E = 422.22 | 0 | 0 | −7 | |
| EGFRPDB_5281119_uff_E = 95.80 | 0 | 0 | −4.8 | |
| EGFRPDB_5280934_uff_E = 142.21 | 0 | 0 | −4.3 | |
| EGFRPDB_5280933_uff_E = 157.30 | 0 | 0 | −5.1 | |
| EGFRPDB_5280899_uff_E = 704.15 | 0 | 0 | −7.6 | |
| EGFRPDB_5280863_uff_E = 362.50 | 0 | 0 | −7.1 | |
| EGFRPDB_5280805_uff_E = 751.59 | 0 | 0 | −8.2 | |
| EGFRPDB_5280794_uff_E = 546.19 | 0 | 0 | −7.7 | |
| EGFRPDB_5280791_uff_E = 556.51 | 0 | 0 | −7.1 | |
| EGFRPDB_5280489_uff_E = 674.37 | 0 | 0 | −7.7 | |
| EGFRPDB_5280450_uff_E = 147.99 | 0 | 0 | −4.7 | |
| EGFRPDB_5280343_uff_E = 380.43 | 0 | 0 | −7.3 | |
| EGFRPDB_689043_uff_E = 98.60 | 0 | 0 | −5.7 | |
| EGFRPDB_638072_uff_E = 248.91 | 0 | 0 | −5.4 | |
| EGFRPDB_637542_uff_E = 90.83 | 0 | 0 | −5.9 | |
| EGFRPDB_637541_uff_E = 189.36 | 0 | 0 | −5.5 | |
| EGFRPDB_637540_uff_E = 280.62 | 0 | 0 | −5.3 | |
| EGFRPDB_493570_uff_E = 317.94 | 0 | 0 | −6.7 | |
| EGFRPDB_445858_uff_E = 177.42 | 0 | 0 | −6 | |
| EGFRPDB_445639_uff_E = 80.35 | 0 | 0 | −4.1 | |
| EGFRPDB_445638_uff_E = 85.31 | 0 | 0 | −4.4 | |
| EGFRPDB_445354_uff_E = 360.80 | 0 | 0 | −6.6 | |
| EGFRPDB_444539_uff_E = 86.61 | 0 | 0 | −5.8 | |
| EGFRPDB_444305_uff_E = 68.76 | 0 | 0 | −4.7 | |
| EGFRPDB_241572_uff_E = 557.95 | 0 | 0 | −7.2 | |
| EGFRPDB_222284_uff_E = 590.88 | 0 | 0 | −7.6 | |
| EGFRPDB_173183_uff_E = 573.30 | 0 | 0 | −7.7 | |
| EGFRPDB_145742_uff_E = 182.22 | 0 | 0 | −4.3 | |
| EGFRPDB_101761_uff_E = 714.50 | 0 | 0 | −8.1 | |
| EGFRPDB_101341_uff_E = 993.36 | 0 | 0 | −8.7 | |
| EGFRPDB_94204_uff_E = 2069.23 | 0 | 0 | −7.7 | |
| EGFRPDB_92110_uff_E = 2128.40 | 0 | 0 | −8 | |
| EGFRPDB_73170_uff_E = 681.31 | 0 | 0 | −8.1 | |
| EGFRPDB_73160_uff_E = 208.21 | 0 | 0 | −7.2 | |
| EGFRPDB_65252_uff_E = 728.67 | 0 | 0 | −7.1 | |
| EGFRPDB_33032_uff_E = 55.55 | 0 | 0 | −4.6 | |
| EGFRPDB_14985_uff_E = 288.57 | 0 | 0 | −5.6 | |
| EGFRPDB_13849_uff_E = 53.38 | 0 | 0 | −4.7 | |
| EGFRPDB_11005_uff_E = 51.08 | 0 | 0 | −4.4 | |
| EGFRPDB_10742_uff_E = 110.08 | 0 | 0 | −5.3 | |
| EGFRPDB_10465_uff_E = 60.66 | 0 | 0 | −4.3 | |
| EGFRPDB_8468_uff_E = 150.75 | 0 | 0 | −5.3 | |
| EGFRPDB_7121_uff_E = 233.36 | 0 | 0 | −5.3 | |
| EGFRPDB_6613_uff_E = 133.81 | 0 | 0 | −4.9 | |
| EGFRPDB_6508_uff_E = 156.41 | 0 | 0 | −5.7 | |
| EGFRPDB_6322_uff_E = 70.50 | 0 | 0 | −4.9 | |
| EGFRPDB_6306_uff_E = 69.94 | 0 | 0 | −4.3 | |
| EGFRPDB_6288_uff_E = 64.57 | 0 | 0 | −4.5 | |
| EGFRPDB_6287_uff_E = 55.97 | 0 | 0 | −4.3 | |
| EGFRPDB_6274_uff_E = 256.87 | 0 | 0 | −5.1 | |
| EGFRPDB_6140_uff_E = 104.20 | 0 | 0 | −5.8 | |
| EGFRPDB_6137_uff_E = 94.18 | 0 | 0 | −4 | |
| EGFRPDB_6106_uff_E = 72.66 | 0 | 0 | −4.6 | |
| EGFRPDB_6057_uff_E = 109.06 | 0 | 0 | −5.6 | |
| EGFRPDB_5962_uff_E = 63.08 | 0 | 0 | −4.3 | |
| EGFRPDB_5960_uff_E = 39.14 | 0 | 0 | −4.8 | |
| EGFRPDB_5951_uff_E = 74.76 | 0 | 0 | −4.2 | |
| EGFRPDB_5950_uff_E = 33.09 | 0 | 0 | −4 | |
| EGFRPDB_5862_uff_E = 55.51 | 0 | 0 | −3.8 | |
| EGFRPDB_4091_uff_E = 136.80 | 0 | 0 | −5 | |
| EGFRPDB_3469_uff_E = 78.01 | 0 | 0 | −5.6 | |
| EGFRPDB_2518_uff_E = 98.61 | 0 | 0 | −5.9 | |
| EGFRPDB_1198_uff_E = 74.39 | 0 | 0 | −5.4 | |
| EGFRPDB_1110_uff_E = 26.16 | 0 | 0 | −4 | |
| EGFRPDB_1060_uff_E = 10.03 | 0 | 0 | −3.8 | |
| EGFRPDB_1054_uff_E = 125.61 | 0 | 0 | −4.8 | |
| EGFRPDB_971_uff_E = 8.52 | 0 | 0 | −4 | |
| EGFRPDB_938_uff_E = 58.73 | 0 | 0 | −4.8 | |
| EGFRPDB_750_uff_E = 27.51 | 0 | 0 | −3.6 | |
| EGFRPDB_525_uff_E = 46.12 | 0 | 0 | −4.5 | |
| EGFRPDB_370_uff_E = 77.82 | 0 | 0 | −5.6 | |
| EGFRPDB_338_uff_E = 73.50 | 0 | 0 | −5.3 | |
| EGFRPDB_311_uff_E = 79.82 | 0 | 0 | −5.1 | |
| EGFRPDB_284_uff_E = 2.48 | 0 | 0 | −2.7 | |
| EGFRPDB_176_uff_E = 4.30 | 0 | 0 | −3.1 |
| Macromolecule | Phytochemical Compound PubChem ID | RMSD Upper Bound | RMSD Down Bound | Molecular Docking: Binding Affinity (kcal/mol) |
|---|---|---|---|---|
| SERPINE1 | SERPINE1PDB_101341_uff_E = 993.36 | 0 | 0 | −8.2 |
| SERPINE1PDB_101761_uff_E = 714.50 | 0 | 0 | −8.3 | |
| SERPINE1PDB_10465_uff_E = 60.66 | 0 | 0 | −4.6 | |
| SERPINE1PDB_1054_uff_E = 125.61 | 0 | 0 | −6 | |
| SERPINE1PDB_1060_uff_E = 10.03 | 0 | 0 | −4 | |
| SERPINE1PDB_10742_uff_E = 110.08 | 0 | 0 | −5.7 | |
| SERPINE1PDB_11005_uff_E = 51.08 | 0 | 0 | −4.9 | |
| SERPINE1PDB_11063337_uff_E = 244.73 | 0 | 0 | −7.1 | |
| SERPINE1PDB_1110_uff_E = 26.16 | 0 | 0 | −4.6 | |
| SERPINE1PDB_1198_uff_E = 74.39 | 0 | 0 | −5.7 | |
| SERPINE1PDB_12795736_uff_E = 643.77 | 0 | 0 | −8 | |
| SERPINE1PDB_13849_uff_E = 53.38 | 0 | 0 | −5.1 | |
| SERPINE1PDB_145742_uff_E = 182.22 | 0 | 0 | −5.2 | |
| SERPINE1PDB_14985_uff_E = 288.57 | 0 | 0 | −6.8 | |
| SERPINE1PDB_157822370_uff_E = 101.29 | 0 | 0 | −5.2 | |
| SERPINE1PDB_173183_uff_E = 573.30 | 0 | 0 | −7.2 | |
| SERPINE1PDB_176_uff_E = 4.30 | 0 | 0 | −3.2 | |
| SERPINE1PDB_222284_uff_E = 590.88 | 0 | 0 | −8.3 | |
| SERPINE1PDB_241572_uff_E = 557.95 | 0 | 0 | −6.9 | |
| SERPINE1PDB_2518_uff_E = 98.61 | 0 | 0 | −6.7 | |
| SERPINE1PDB_25203368_uff_E = 541.72 | 0 | 0 | −7.9 | |
| SERPINE1PDB_25244964_uff_E = 572.46 | 0 | 0 | −9 | |
| SERPINE1PDB_284_uff_E = 2.48 | 0 | 0 | −3 | |
| SERPINE1PDB_311_uff_E = 79.82 | 0 | 0 | −5.4 | |
| SERPINE1PDB_33032_uff_E = 55.55 | 0 | 0 | −5.1 | |
| SERPINE1PDB_338_uff_E = 73.50 | 0 | 0 | −6 | |
| SERPINE1PDB_3469_uff_E = 78.01 | 0 | 0 | −5.8 | |
| SERPINE1PDB_370_uff_E = 77.82 | 0 | 0 | −6.5 | |
| SERPINE1PDB_4091_uff_E = 136.80 | 0 | 0 | −5.9 | |
| SERPINE1PDB_444305_uff_E = 68.76 | 0 | 0 | −5 | |
| SERPINE1PDB_444539_uff_E = 86.61 | 0 | 0 | −6.5 | |
| SERPINE1PDB_445354_uff_E = 360.80 | 0 | 0 | −6.4 | |
| SERPINE1PDB_445638_uff_E = 85.31 | 0 | 0 | −5.3 | |
| SERPINE1PDB_445639_uff_E = 80.35 | 0 | 0 | −5.2 | |
| SERPINE1PDB_445858_uff_E = 177.42 | 0 | 0 | −6 | |
| SERPINE1PDB_493570_uff_E = 317.94 | 0 | 0 | −8.8 | |
| SERPINE1PDB_525_uff_E = 46.12 | 0 | 0 | −5.1 | |
| SERPINE1PDB_5280343_uff_E = 380.43 | 0 | 0 | −8.2 | |
| SERPINE1PDB_5280450_uff_E = 147.99 | 0 | 0 | −5.1 | |
| SERPINE1PDB_5280489_uff_E = 674.37 | 0 | 0 | −7.9 | |
| SERPINE1PDB_5280791_uff_E = 556.51 | 0 | 0 | −6.7 | |
| SERPINE1PDB_5280794_uff_E = 546.19 | 0 | 0 | −8.6 | |
| SERPINE1PDB_5280805_uff_E = 751.59 | 0 | 0 | −8.2 | |
| SERPINE1PDB_5280863_uff_E = 362.50 | 0 | 0 | −7.8 | |
| SERPINE1PDB_5280899_uff_E = 704.15 | 0 | 0 | −7.3 | |
| SERPINE1PDB_5280933_uff_E = 157.30 | 0 | 0 | −5.2 | |
| SERPINE1PDB_5280934_uff_E = 142.21 | 0 | 0 | −5.6 | |
| SERPINE1PDB_5281119_uff_E = 95.80 | 0 | 0 | −5.4 | |
| SERPINE1PDB_5281230_uff_E = 422.22 | 0 | 0 | −6.9 | |
| SERPINE1PDB_5281235_uff_E = 709.09 | 0 | 0 | −7.9 | |
| SERPINE1PDB_5281243_uff_E = 655.10 | 0 | 0 | −7.1 | |
| SERPINE1PDB_5281654_uff_E = 450.92 | 0 | 0 | −8 | |
| SERPINE1PDB_5282761_uff_E = 74.80 | 0 | 0 | −4.7 | |
| SERPINE1PDB_5318645_uff_E = 688.07 | 0 | 0 | −7.6 | |
| SERPINE1PDB_54670067_uff_E = 200.65 | 0 | 0 | −5.9 | |
| SERPINE1PDB_5481663_uff_E = 826.93 | 0 | 0 | −8.3 | |
| SERPINE1PDB_5862_uff_E = 55.51 | 0 | 0 | −4.2 | |
| SERPINE1PDB_5950_uff_E = 33.09 | 0 | 0 | −4.4 | |
| SERPINE1PDB_5951_uff_E = 74.76 | 0 | 0 | −4.6 | |
| SERPINE1PDB_5960_uff_E = 39.14 | 0 | 0 | −4.5 | |
| SERPINE1PDB_5962_uff_E = 63.08 | 0 | 0 | −5 | |
| SERPINE1PDB_6057_uff_E = 109.06 | 0 | 0 | −6.1 | |
| SERPINE1PDB_6106_uff_E = 72.66 | 0 | 0 | −4.9 | |
| SERPINE1PDB_6137_uff_E = 94.18 | 0 | 0 | −4.6 | |
| SERPINE1PDB_6140_uff_E = 104.20 | 0 | 0 | −5.7 | |
| SERPINE1PDB_6274_uff_E = 256.87 | 0 | 0 | −5.2 | |
| SERPINE1PDB_6287_uff_E = 55.97 | 0 | 0 | −4.8 | |
| SERPINE1PDB_6288_uff_E = 64.57 | 0 | 0 | −4.7 | |
| SERPINE1PDB_6306_uff_E = 69.94 | 0 | 0 | −5.1 | |
| SERPINE1PDB_6322_uff_E = 70.50 | 0 | 0 | −5.6 | |
| SERPINE1PDB_637540_uff_E = 280.62 | 0 | 0 | −5.6 | |
| SERPINE1PDB_637541_uff_E = 189.36 | 0 | 0 | −5.8 | |
| SERPINE1PDB_637542_uff_E = 90.83 | 0 | 0 | −6.9 | |
| SERPINE1PDB_638072_uff_E = 248.91 | 0 | 0 | −5.3 | |
| SERPINE1PDB_6419725_uff_E = 609.04 | 0 | 0 | −7.7 | |
| SERPINE1PDB_6508_uff_E = 156.41 | 0 | 0 | −5.9 | |
| SERPINE1PDB_65252_uff_E = 728.67 | 0 | 0 | −8.1 | |
| SERPINE1PDB_6613_uff_E = 133.81 | 0 | 0 | −6.2 | |
| SERPINE1PDB_689043_uff_E = 98.60 | 0 | 0 | −6.9 | |
| SERPINE1PDB_7121_uff_E = 233.36 | 0 | 0 | −5.6 | |
| SERPINE1PDB_73160_uff_E = 208.21 | 0 | 0 | −7.5 | |
| SERPINE1PDB_73170_uff_E = 681.31 | 0 | 0 | −8.5 | |
| SERPINE1PDB_750_uff_E = 27.51 | 0 | 0 | −3.8 | |
| SERPINE1PDB_8468_uff_E = 150.75 | 0 | 0 | −6.1 | |
| SERPINE1PDB_92110_uff_E = 2128.40 | 0 | 0 | −7.7 | |
| SERPINE1PDB_938_uff_E = 58.73 | 0 | 0 | −5.7 | |
| SERPINE1PDB_94204_uff_E = 2069.23 | 0 | 0 | −8 | |
| SERPINE1PDB_9548595_uff_E = 638.65 | 0 | 0 | −7.4 | |
| SERPINE1PDB_971_uff_E = 8.52 | 0 | 0 | −4.2 | |
| SERPINE1PDB_9828626_uff_E = 669.99 | 0 | 0 | −7.9 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Safavi, K.; Abedpoor, N.; Hajibabaie, F.; Kaviani, E. Mitigating Diabetic Cardiomyopathy: The Synergistic Potential of Sea Buckthorn and Metformin Explored via Bioinformatics and Chemoinformatics. Biology 2025, 14, 361. https://doi.org/10.3390/biology14040361
Safavi K, Abedpoor N, Hajibabaie F, Kaviani E. Mitigating Diabetic Cardiomyopathy: The Synergistic Potential of Sea Buckthorn and Metformin Explored via Bioinformatics and Chemoinformatics. Biology. 2025; 14(4):361. https://doi.org/10.3390/biology14040361
Chicago/Turabian StyleSafavi, Kamran, Navid Abedpoor, Fatemeh Hajibabaie, and Elina Kaviani. 2025. "Mitigating Diabetic Cardiomyopathy: The Synergistic Potential of Sea Buckthorn and Metformin Explored via Bioinformatics and Chemoinformatics" Biology 14, no. 4: 361. https://doi.org/10.3390/biology14040361
APA StyleSafavi, K., Abedpoor, N., Hajibabaie, F., & Kaviani, E. (2025). Mitigating Diabetic Cardiomyopathy: The Synergistic Potential of Sea Buckthorn and Metformin Explored via Bioinformatics and Chemoinformatics. Biology, 14(4), 361. https://doi.org/10.3390/biology14040361








