The Abundance of Harmful Rare Homozygous Variants in Children of Consanguineous Parents
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Genome Data
2.2. Data Analysis
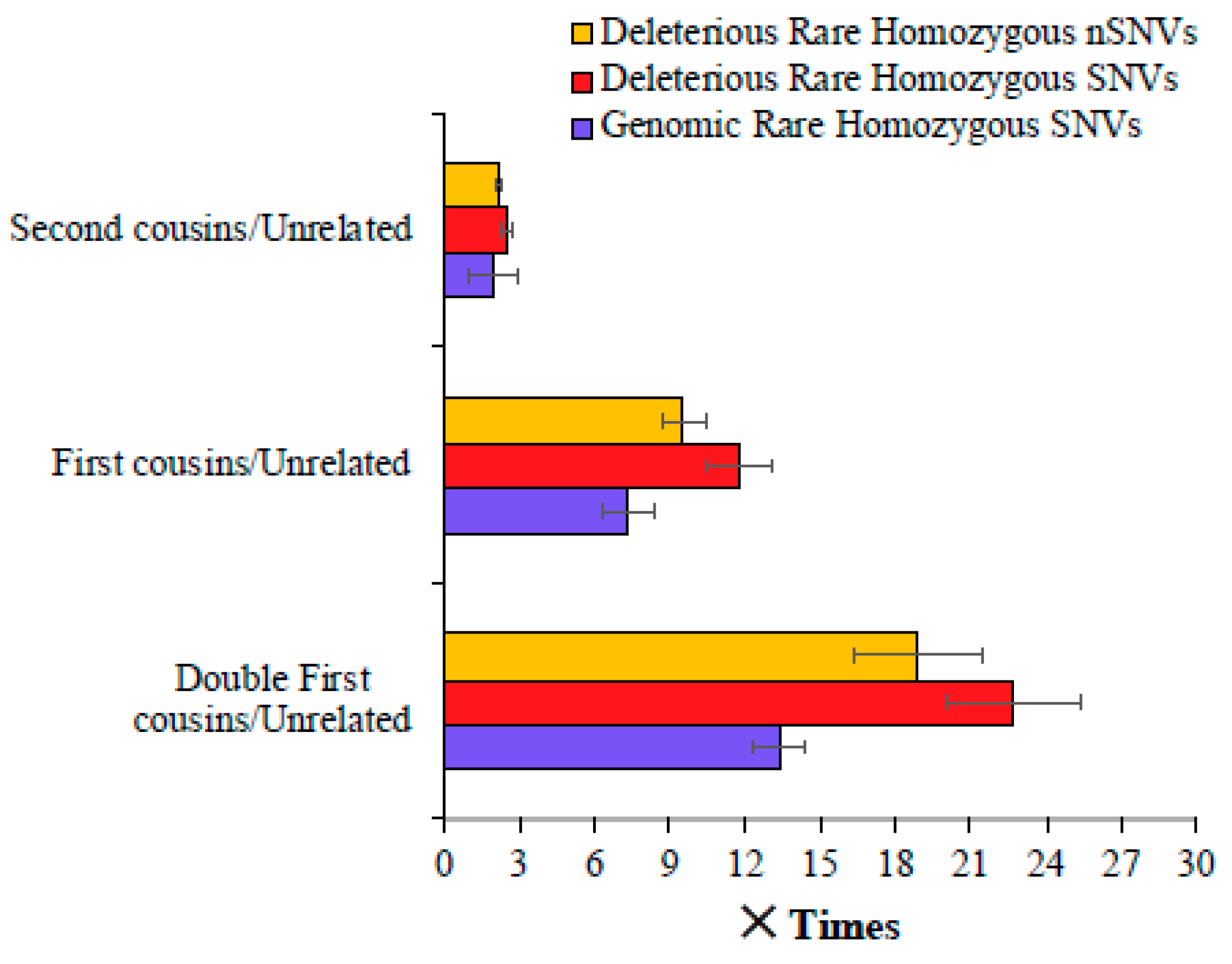
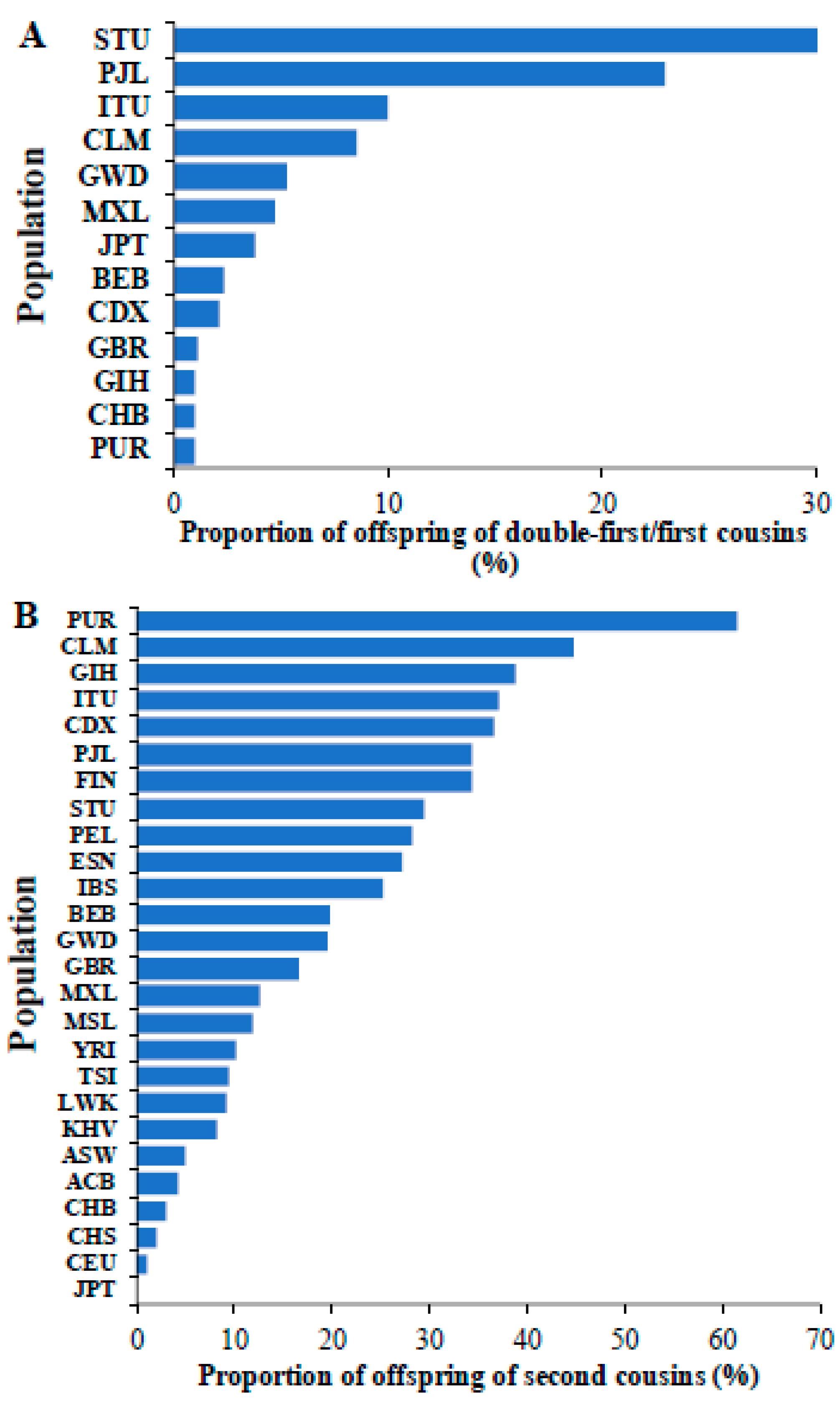
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bittles, A.H.; Black, M.L. Evolution in health and medicine Sackler colloquium: Consanguinity, human evolution, and complex diseases. Proc. Natl. Acad. Sci. USA 2010, 107 (Suppl. 1), 1779–1786. [Google Scholar] [CrossRef] [PubMed]
- Hamamy, H. Consanguineous marriages: Preconception consultation in primary health care settings. J. Community Genet. 2012, 3, 185–192. [Google Scholar] [CrossRef] [PubMed]
- Hamamy, H.; Antonarakis, S.E.; Cavalli-Sforza, L.L.; Temtamy, S.; Romeo, G.; Kate, L.P.; Bennett, R.L.; Shaw, A.; Megarbane, A.; van Duijn, C.; et al. Consanguineous marriages, pearls and perils: Geneva International Consanguinity Workshop Report. Genet. Med. 2011, 13, 841–847. [Google Scholar] [CrossRef] [PubMed]
- Bener, A.; Dafeeah, E.E.; Samson, N. The impact of consanguinity on risk of schizophrenia. Psychopathology 2012, 45, 399–400. [Google Scholar] [CrossRef]
- Bener, A.; El Ayoubi, H.R.; Chouchane, L.; Ali, A.I.; Al-Kubaisi, A.; Al-Sulaiti, H.; Teebi, A.S. Impact of consanguinity on cancer in a highly endogamous population. Asian Pac. J. Cancer Prev. 2009, 10, 35–40. [Google Scholar]
- Bhasin, P.; Kapoor, S. Impact of consanguinity on cardio-metabolic health and other diseases: Findings from an Afro-Indian tribal community. J. Community Genet. 2015, 6, 129–135. [Google Scholar] [CrossRef]
- Bittles, A.H.; Black, M.L. The impact of consanguinity on neonatal and infant health. Early Hum. Dev. 2010, 86, 737–741. [Google Scholar] [CrossRef]
- Hussain, R. The impact of consanguinity and inbreeding on perinatal mortality in Karachi, Pakistan. Paediatr. Perinat. Epidemiol. 1998, 12, 370–382. [Google Scholar] [CrossRef]
- Jabaiti, S.; Salah, B.; Al-Lawama, M.; AlRyalat, S.A.; Jabaiti, O.; Al-Mikhi, B.; Alsmady, D.M.; Al-Basti, H. Impact of Parental Consanguinity on the Frequency of Orofacial Clefts in Jordan. J. Craniofac. Surg. 2022, 33, e203–e206. [Google Scholar] [CrossRef]
- Gonzales, P.R.; Andersen, E.F.; Brown, T.R.; Horner, V.L.; Horwitz, J.; Rehder, C.W.; Rudy, N.L.; Robin, N.H.; Thorland, E.C.; on Behalf of The Acmg Laboratory Quality Assurance, C. Interpretation and reporting of large regions of homozygosity and suspected consanguinity/uniparental disomy, 2021 revision: A technical standard of the American College of Medical Genetics and Genomics (ACMG). Genet. Med. 2022, 24, 255–261. [Google Scholar] [CrossRef]
- Lander, E.S.; Botstein, D. Homozygosity mapping: A way to map human recessive traits with the DNA of inbred children. Science 1987, 236, 1567–1570. [Google Scholar] [CrossRef] [PubMed]
- Woods, C.G.; Cox, J.; Springell, K.; Hampshire, D.J.; Mohamed, M.D.; McKibbin, M.; Stern, R.; Raymond, F.L.; Sandford, R.; Malik Sharif, S.; et al. Quantification of homozygosity in consanguineous individuals with autosomal recessive disease. Am. J. Hum. Genet. 2006, 78, 889–896. [Google Scholar] [CrossRef] [PubMed]
- Wright, S. Coefficients of inbreeding and relationship. Am. Nat. 1922, 56, 330–338. [Google Scholar] [CrossRef]
- Modell, B.; Darr, A. Science and society: Genetic counselling and customary consanguineous marriage. Nat. Rev. Genet. 2002, 3, 225–229. [Google Scholar] [CrossRef]
- Shawky, R.M.; Elsayed, S.M.; Zaki, M.E.; Nour El-Din, S.M.; Kamal, F.M. Consanguinity and its relevance to clinical genetics. Egypt. J. Med. Hum. Genet. 2013, 14, 157–164. [Google Scholar] [CrossRef]
- Fridman, H.; Yntema, H.G.; Magi, R.; Andreson, R.; Metspalu, A.; Mezzavila, M.; Tyler-Smith, C.; Xue, Y.; Carmi, S.; Levy-Lahad, E.; et al. The landscape of autosomal-recessive pathogenic variants in European populations reveals phenotype-specific effects. Am. J. Hum. Genet. 2021, 108, 608–619. [Google Scholar] [CrossRef]
- Temaj, G.; Nuhii, N.; Sayer, J.A. The impact of consanguinity on human health and disease with an emphasis on rare diseases. J. Rare Dis. 2022, 1, 2. [Google Scholar] [CrossRef]
- Aamer, W.; Al-Maraghi, A.; Syed, N.; Gandhi, G.D.; Aliyev, E.; Al-Kurbi, A.A.; Al-Saei, O.; Kohailan, M.; Krishnamoorthy, N.; Palaniswamy, S.; et al. Burden of Mendelian disorders in a large Middle Eastern biobank. Genome Med. 2024, 16, 46. [Google Scholar] [CrossRef]
- Albanghali, M.A. Prevalence of Consanguineous Marriage among Saudi Citizens of Albaha, a Cross-Sectional Study. Int. J. Env. Res. Public Health 2023, 20, 3767. [Google Scholar] [CrossRef]
- Monies, D.; Abouelhoda, M.; Assoum, M.; Moghrabi, N.; Rafiullah, R.; Almontashiri, N.; Alowain, M.; Alzaidan, H.; Alsayed, M.; Subhani, S.; et al. Lessons Learned from Large-Scale, First-Tier Clinical Exome Sequencing in a Highly Consanguineous Population. Am. J. Hum. Genet. 2019, 105, 879. [Google Scholar] [CrossRef]
- Ben-Omran, T.; Al Ghanim, K.; Yavarna, T.; El Akoum, M.; Samara, M.; Chandra, P.; Al-Dewik, N. Effects of consanguinity in a cohort of subjects with certain genetic disorders in Qatar. Mol. Genet. Genom. Med. 2020, 8, e1051. [Google Scholar] [CrossRef] [PubMed]
- Kundu, S.; Jana, A. Consanguineous marriage and associated diseases among their children and grandchildren in India: Evidence from large-scale data. J. Biosoc. Sci. 2024, 56, 796–808. [Google Scholar] [CrossRef] [PubMed]
- Dahbi, N.; El khair, A.; Cheffi, K.; Habibeddine, L.; Talbi, J.; Hilali, A.; El ossmani, H. Consanguinity, complex diseases and congenital disabilities in the Souss population (Southern Morocco): A cross-sectional survey. Egypt. J. Med. Hum. Genet. 2024, 25, 27. [Google Scholar] [CrossRef]
- Gazal, S.; Sahbatou, M.; Babron, M.C.; Genin, E.; Leutenegger, A.L. High level of inbreeding in final phase of 1000 Genomes Project. Sci. Rep. 2015, 5, 17453. [Google Scholar] [CrossRef]
- Auton, A.; Brooks, L.D.; Durbin, R.M.; Garrison, E.P.; Kang, H.M.; Korbel, J.O.; Marchini, J.L.; McCarthy, S.; McVean, G.A. A global reference for human genetic variation. Nature 2015, 526, 68–74. [Google Scholar] [CrossRef]
- Paten, B.; Herrero, J.; Fitzgerald, S.; Beal, K.; Flicek, P.; Holmes, I.; Birney, E. Genome-wide nucleotide-level mammalian ancestor reconstruction. Genome Res. 2008, 18, 1829–1843. [Google Scholar] [CrossRef]
- Kircher, M.; Witten, D.M.; Jain, P.; O’Roak, B.J.; Cooper, G.M.; Shendure, J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet. 2014, 46, 310–315. [Google Scholar] [CrossRef]
- Erzurumluoglu, A.M.; Shihab, H.A.; Rodriguez, S.; Gaunt, T.R.; Day, I.N. Importance of Genetic Studies in Consanguineous Populations for the Characterization of Novel Human Gene Functions. Ann. Hum. Genet. 2016, 80, 187–196. [Google Scholar] [CrossRef]
- Salazar-Silva, R.; Dantas, V.L.G.; Alves, L.U.; Batissoco, A.C.; Oiticica, J.; Lawrence, E.A.; Kawafi, A.; Yang, Y.; Nicastro, F.S.; Novaes, B.C.; et al. NCOA3 identified as a new candidate to explain autosomal dominant progressive hearing loss. Hum. Mol. Genet. 2021, 29, 3691–3705. [Google Scholar] [CrossRef]
- Lakshminarayanan, R.; Chaurasia, S.S.; Anandalakshmi, V.; Chai, S.M.; Murugan, E.; Vithana, E.N.; Beuerman, R.W.; Mehta, J.S. Clinical and genetic aspects of the TGFBI-associated corneal dystrophies. Ocul. Surf. 2014, 12, 234–251. [Google Scholar] [CrossRef]
- Runhart, E.H.; Khan, M.; Cornelis, S.S.; Roosing, S.; Del Pozo-Valero, M.; Lamey, T.M.; Liskova, P.; Roberts, L.; Stohr, H.; Klaver, C.C.W.; et al. Association of Sex With Frequent and Mild ABCA4 Alleles in Stargardt Disease. JAMA Ophthalmol. 2020, 138, 1035–1042. [Google Scholar] [CrossRef] [PubMed]
- Schwarz, N.; Seiffert, S.; Pendziwiat, M.; Rademacher, A.V.; Brunger, T.; Hedrich, U.B.S.; Augustijn, P.B.; Baier, H.; Bayat, A.; Bisulli, F.; et al. Spectrum of Phenotypic, Genetic, and Functional Characteristics in Patients with Epilepsy with KCNC2 Pathogenic Variants. Neurology 2022, 98, e2046–e2059. [Google Scholar] [CrossRef] [PubMed]
- Canda, E.; Yazici, H.; Er, E.; Kose, M.; Basol, G.; Onay, H.; Ucar, S.K.; Habif, S.; Ozkinay, F.; Coker, M. Single center experience of biotinidase deficiency: 259 patients and six novel mutations. J. Pediatr. Endocrinol. Metab. 2018, 31, 917–926. [Google Scholar] [CrossRef] [PubMed]
- Manoharan, I.; Wieseler, S.; Layer, P.G.; Lockridge, O.; Boopathy, R. Naturally occurring mutation Leu307Pro of human butyrylcholinesterase in the Vysya community of India. Pharmacogenet. Genom. 2006, 16, 461–468. [Google Scholar] [CrossRef]
- Lasseaux, E.; Plaisant, C.; Michaud, V.; Pennamen, P.; Trimouille, A.; Gaston, L.; Monferme, S.; Lacombe, D.; Rooryck, C.; Morice-Picard, F.; et al. Molecular characterization of a series of 990 index patients with albinism. Pigment Cell Melanoma Res. 2018, 31, 466–474. [Google Scholar] [CrossRef]
- Banka, S.; Veeramachaneni, R.; Reardon, W.; Howard, E.; Bunstone, S.; Ragge, N.; Parker, M.J.; Crow, Y.J.; Kerr, B.; Kingston, H.; et al. How genetically heterogeneous is Kabuki syndrome?: MLL2 testing in 116 patients, review and analyses of mutation and phenotypic spectrum. Eur. J. Hum. Genet. 2012, 20, 381–388. [Google Scholar] [CrossRef]
- Muller, Y.L.; Piaggi, P.; Hoffman, D.; Huang, K.; Gene, B.; Kobes, S.; Thearle, M.S.; Knowler, W.C.; Hanson, R.L.; Baier, L.J.; et al. Common genetic variation in the glucokinase gene (GCK) is associated with type 2 diabetes and rates of carbohydrate oxidation and energy expenditure. Diabetologia 2014, 57, 1382–1390. [Google Scholar] [CrossRef]
- Warthen, D.M.; Moore, E.C.; Kamath, B.M.; Morrissette, J.J.; Sanchez-Lara, P.A.; Piccoli, D.A.; Krantz, I.D.; Spinner, N.B. Jagged1 (JAG1) mutations in Alagille syndrome: Increasing the mutation detection rate. Hum. Mutat. 2006, 27, 436–443. [Google Scholar] [CrossRef]
- Bener, A.; Mohammadc, R.R. Global distribution of consanguinity and their impact on complex diseases: Genetic disorders from an endogamous population. Egypt. J. Med. Hum. Genet. 2017, 18, 315–320. [Google Scholar] [CrossRef]
- Pritchard, J.K. Are rare variants responsible for susceptibility to complex diseases? Am. J. Hum. Genet. 2001, 69, 124–137. [Google Scholar] [CrossRef]
- Bittles, A. Consanguinity and its relevance to clinical genetics. Clin. Genet. 2001, 60, 89–98. [Google Scholar] [CrossRef] [PubMed]
- Bener, A.; Hussain, R. Consanguineous unions and child health in the State of Qatar. Paediatr. Perinat. Epidemiol. 2006, 20, 372–378. [Google Scholar] [CrossRef] [PubMed]
- Khayat, A.M.; Alshareef, B.G.; Alharbi, S.F.; AlZahrani, M.M.; Alshangity, B.A.; Tashkandi, N.F. Consanguineous Marriage and Its Association With Genetic Disorders in Saudi Arabia: A Review. Cureus 2024, 16, e53888. [Google Scholar] [CrossRef]
- Bhinder, M.A.; Sadia, H.; Mahmood, N.; Qasim, M.; Hussain, Z.; Rashid, M.M.; Zahoor, M.Y.; Bhatti, R.; Shehzad, W.; Waryah, A.M.; et al. Consanguinity: A blessing or menace at population level? Ann. Hum. Genet. 2019, 83, 214–219. [Google Scholar] [CrossRef]
- Saadat, M. Association between consanguinity and survival of marriages. Egypt. J. Med. Hum. Genet. 2015, 16, 67–70. [Google Scholar] [CrossRef]
- Khoury, M.J.; Cohen, B.H.; Diamond, E.L.; Chase, G.A.; McKusick, V.A. Inbreeding and prereproductive mortality in the Old Order Amish. I. Genealogic epidemiology of inbreeding. Am. J. Epidemiol. 1987, 125, 453–461. [Google Scholar] [CrossRef]
- Stenson, P.D.; Mort, M.; Ball, E.V.; Chapman, M.; Evans, K.; Azevedo, L.; Hayden, M.; Heywood, S.; Millar, D.S.; Phillips, A.D.; et al. The Human Gene Mutation Database (HGMD((R))): Optimizing its use in a clinical diagnostic or research setting. Hum. Genet. 2020, 139, 1197–1207. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Subramanian, S. The Abundance of Harmful Rare Homozygous Variants in Children of Consanguineous Parents. Biology 2025, 14, 310. https://doi.org/10.3390/biology14030310
Subramanian S. The Abundance of Harmful Rare Homozygous Variants in Children of Consanguineous Parents. Biology. 2025; 14(3):310. https://doi.org/10.3390/biology14030310
Chicago/Turabian StyleSubramanian, Sankar. 2025. "The Abundance of Harmful Rare Homozygous Variants in Children of Consanguineous Parents" Biology 14, no. 3: 310. https://doi.org/10.3390/biology14030310
APA StyleSubramanian, S. (2025). The Abundance of Harmful Rare Homozygous Variants in Children of Consanguineous Parents. Biology, 14(3), 310. https://doi.org/10.3390/biology14030310





