Complete Chloroplast Genome Characterization, and Phylogenetic Analyses of the Rare and Endangered Plant Platycrater arguta
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. The Sequence, Assembly and Annotation of Chloroplast Genome Sequence in P. arguta
2.3. Chloroplast Genome Analysis
2.4. Phylogenetic Tree Was Conducted by the Chloroplast Genome Sequence
3. Results
3.1. Characteristics of the Chloroplast Genome in P. arguta
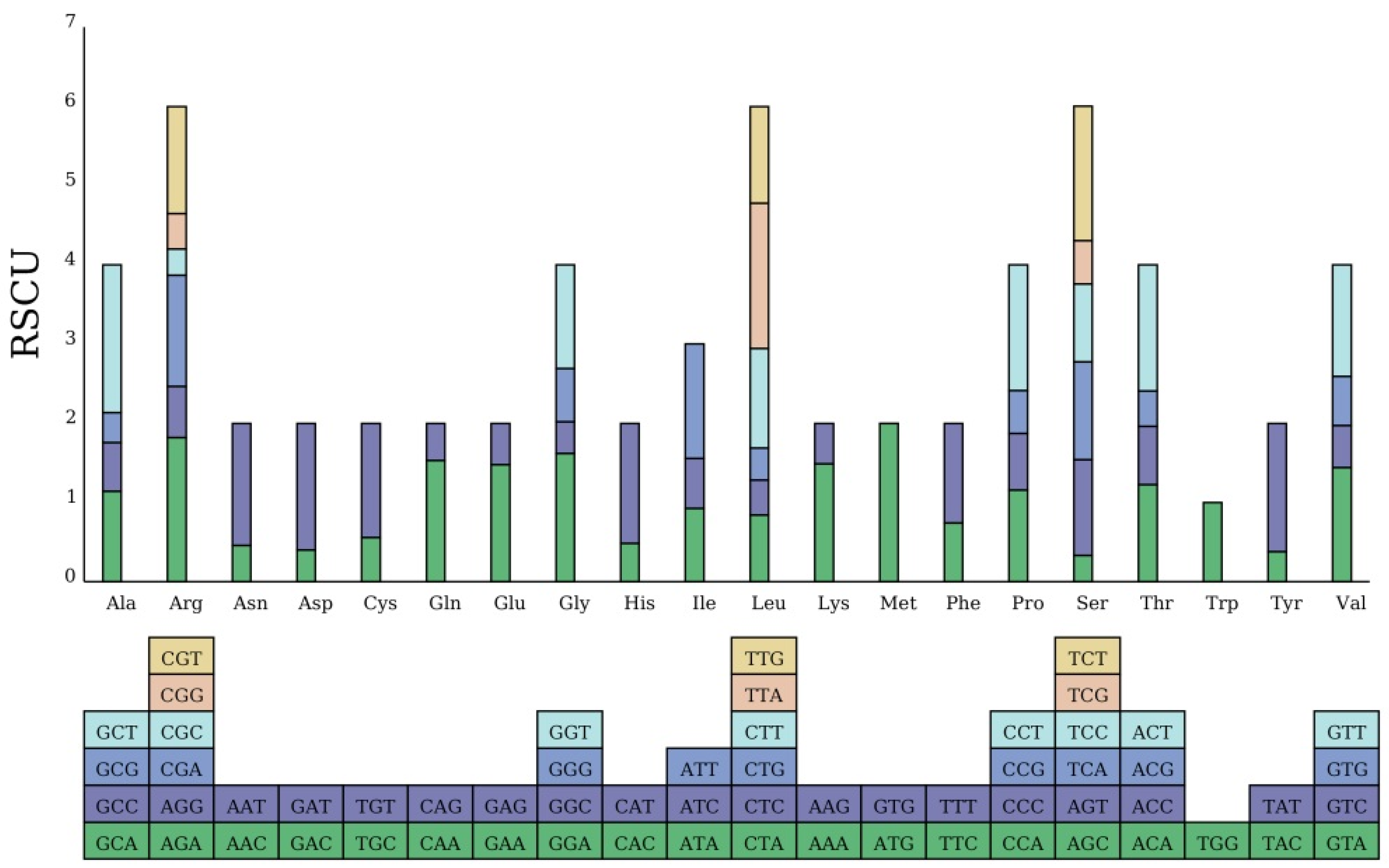
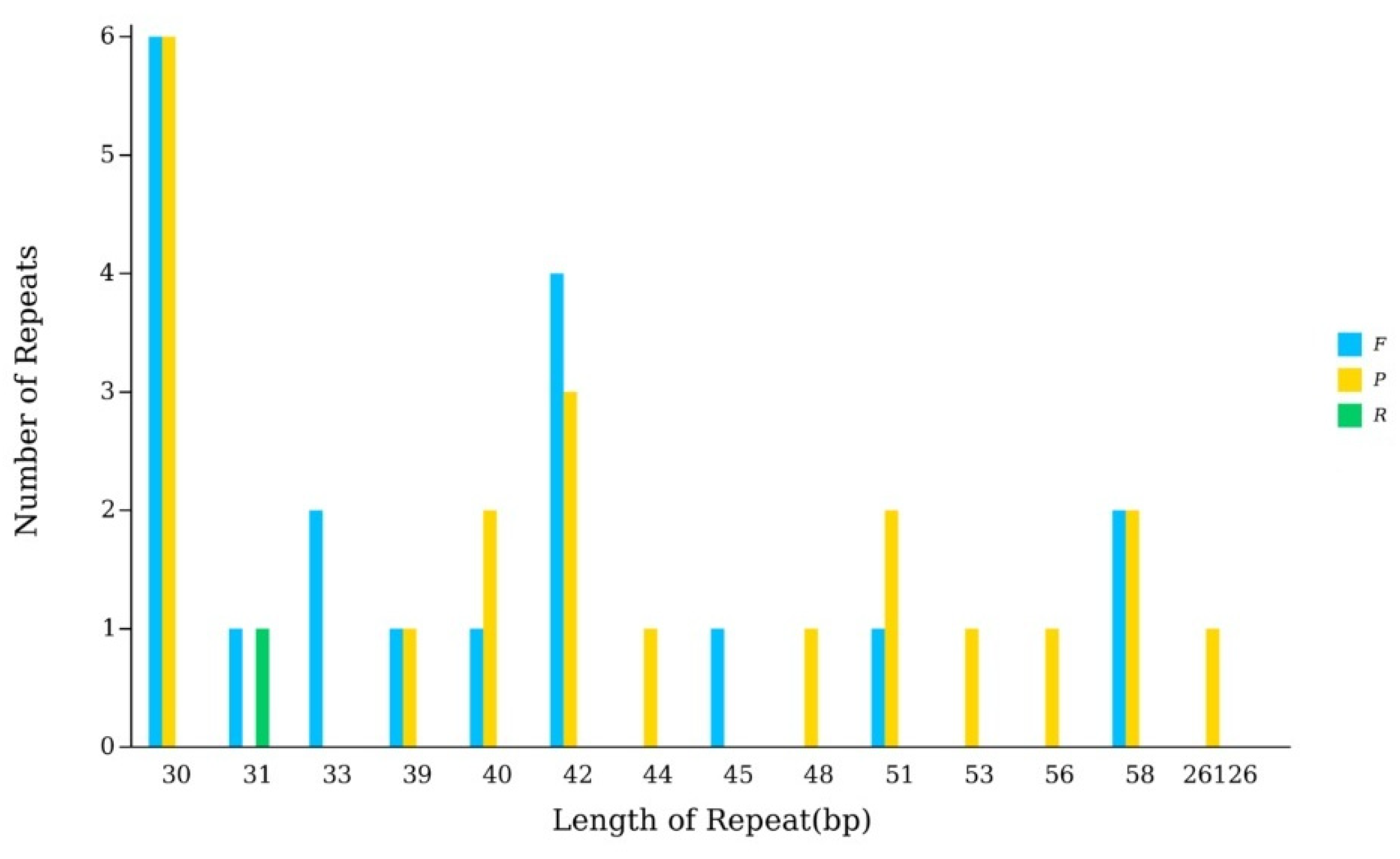
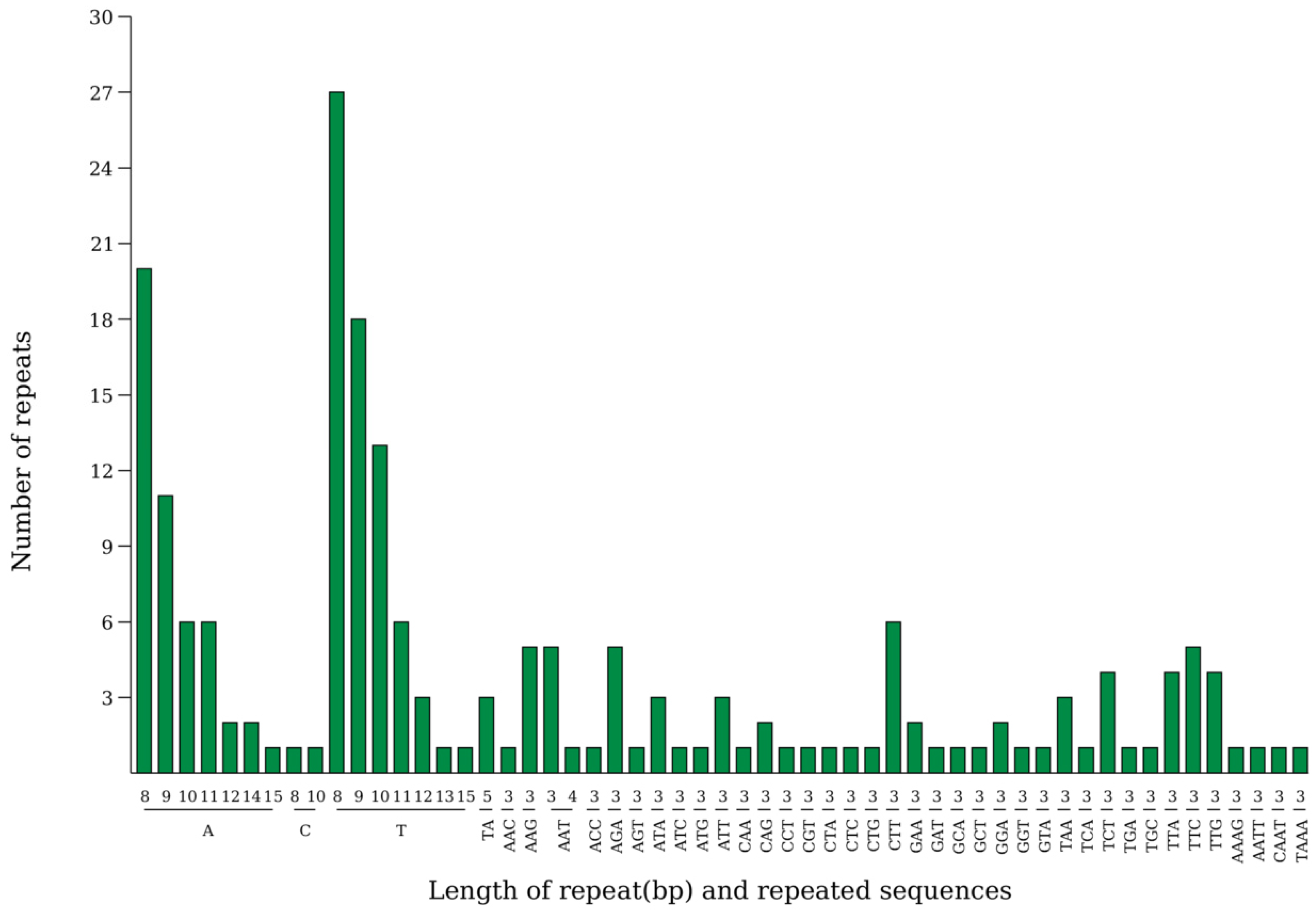
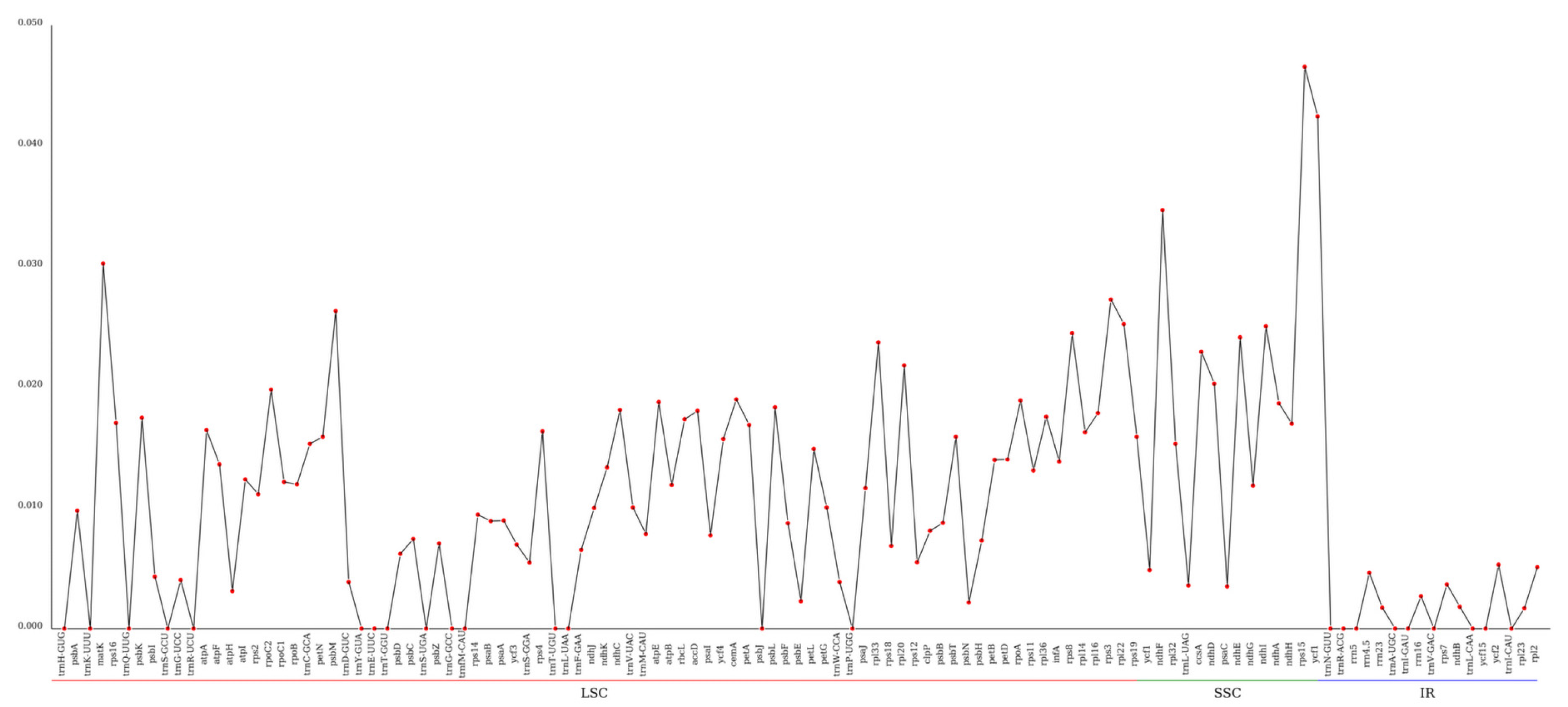
3.2. Sequence Analysis
3.3. Expansion and Contraction of IR Regions
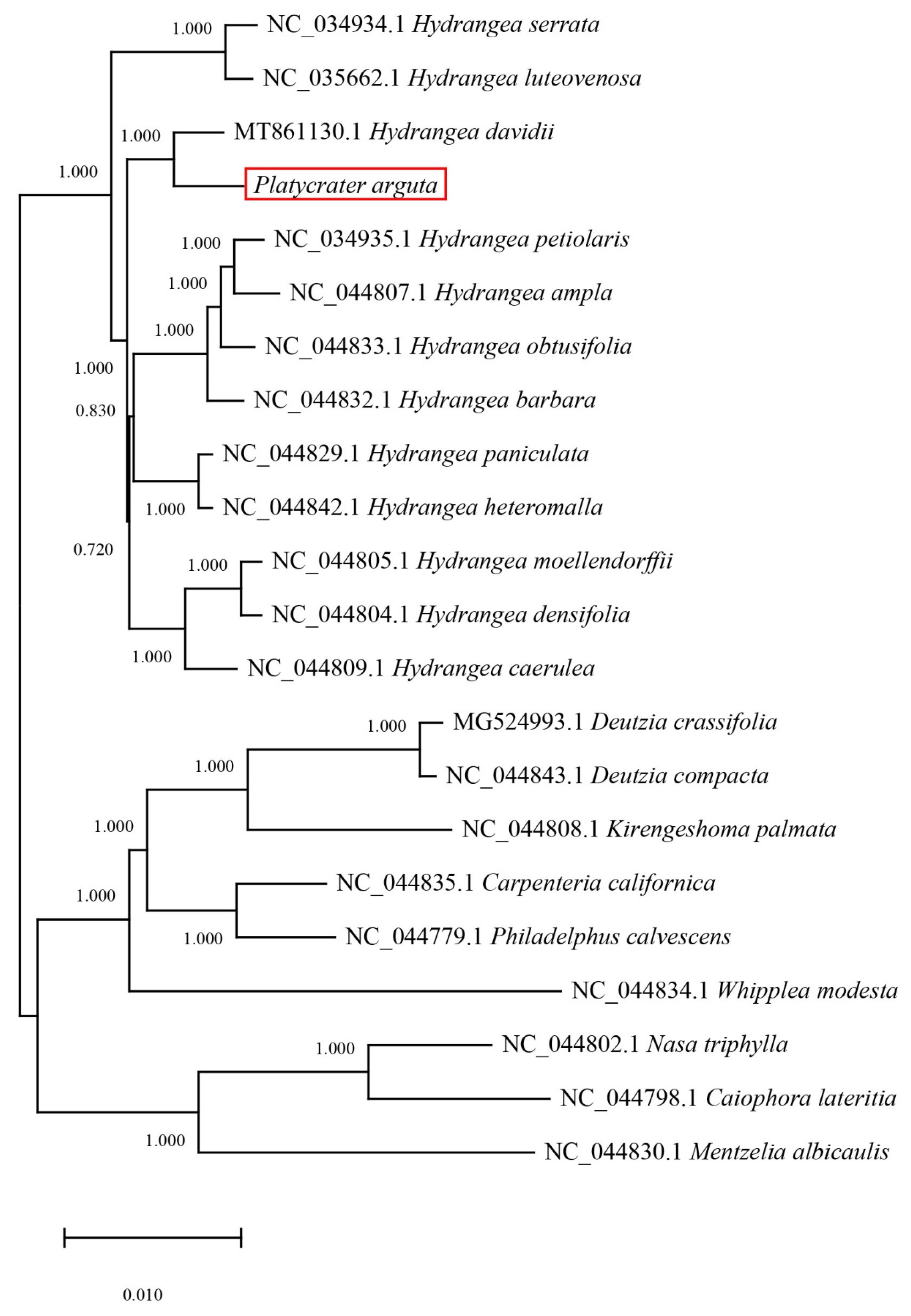
3.4. Phylogenetic Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| Bp | Base pair |
| LSC | Large single copy |
| SSC | Small single copy |
| IR | Inverted repeat |
| IUCN | International Union for Conservation of Nature |
| A | Adenine |
| T | Thymine |
| G | Guanine |
| C | Cytosine |
| Leu | Leucine |
| Cys | Cysteine |
| RSCU | Relative synonymous codon usage |
| SSR | Single sequence repeat |
| PEP | Plastid-encoded plastid RNA polymerase |
| NEP | Nucleus-encoded plastid RNA polymerase |
| DNA | Deoxyribonucleic acid |
References
- Heath, R.L.; Packer, L. Photoperoxidation in isolated chloroplast i. kinetics and stoichiometry of fatty acid peroxidation. Arch. Biochem. Biophys. 1968, 125, 89–198. [Google Scholar] [CrossRef] [PubMed]
- Daniell, H.; Lin, C.S.; Yu, M.; Chang, W. Chloroplast genomes: Diversity, evolution, and applications in genetic engineering. Genome Biol. 2016, 17, 134. [Google Scholar] [CrossRef] [PubMed]
- Schubert, M.; Petersson, U.; Haas, B.; Funk, C.; Schroder, W.; Kieselbach, T. Proteome Map of the Chloroplast Lumen of Arabidopsis thaliana. J. Biol. Chem. 2002, 277, 8354–8365. [Google Scholar] [CrossRef] [PubMed]
- Sugiura, M. The chloroplast genome. Plant Mol. Biol. 1992, 19, 149–168. [Google Scholar] [CrossRef]
- Martin, W.; Rujan, T.; Richly, E.; Cornelsen, S.; Lins, T.; Leister, D.; Stoebe, B.; Hasegawa, M.; Penny, D. Evolutionary analysis of Arabidopsis, cyanobacterial, and chloroplast genomes reveals plastid phylogeny and thousands of cyanobacterial genes in the nucleus. Proc. Natl. Acad. Sci. USA 2002, 99, 12246. [Google Scholar] [CrossRef]
- Shaw, J.; Lickey, E.; Schilling, E.; Small, R. Comparison of whole chloroplast genome sequences to choose noncoding regions for phylogenetic studies in angiosperms: The tortoise and the hare III. Am. J. Bot. 2007, 94, 3. [Google Scholar] [CrossRef]
- Green, B. Chloroplast genomes of photosynthetic eukaryotes. Plant J. 2011, 66, 34–44. [Google Scholar] [CrossRef]
- Keeling, P.J. Plastid genomes. Curr. Biol. 2018, 28, R336–R337. [Google Scholar] [CrossRef]
- Martin, W.; Deusch, O.; Stawski, N.; Grünheit, N.; Goremykin, V. Chloroplast genome phylogenetics: Why we need independent approaches to plant molecular evolution. Trends Plant Sci. 2005, 10, 203–209. [Google Scholar] [CrossRef]
- Hu, Q.; Qian, R.; Zhang, Y.; Ma, X.; Ye, Y.; Zhang, X.; Lin, L.; Liu, H.; Zheng, J. Complete chloroplast genome molecular structure, comparative and phylogenetic analyses of Sphaeropteris lepifera of Cyatheaceae family: A tree fern from China. Sci. Rep. 2023, 13, 1356. [Google Scholar] [CrossRef]
- Shinozaki, K.; Ohme, M.; Tanaka, M.; Wakasugi, T.; Hayashida, N.; Matsubayashi, T. The complete nucleotide sequence of the tobacco chloroplast genome: Its gene organization and expression. EMBO J. 1986, 5, 2043–2049. [Google Scholar] [CrossRef] [PubMed]
- Ohyama, K.; Fukuzawa, H.; Kohchi, T.; Shirai, H.; Sano, T.; Sano, S.; Umesono, K.; Shiki, Y.; Takeuchi, M.; Chang, Z. Chloroplast gene organization deduced from complete sequence of liverwort Marchantia polymorpha chloroplast DNA. Nature 1986, 322, 572–574. [Google Scholar] [CrossRef]
- Hiratsuka, J.; Shimada, H.; Whittier, R.; Ishibashi, T.; Sakamoto, M.; Mori, M. The complete sequence of the rice (Oryza sativa) chloroplast genome: Intermolecular recombination between distinct tRNA genes accounts for a major plastid DNA inversion during the evolution of the cereals. Mol. Gen. Genet. 1989, 217, 185–194. [Google Scholar] [CrossRef] [PubMed]
- Hallick, R.B.; Hong, L.; Drager, R.G.; Favreau, M.R.; Monfort, A.; Orsat, B.; Spielmann, A.; Stutz, E. Complete sequence of Euglena gracilis chloroplast DNA. Nucleic Acid Res. 1993, 21, 3537–3544. [Google Scholar] [CrossRef]
- Wakasugi, T.; Tsudzuki, J.; Ito, S.; Nakashima, K.; Tsudzuki, T.; Sugiura, M. Loss of all ndh genes as determined by sequencing the entire chloroplast genome of the black pine Pinus thunbergii. Proc. Natl. Acad. Sci. USA 1994, 91, 9794–9798. [Google Scholar] [CrossRef]
- Wambugu, P.W.; Brozynska, M.; Furtado, A.; Waters, D.L.; Henry, R.J. Relationships of wild and domesticated rices (Oryza AA genome species) based upon whole chloroplast genome sequences. Sci. Rep. 2015, 5, 13957. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, S.; Liu, Y.; Yuan, Q.; Guo, L. Chloroplast genome variation and phylogenetic relationships of Atractylodes species. BMC Genom. 2021, 22, 103. [Google Scholar] [CrossRef]
- Huo, Y.; Gao, L.; Liu, B.; Yang, Y.; Kong, S.; Sun, Y.; Yang, Y.; Wu, X. Complete chloroplast genome sequences of four Allium species: Comparative and phylogenetic analyses. Sci. Rep. 2019, 9, 12250. [Google Scholar] [CrossRef]
- Wang, Y.; Lv, K.; Wu, Y.; Chen, F. Changes in the functional traits of the Platycrater arguta leaves with plant growth development. Plant Sci. J. 2021, 39, 526–534. [Google Scholar]
- Wang, J. FastPval: A fast and memory efficient program to calculate very low P-values from empirical distribution. Bioinformatics 2010, 26, 2897–2899. [Google Scholar]
- Langmead, B.; Salzberg, S. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Gao, J.; Zhang, M.; Guan, S.; Chen, Y.; Liu, A.; Yan, Y.; Zhang, G. The complete chloroplast genome of Tradescantia pallida (Rose) DR Hunt. Mitochondrial DNA Part B 2020, 5, 2932–2933. [Google Scholar] [CrossRef]
- Boetzer, M.; Pirovano, W. Toward almost closed genomes with GapFiller. Genome Biol. 2012, 13, R56. [Google Scholar] [CrossRef]
- Altschul, S. Basic local alignment search tool (BLAST). J. Mol. Biol. 2012, 215, 403–410. [Google Scholar] [CrossRef] [PubMed]
- Goecks, J.; Nekrutenko, A.; Taylor, J. Galaxy: A comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol. 2010, 11, R86. [Google Scholar] [CrossRef] [PubMed]
- Powell, W.; Machray, G.; Provan, J. Polymorphism revealed by simple sequence repeats. Trends Plant Sci. 1996, 1, 215–222. [Google Scholar] [CrossRef]
- Fenton, B.; Malloch, G.; Moxey, E. Analysis of eriophyid mite rDNA internal transcribed spacer sequences reveals variable simple sequence repeats. Insect Mol. Biol. 2010, 6, 23–32. [Google Scholar] [CrossRef]
- Khayi, S.; Gaboun, F.; Pirro, S.; Tatusova, T.; Mentag, R. Complete Chloroplast Genome of Argania spinosa: Structural Organization and Phylogenetic Relationships in Sapotaceae. Plants 2020, 9, 1354. [Google Scholar] [CrossRef]
- Ali, A.; Jaakko, H.; Peter, P. IRscope: An online program to visualize the junction sites of chloroplast genomes. Bioinformatics 2018, 17, 17. [Google Scholar]
- Soltis, P.S.; Folk, R.A.; Soltis, D.E. Darwin review: Angiosperm phylogeny and evolutionary radiations. Proc. R. Soc. B 2019, 286, 20190099. [Google Scholar] [CrossRef]
- Shaw, A.; Allen, B. Phylogenetic Relationships, Morphological Incongruence, and Geographic Speciation in the Fontinalaceae (Bryophyta). Mol. Phylogenet. Evol. 2000, 16, 225–237. [Google Scholar]
- Ivanova, Z.; Sablok, G.; Daskalova, E.; Zahmanova, G.; Apostolova, E.; Yahubyan, G.; Baev, V. Chloroplast genome analysis of resurrection tertiary relict Haberlea rhodopensis highlights genes important for desiccation stress response. Front. Plant Sci. 2017, 8, 204. [Google Scholar] [CrossRef]
- Chakraborty, S.; Yengkhom, S.; Uddin, A. Analysis of codon usage bias of chloroplast genes in Oryza species. Planta 2020, 252, 67. [Google Scholar] [CrossRef]
- Yao, X.H.; Tang, P.; Li, Z.Z.; Li, D.; Liu, Y.F.; Huang, H.W. The first complete chloroplast genome sequences in Actinidiaceae: Genome structure and comparative analysis. PLoS ONE 2015, 10, e0129347. [Google Scholar] [CrossRef] [PubMed]
- He, L.F.; Qiang, S.J.; Zhang, Y.H.; Li, H.F. The complete chloroplast genome of Hydrangea davidii (Hydrangeaceae). Mitochondrial DNA Part B 2020, 5, 3605–3607. [Google Scholar] [CrossRef] [PubMed]
- Powell, W.; Morgante, M.; McDevitt, R.; Vendramin, G.G.; A Rafalski, J. Polymorphic simple sequence repeat regions in chloroplast genomes: Applications to the population genetics of pines. Proc. Natl. Acad. Sci. USA 1995, 92, 7759–7763. [Google Scholar] [CrossRef]
- Yuan, Q.J.; Zhang, Z.Y.; Hu, J.; Guo, L.P.; Shao, A.J.; Huang, L.Q. Impacts of recent cultivation on genetic diversity pattern of a medicinal plant, Scutellaria baicalensis (Lamiaceae). BMC Genet. 2010, 11, 29. [Google Scholar] [CrossRef]
- Du, Q.Z.; Wang, B.W.; Wei, Z.; Zhang, D.; Li, B. Genetic diversity and population structure of Chinese white poplar (Populus tomentosa) revealed by SSR markers. J. Hered. 2012, 103, 853–862. [Google Scholar] [CrossRef]
- Chmielewski, M.; Meyza, K.; Chybicki, I.; Dzialuk, A.; Litkowiec, M.; Burczyk, J. Chloroplast microsatellites as a tool for phylogeographic studies: The case of white oaks in Poland. iForest—Biogeosci. For. 2015, 8, 765–771. [Google Scholar]
- Tatarinova, T.V.; Alexandrov, N.N.; Bouck, J.B.; Feldmann, K.A. GC3 biology in corn, rice, sorghum and other grasses. BMC Genom. 2010, 11, 308. [Google Scholar] [CrossRef]
- Im, E.H.; Choi, S.S. Synonymous codon usage controls various molecular aspects. Genom. Inform. 2017, 15, 123. [Google Scholar] [CrossRef]
- Bu, Y.; Wu, X.; Sun, N.; Man, Y.; Jing, Y. Codon usage bias predicts the functional MYB10 gene in Populus. J. Plant Physiol. 2021, 265, 153491. [Google Scholar] [CrossRef]
- Lilly, J.W.; Havey, M.J.; Jackson, S.A.; Jiang, J. Cytogenomic analyses reveal the structural plasticity of the chloroplast genome in higher plants. Plant Cell 2001, 13, 245–254. [Google Scholar] [CrossRef]
- Li, L.; Hu, Y.; He, M.; Zhang, B.; Wu, W.; Cai, P.; Huo, D.; Hong, Y. Comparative chloroplast genomes: Insights into the evolution of the chloroplast genome of Camellia sinensis and the phylogeny of Camellia. BMC Genom. 2021, 22, 138. [Google Scholar] [CrossRef]
- Zhang, R.; Liu, S.; Liu, Y.; Wei, P.; Xiang, N.; Zhao, Y.; Gao, X.; Yin, Y.; Qin, L.; Yuan, T. Comparative analysis of the organelle genomes of seven Rosa species (Rosaceae): Insights into structural variation and phylogenetic position. Front. Plant Sci. 2025, 16, 1584289. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Zheng, Y. Dynamic evolution and phylogenomic analysis of the chloroplast genome in Schisandraceae. Sci. Rep. 2018, 8, 9285. [Google Scholar] [CrossRef] [PubMed]
- Kuroda, H.; Bock, R. Essentiality of the chloroplast-encoded open reading frames ycf1 and ycf2 in tobacco. Plant J. 2008, 55, 681–690. [Google Scholar] [CrossRef]
- de Vries, J.; Gould, S.B.; Archibald, J.M. The carboxy terminus of YCF1 contains a motif conserved throughout >500 million years of streptophyte evolution. Genome Biol. Evol. 2017, 9, 345–356. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Wang, Y.; He, P.; Lee, J.; Douglas, E.; Fu, C. Chloroplast genome analyses and genomic resource development for epilithic sister genera Oresitrophe and Mukdenia (Saxifragaceae), using genome skimming data. BMC Genom. 2018, 19, 235. [Google Scholar] [CrossRef]
- Wu, Z.; Liao, R.; Yang, T.; Dong, X.; Liu, H. Analysis of six chloroplast genomes provides insight into the evolution of Chrysosplenium (Saxifragaceae). BMC Genom. 2020, 21, 621. [Google Scholar] [CrossRef]
- Liu, R.; Xia, M.; Liu, D.; Jiang, L.; Shen, J.; Chen, W. Analysis of the maternal genome of Elymus nutans from the Qinghai-Tibet Plateau based on chloroplast genomes. Grassl. Sci. 2022, 6, 114–123. [Google Scholar] [CrossRef]
- Yuan, W.; He, S.; Zhang, S.; Chang, D.; He, Y. The complete chloroplast genome sequence of Cornus alba L. (Cornaceae). Mitochondrial DNA Part B 2021, 6, 1997–1998. [Google Scholar] [CrossRef]
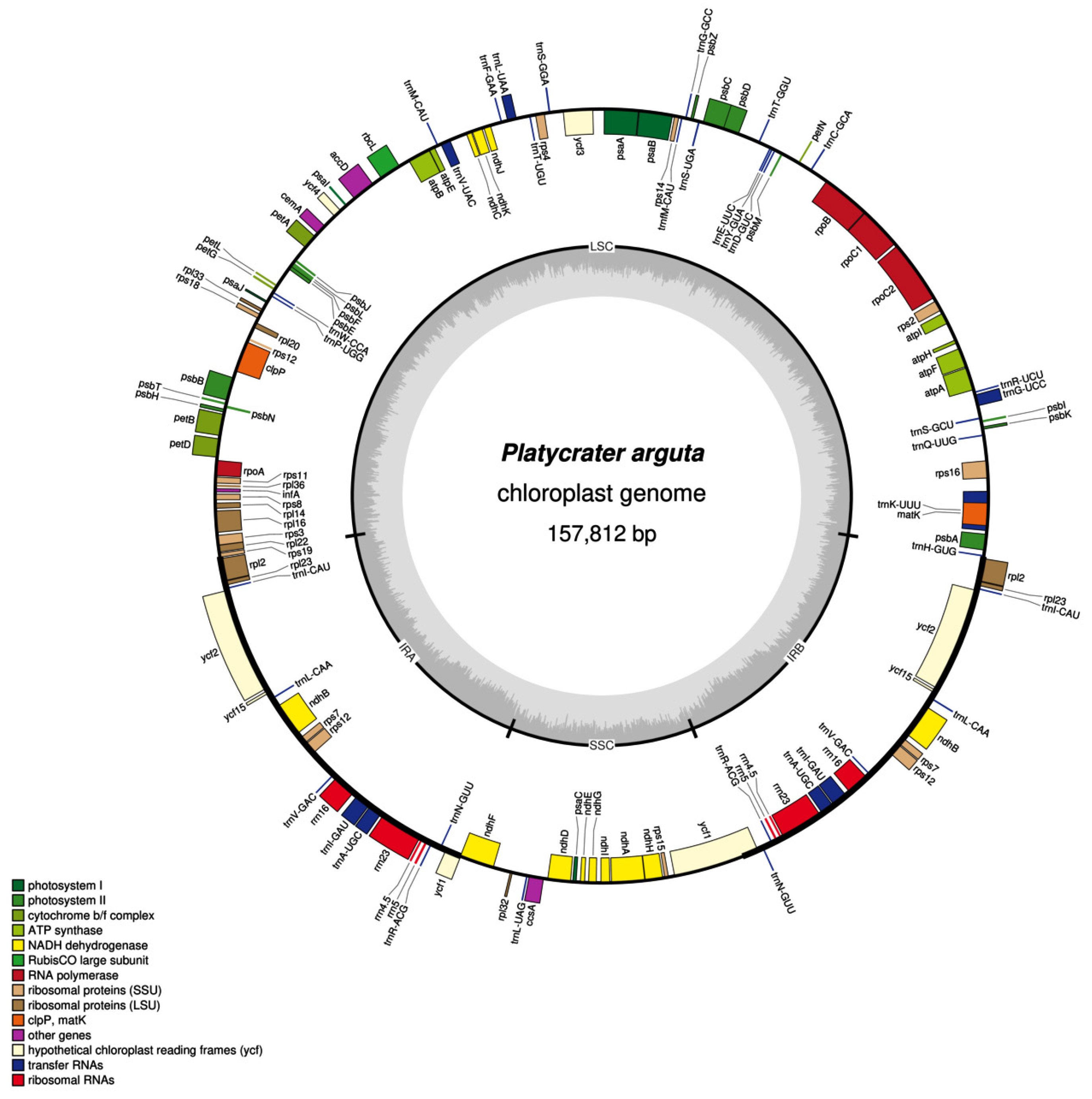

| Category | Gene Group | Gene Name |
|---|---|---|
| Photosynthesis | Subunits of photosystem I | psaA, psaB, psaC, psaI, psaJ |
| Subunits of photosystem II | psbA, psbB, psbC, psbD, psbE, psbF, psbH, psbI, psbJ, psbK, psbL, psbM, psbN, psbT, psbZ | |
| Subunits of NADH dehydrogenase | ndhA, ndhB (2), ndhC, ndhD, ndhE, ndhF, ndhG, ndhH, ndhI, ndhJ, ndhK | |
| Subunits of cytochrome b/f complex | petA, petB, petD, petG, petL, petN | |
| Subunits of ATP synthase | atpA, atpB, atpE, atpF, atpH, atpI | |
| Large subunit of rubisco | rbcL | |
| Self-replication | Proteins of large ribosomal subunit | rpl14, rpl16, rpl2 (2), rpl20, rpl22, rpl23(2), rpl32, rpl33, rpl36 |
| Proteins of small ribosomal subunit | rps11, rps12 (2), rps14, rps15, rps16, rps18, rps19, rps2, rps3, rps4, rps7(2), rps8 | |
| Subunits of RNA polymerase | rpoA, rpoB, rpoC1, rpoC2 | |
| Ribosomal RNAs | rrn16(2), rrn23(2), rrn4.5(2), rrn5(2) | |
| Transfer RN | trnA-UGC (2), trnC-GCA, trnD-GUC, trnE-UUC, trnF-GAA, trnG-GCC, trnG-UCC, trnH-GUG, trnI-CAU(2), trnI-GAU (2), trnK-UUU, trnL-CAA(2), trnL-UAA, trnL-UAG, trnM-CAU, trnN-GUU(2), trnP-UGG, trnQ-UUG, trnR-ACG(2), trnR-UCU, trnS-GCU, trnS-GGA, trnS-UGA, trnT-GGU, trnT-UGU, trnV-GAC(2), trnV-UAC, trnW-CCA, trnY-GUA, trnfM-CAU | |
| Other genes | Maturase | matK |
| Protease | clpP | |
| Envelope membrane protein | cemA | |
| Acetyl-CoA carboxylase | accD | |
| c-type cytochrome synthesis gene | ccsA | |
| Translation initiation factor | infA | |
| Genes of unknown function | Conserved hypothetical chloroplast ORF | ycf1(2), ycf15(2), ycf2(2), ycf3, ycf4 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ma, X.; Ye, Y.; Lin, R.; Hu, Q.; Zhang, X.; Hu, Y.; Feng, L.; Qian, R.; Zheng, J. Complete Chloroplast Genome Characterization, and Phylogenetic Analyses of the Rare and Endangered Plant Platycrater arguta. Biology 2025, 14, 1726. https://doi.org/10.3390/biology14121726
Ma X, Ye Y, Lin R, Hu Q, Zhang X, Hu Y, Feng L, Qian R, Zheng J. Complete Chloroplast Genome Characterization, and Phylogenetic Analyses of the Rare and Endangered Plant Platycrater arguta. Biology. 2025; 14(12):1726. https://doi.org/10.3390/biology14121726
Chicago/Turabian StyleMa, Xiaohua, Youju Ye, Ren’an Lin, Qingdi Hu, Xule Zhang, Yaping Hu, Lei Feng, Renjuan Qian, and Jian Zheng. 2025. "Complete Chloroplast Genome Characterization, and Phylogenetic Analyses of the Rare and Endangered Plant Platycrater arguta" Biology 14, no. 12: 1726. https://doi.org/10.3390/biology14121726
APA StyleMa, X., Ye, Y., Lin, R., Hu, Q., Zhang, X., Hu, Y., Feng, L., Qian, R., & Zheng, J. (2025). Complete Chloroplast Genome Characterization, and Phylogenetic Analyses of the Rare and Endangered Plant Platycrater arguta. Biology, 14(12), 1726. https://doi.org/10.3390/biology14121726






