Environmental DNA as a Tool for the Preliminary Assessment of Vertebrate Biodiversity: A Case Study from Sicilian Freshwater Ecosystems
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
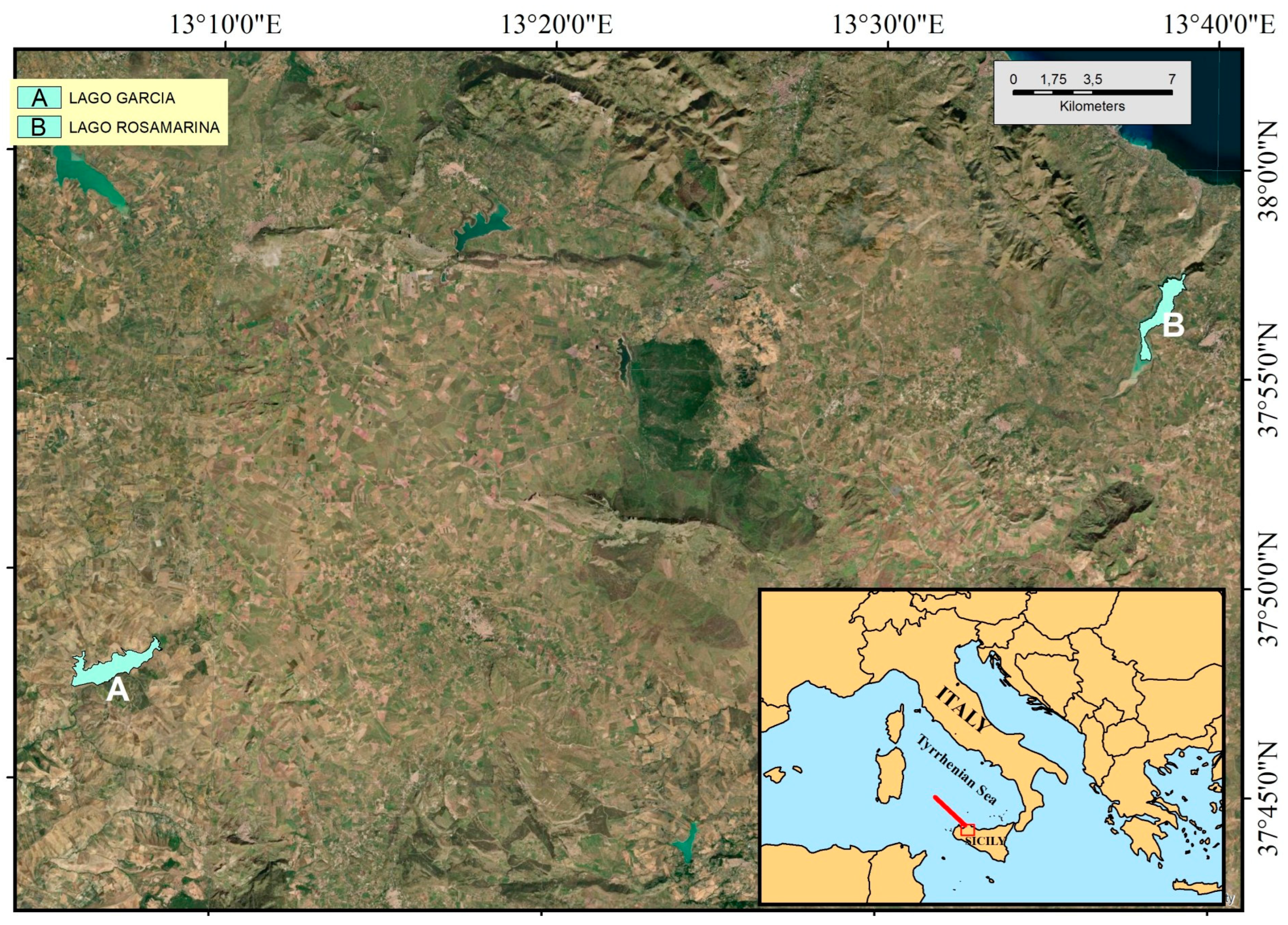
2.1. Study Area Lakes
2.2. Water Sampling and eDNA Extraction
2.3. eDNA Library Preparation and Bioinformatics Analysis
2.4. Data Analysis
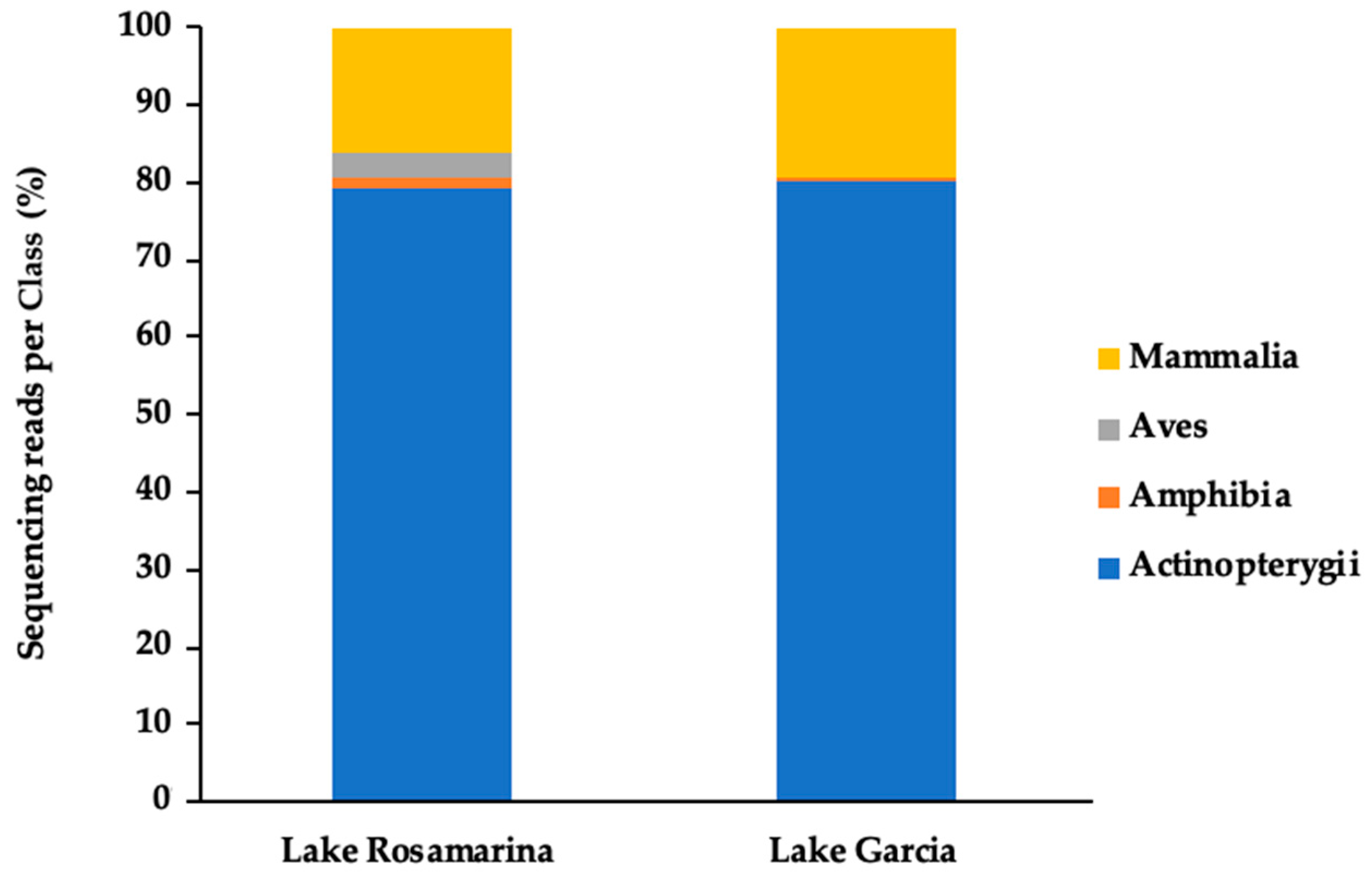
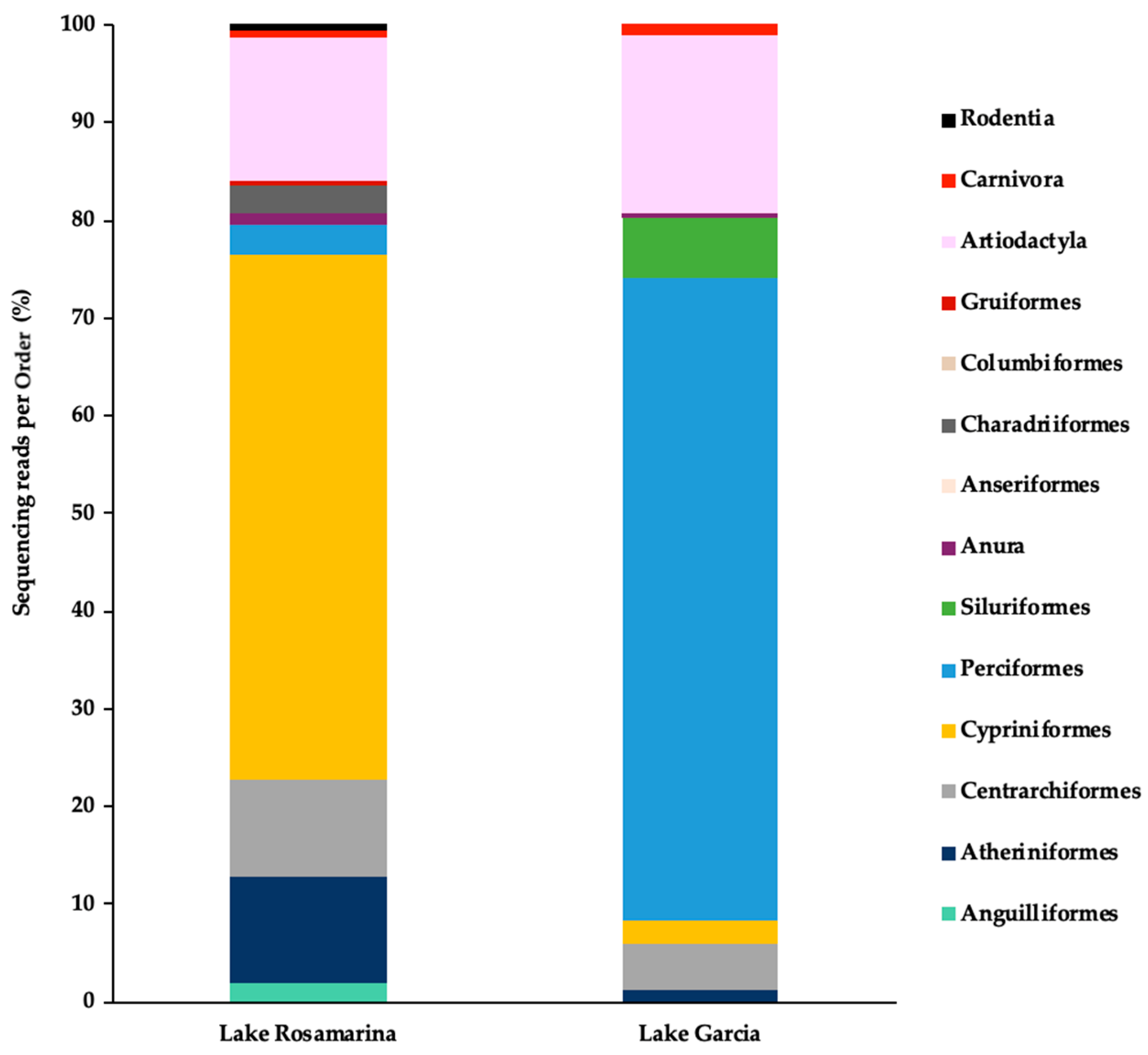
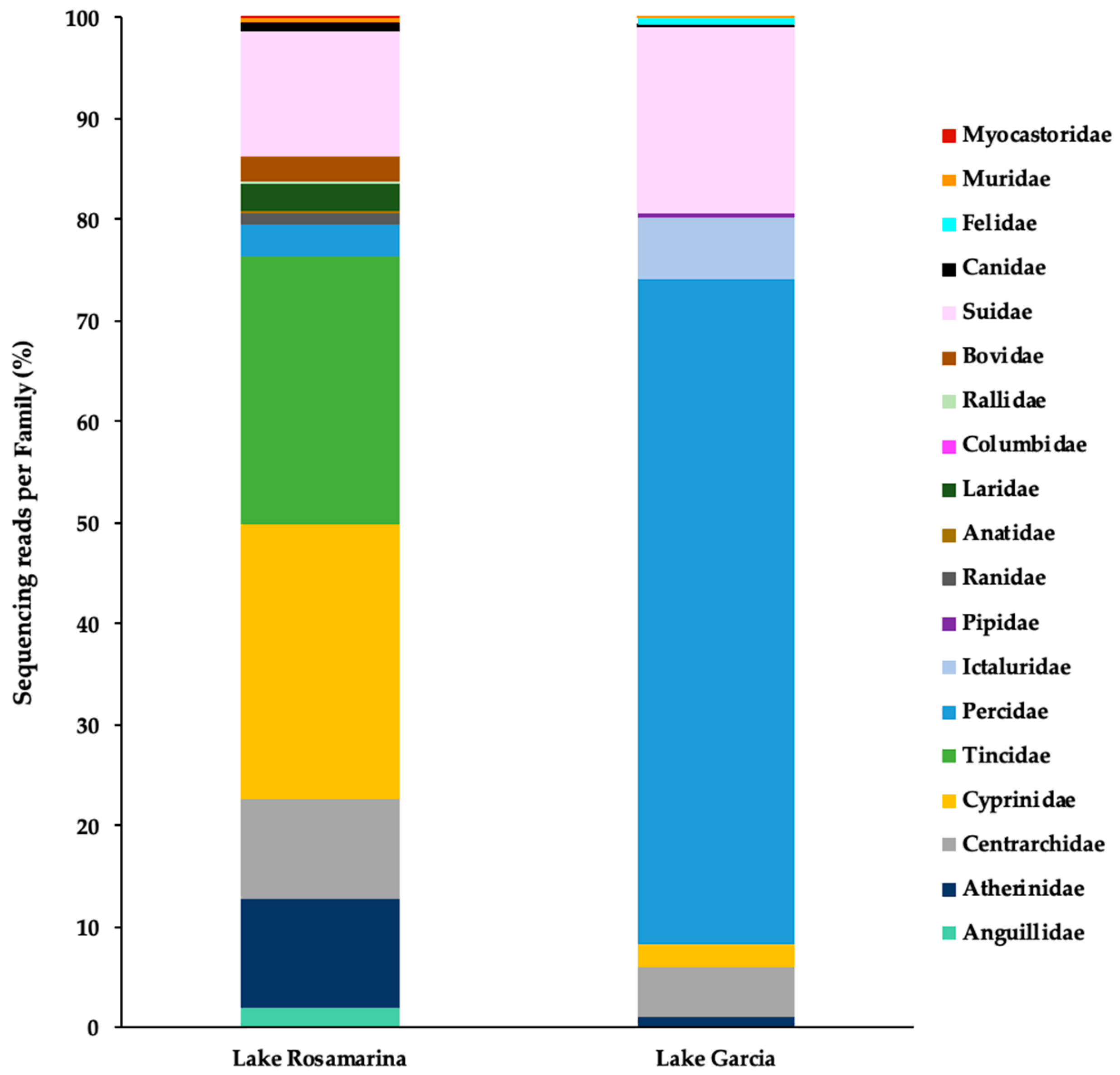
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Diaz, S.; Settele, J.; Brondízio, E.; Ngo, H.; Agard, J.; Arneth, A.; Balvanera, P.; Brauman, K. Pervasive human-driven decline of life on Earth points to the need for transformative change. Science 2019, 366, 6471. [Google Scholar] [CrossRef]
- Barnosky, A.D.; Matzke, N.; Tomiya, S.; Wogan, G.O.; Swartz, B.; Quental, T.B.; Marshall, C.; McGuire, J.L.; Lindsey, E.L.; Maguire, K.C. Has the Earth’s sixth mass extinction already arrived? Nature 2011, 471, 51–57. [Google Scholar] [CrossRef]
- Tilman, D.; Clark, M.; Williams, D.R.; Kimmel, K.; Polasky, S.; Packer, C. Future Threats to Biodiversity and Pathways to Their Prevention. Nature 2017, 546, 73–81. [Google Scholar] [CrossRef]
- Dudgeon, D.; Arthington, A.H.; Gessner, M.O.; Kawabata, Z.I.; Knowler, D.J.; Lévêque, C.; Naiman, R.J.; Prieur-Richard, A.H.; Soto, D.; Stiassny, M.L.J. Freshwater Biodiversity: Importance, Threats, Status and Conservation Challenges. Biol. Rev. Camb. Philos. Soc. 2006, 81, 163–182. [Google Scholar] [CrossRef]
- Reid, A.J.; Carlson, A.K.; Creed, I.F.; Eliason, E.J.; Gell, P.A.; Johnson, P.T.J.; Kidd, K.A.; MacCormack, T.J.; Olden, J.D.; Ormerod, S.J.; et al. Emerging Threats and Persistent Conservation Challenges for Freshwater Biodiversity. Biol. Rev. 2019, 94, 849–873. [Google Scholar] [CrossRef] [PubMed]
- Cantonati, M.; Poikane, S.; Pringle, C.M.; Stevens, L.E.; Turak, E.; Heino, J.; Richardson, J.S.; Bolpagni, R.; Borrini, A.; Cid, N. Characteristics, Main Impacts, and Stewardship of Natural and Artificial Freshwater Environments: Consequences for Biodiversity Conservation. Water 2020, 12, 260. [Google Scholar] [CrossRef]
- Heino, J.; Alahuhta, J.; Bini, L.M.; Cai, Y.; Heiskanen, A.S.; Hellsten, S.; Kortelainen, P.; Kotamäki, N.; Tolonen, K.T.; Vihervaara, P. Lakes in the Era of Global Change: Moving beyond Single-Lake Thinking in Maintaining Biodiversity and Ecosystem Services. Biol. Rev. 2021, 96, 89–106. [Google Scholar] [CrossRef] [PubMed]
- Vecchioni, L.; Chirco, P.; Bazan, G.; Marrone, F.; Arizza, V.; Arculeo, M. First Record of Temnosewellia minor (Platyhelminthes, Temnocephalidae) in Sicily, with a Plea for a Re-Examination of the Identity of the Publicly Available Molecular Sequences of the Genus. Biogeographia 2021, 36, a003. [Google Scholar] [CrossRef]
- Marrone, F.; Naselli-Flores, L. A Review on the Animal Xenodiversity in Sicilian Inland Waters (Italy). Adv. Oceanogr. Limnol. 2015, 6, 2–12. [Google Scholar] [CrossRef]
- Williams, M.A.; O’Grady, J.; Ball, B.; Carlsson, J.; de Eyto, E.; McGinnity, P.; Parle- McDermott, A. The application of CRISPR-Cas for single species identification from environmental DNA. Mol. Ecol. Resour. 2019, 19, 1106–1114. [Google Scholar] [CrossRef]
- Adamo, M.; Voyron, S.; Chialva, M.; Marmeisse, R.; Girlanda, M. Metabarcoding on both environmental DNA and RNA highlights differences between fungal communities sampled in different habitats. PLoS ONE 2020, 15, e0244682. [Google Scholar] [CrossRef]
- Ruppert, K.M.; Kline, R.J.; Rahman, M.S. Past, present, and future perspectives of environmental DNA (eDNA) metabarcoding: A systematic review in methods, monitoring, and applications of global eDNA. Glob. Ecol. Conserv. 2019, 17, e00547. [Google Scholar] [CrossRef]
- Sahu, A.; Kumar, N.; Singh, C.P.; Singh, M. Environmental DNA (eDNA): Powerful Technique for Biodiversity Conservation. J. Nat. Conserv. 2023, 71, 126325. [Google Scholar] [CrossRef]
- Frøslev, T.G.; Nielsen, I.B.; Santos, S.S.; Barnes, C.J.; Bruun, H.H.; Ejrnæs, R. The biodiversity effect of reduced tillage on soil microbiota. Ambio 2022, 51, 1022–1033. [Google Scholar] [CrossRef] [PubMed]
- Redondo, M.A.; Berlin, A.; Boberg, J.; Oliva, J. Vegetation type determines spore deposition within a forest–agricultural mosaic landscape. FEMS Microbiol. Ecol. 2020, 96, fiaa082. [Google Scholar] [CrossRef]
- Yang, J.; Zhang, L.; Mu, Y.; Zhang, X. Small changes make big progress: A more efficient eDNA monitoring method for freshwater fish. Environ. DNA 2023, 5, 363–374. [Google Scholar] [CrossRef]
- Nelson–Chorney, H.T.; Davis, C.S.; Poesch, M.S.; Vinebrooke, R.D.; Carli, C.M.; Taylor, M.K. Environmental DNA in lake sediment reveals biogeography of native genetic diversity. Front. Ecol. Environ. 2019, 17, 313–318. [Google Scholar] [CrossRef]
- Khalsa, N.S.; Smith, J.; Jochum, K.A.; Savory, G.; L’opez, J.A. Identifying under–ice overwintering locations of juvenile Chinook salmon by using environmental DNA. North Am. J. Fish. Manag. 2020, 40, 762–772. [Google Scholar] [CrossRef]
- Kinoshita, G.; Yonezawa, S.; Murakami, S.; Isagi, Y. Environmental DNA collected from snow tracks is useful for identification of mammalian species. Zoolog. Sci. 2019, 36, 198–207. [Google Scholar] [CrossRef]
- Günther, B.; Jourdain, E.; Rubincam, L.; Karoliussen, R.; Cox, S.L.; Arnaud Haond, S. Feces DNA analyses track the rehabilitation of a free-ranging beluga whale. Sci. Rep. 2022, 12, 6412. [Google Scholar] [CrossRef]
- Hänfling, B.; Lawson Handley, L.; Read, D.S.; Hahn, C.; Li, J.; Nichols, P.; Winfield, I.J. Environmental DNA metabarcoding of lake fish communities reflects long-term data from established survey methods. Mol. Ecol. 2016, 25, 3101–3119. [Google Scholar] [CrossRef]
- Valentini, A.; Taberlet, P.; Miaud, C.; Civade, R.; Herder, J.; Thomsen, P.F.; Dejean, T. Next-generation monitoring of aquatic biodiversity using environmental DNA metabarcoding. Mol. Ecol. 2016, 25, 929–942. [Google Scholar] [CrossRef]
- Sard, N.M.; Herbst, S.J.; Nathan, L.; Uhrig, G.; Kanefsky, J.; Robinson, J.D.; Scribner, K.T. Comparison of fish detections, community diversity, and relative abundance using environmental DNA metabarcoding and traditional gears. Enviro. DNA 2019, 1, 368–384. [Google Scholar] [CrossRef]
- Coutant, O.; Richard-Hansen, C.; de Thoisy, B.; Decotte, J.B.; Valentini, A.; Dejean, T.; Brosse, S. Amazonian mammal monitoring using aquatic environmental DNA. Mol. Ecol. Resour. 2021, 21, 1875–1888. [Google Scholar] [CrossRef]
- Dal Pont, G.; Duarte Ritter, C.; Agostinis, A.O.; Stica, P.V.; Horodesky, A.; Cozer, N.; Balsanelli, E.; Netto, O.S.M.; Henn, C.; Ostrensky, A.; et al. Monitoring fish communities through environmental DNA metabarcoding in the fish pass system of the second largest hydropower plant in the world. Sci. Rep. 2021, 11, 23167. [Google Scholar] [CrossRef]
- Mena, J.L.; Yagui, H.; Tejeda, V.; Bonifaz, E.; Bellemain, E.; Valentini, A.; Tobler, M.W.; Sánchez-Vendizú, P.; Lyet, A. Environmental DNA metabarcoding as a useful tool for evaluating terrestrial mammal diversity in tropical forests. Ecol. Appl. 2021, 31, e02335. [Google Scholar] [CrossRef]
- Pawlowski, J.; Bonin, A.; Boyer, F.; Cordier, T.; Taberlet, P. Environmental DNA for biomonitoring. Mol. Ecol. 2021, 30, 2931. [Google Scholar] [CrossRef]
- Jia, H.; Zhang, H.; Xian, W. Fish diversity monitored by environmental DNA in the Yangtze River mainstream. Fishes 2021, 7, 1. [Google Scholar] [CrossRef]
- Mas-Carrió, E.; Schneider, J.; Nasanbat, B.; Ravchig, S.; Buxton, M.; Nyamukondiwa, C.; Fumagalli, L. Assessing environmental DNA metabarcoding and camera trap surveys as complementary tools for biomonitoring of remote desert water bodies. Environ. DNA 2022, 4, 580–595. [Google Scholar] [CrossRef]
- Ficetola, G.F.; Miaud, C.; Pompanon, F.; Taberlet, P. Species detection using environmental DNA from water samples. Biol. Lett. 2008, 4, 423–425. [Google Scholar] [CrossRef] [PubMed]
- Schütz, R.; Fukaya, K.; Beermann, A.J.; Díaz, C.; Lenz, A.C.; Macher, T.H.; Leese, F. Optimizing Environmental DNA metabarcoding of archived suspended particulate matter for river biodiversity monitoring. Environ. DNA 2025, 7, e70130. [Google Scholar] [CrossRef]
- Adrian-Kalchhauser, I.; Burkhardt-Holm, P. An eDNA assay to monitor a globally invasive fish species from flowing freshwater. PLoS ONE 2016, 11, e0147558. [Google Scholar] [CrossRef]
- Sugawara, K.; Sasaki, Y.; Okano, K.; Watanabe, M.; Miyata, N. Application of eDNA for monitoring freshwater bivalve Nodularia nipponensis and its glochidium larvae. Environ. DNA 2022, 4, 908–919. [Google Scholar] [CrossRef]
- Ratsch, R.; Kingsbury, B.A.; Jordan, M.A. Exploration of environmental DNA (eDNA) to detect Kirtland’s snake (Clonophis kirtlandii). Animals 2020, 10, 1057. [Google Scholar] [CrossRef]
- Allen, M.C.; Kwait, R.; Vastano, A.; Kisurin, A.; Zoccolo, I.; Jaffe, B.D.; Lockwood, J.L. Sampling environmental DNA from trees and soil to detect cryptic arboreal mammals. Sci. Rep. 2023, 13, 180. [Google Scholar] [CrossRef]
- Sander, M.; Beermann, A.J.; Buchner, D.; Madge Pimentel, I.; Sinclair, J.S.; Weiss, M.; Leese, F. Environmental DNA time series analysis of a temperate stream reveals distinct seasonal community and functional shifts. Riv. Res. Appl. 2024, 40, 850–862. [Google Scholar] [CrossRef]
- Mauro, M.; Lo Valvo, M.; Vazzana, M.; Radovic, S.; Vizzini, A.; Badalamenti, R.; Hornsby, L.B.; Arizza, V. Environmental DNA: The First Snapshot of the Vertebrate Biodiversity in Three Sicilian Lakes. Animals 2023, 13, 3687. [Google Scholar] [CrossRef]
- Mauro, M.; Longo, F.; Lo Valvo, M.; Vizzini, A.; Di Grigoli, A.; Radovic, S.; Arizza, V.; Vecchioni, L.; La Paglia, L.; Queiroz, V.; et al. The Use of Environmental DNA as Preliminary Description of Invertebrate Diversity in Three Sicilian Lakes. Animals 2025, 15, 355. [Google Scholar] [CrossRef]
- Thomsen, P.F.; Kielgast, J.; Iversen, L.L.; Møller, P.R.; Rasmussen, M.; Willerslev, E. Detection of a diverse marine fish fauna using environmental DNA from seawater samples. PLoS ONE 2012, 7, e41732. [Google Scholar] [CrossRef] [PubMed]
- Riaz, T.; Shehzad, W.; Viari, A.; Pompanon, F.; Taberlet, P.; Coissac, E. ecoPrimers: Inference of new DNA barcode markers from whole genome sequence analysis. Nucl. Acids Res. 2011, 39, e145. [Google Scholar] [CrossRef]
- Liu, Q.; Zhang, H.; Chang, F.; Xie, P.; Zhang, Y.; Wu, H.; Zhang, X.; Peng, W.; Liu, F. eDNA revealed in situ microbial community changes in response to Trapa japonica in Lake Qionghai and Lake Erhai, southwestern China. Chemosphere 2022, 288, 132605. [Google Scholar] [CrossRef]
- Kumar, G.; Reaume, A.M.; Farrell, E.; Gaither, M.R. Comparing eDNA metabarcoding primers for assessing fish communities in a biodiverse estuary. PLoS ONE 2022, 17, e0266720. [Google Scholar] [CrossRef]
- Magoč, T.; Salzberg, S.L. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 2011, 27, 2957–2963. [Google Scholar] [CrossRef] [PubMed]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Del Fabbro, C.; Scalabrin, S.; Morgante, M.; Giorgi, F.M. An extensive evaluation of read trimming effects on Illumina NGS data analysis. PLoS ONE 2013, 8, e85024. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef]
- Rognes, T.; Flouri, T.; Nichols, B.; Quince, C.; Mahé, F. VSEARCH: A versatile open source tool for metagenomics. Peer J. 2016, 4, e2584. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef]
- Sato, Y.; Miya, M.; Fukunaga, T.; Sado, T.; Iwasaki, W.; Kumar, S. MitoFish and MiFish Pipeline: A mitochondrial genome database of fish with an analysis pipeline for environmental DNA metabarcoding. Mol. Biol. Evol. 2018, 35, 1553. [Google Scholar] [CrossRef]
- Porter, T.M. Terrimporter/12SvertebrateClassifier: 12S Vertebrate Classifier v2.0.0-ref (v2.0.0-ref) [Computer Software]. Zenodo. 2021. Available online: https://github.com/terrimporter/12SvertebrateClassifier (accessed on 20 November 2023).
- Tierno de Figueroa, J.M.; López-Rodríguez, M.J.; Fenoglio, S.; Sánchez-Castillo, P.; Fochetti, R. Freshwater biodiversity in the rivers of the Mediterranean Basin. Hydrobiologia 2013, 719, 137–186. [Google Scholar] [CrossRef]
- Skoulikidis, N.T.; Sabater, S.; Datry, T.; Morais, M.M.; Buffagni, A.; Dörflinger, G.; Tockner, K. Non-perennial Mediterranean rivers in Europe: Status, pressures, and challenges for research and management. Sci. Total Environ. 2017, 577, 1–18. [Google Scholar] [CrossRef]
- Wang, B.; Jiao, L.; Ni, L.; Wang, M.; You, P. Bridging the gap: The integration of eDNA techniques and traditional sampling in fish diversity analysis. Front. Mar. Sci. 2024, 11, 1289589. [Google Scholar] [CrossRef]
- Rees, H.C.; Maddison, B.C.; Middleditch, D.J.; Patmore, J.R.; Gough, K.C. The detection of aquatic animal species using environmental DNA a review of eDNA as a survey tool in ecology. J. Appl. Ecol. 2014, 51, 1450–1459. [Google Scholar] [CrossRef]
- Deiner, K.; Fronhofer, E.; Mächler, E.; Walser, J.C.; Altermatt, F. Environmental DNA reveals that rivers are conveyer belts of biodiversity information. Nat. Commun. 2016, 7, 12544. [Google Scholar] [CrossRef]
- Strickler, K.M.; Fremier, A.K.; Goldberg, C.S. Quantifying effects of UV-B, temperature, and pH on eDNA degradation in aquatic microcosms. Biol. Conserv. 2015, 183, 85–92. [Google Scholar] [CrossRef]
- Zulkefli, N.S.; Kim, K.-H.; Hwang, S.-J. Effects of Microbial Activity and Environmental Parameters on the Degradation of Extracellular Environmental DNA from a Eutrophic Lake. Int. J. Environ. Res. Public Health 2019, 16, 3339. [Google Scholar] [CrossRef]
- Xia, Z.; Zhan, A.; Johansson, M.L.; DeRoy, E.; Haffner, G.D.; MacIsaac, H.J. Screening marker sensitivity: Optimizing eDNA-based rare species detection. Divers. Distrib. 2021, 27, 1981–1988. [Google Scholar] [CrossRef]
- Garlaschè, G.; Borgomaneiro, G.; Piscia, R.; Manca, M.; Eckert, E.M.; Fontaneto, D. Metabarcoding to monitor the crustacean zooplankton of a lake improves when using a reference DNA library from local samples. J. Limnol. 2023, 82, 1. [Google Scholar] [CrossRef]
- Lamb, P.D.; Hunter, E.; Pinnegar, J.K.; Creer, S.; Davies, R.G.; Taylor, M.I. How quantitative is metabarcoding: A meta-analytical approach. Mol. Ecol. 2019, 28, 420–430. [Google Scholar] [CrossRef]
- Duchi, A. Tench (Tinca tinca) in Sicily: Current knowledge and research needs for conservation and management. CYBIUM 2016, 40, 329–332. [Google Scholar]
- Tortonese, E. Fauna d’Italia; Pisces Osteichthyes: Bologna, Italy, 1970; Volume 10. [Google Scholar]
- Vinciguerra, D. Relazione intorno alla pesca di acqua dolce e di mare in Sicilia e dei modi per aumentare il prodotto. Boll. Not. Agrar. 1896, 29, 105–128. [Google Scholar]
- Ferrito, V.; Tigano, C. The distribution of the Icththyofauna in the Simeto basins (Sicily). Cybium 1995, 19, 187–192. [Google Scholar]
- Duchi, A. Distribuzione della fauna ittica nelle acque interne dell’areale ibleo: La provincia di Ragusa. Biol. Ambient. 2006, 20, 291–294. [Google Scholar]
- Duchi, A.; Milano, A. Il progetto P. Int. 2005–2007 “Utilizzo dei grandi invasi siciliani per fini produttivi, ambientali e ricreativi” dell’Assessorato Regionale Risorse Agricole ed Alimentari: Un’occasione per l’ampliamento della conoscenza sulla fauna ittica siciliana. Ital. J. Freshw. Ichthyol. 2014, 2014, 215–219. [Google Scholar]
- Duchi, A.; Divincenzo, S. First data on the fish fauna of the saint elia stream (troina, en, sicily, italy). Ital. J. Freshw. Ichthyol. 2017, 4, 183–187. [Google Scholar]
- Tekin-Özan, S.; Kir, İ. Comparative study on the accumulation of heavy metals in different organs of tench (Tinca tinca L. 1758) and plerocercoids of its endoparasite Ligula intestinalis. Parasitol. Res. 2005, 97, 156–159. [Google Scholar] [CrossRef]
- Tekin-Özan, S.; Kir, I. Concentrations of Some Heavy Metals in Tench (Tinca tinca L., 1758), Its Endoparasite (Ligula intestinalis L., 1758), Sediment and Water in Beyşehir Lake, Turkey. Polish J. of Environ. Stud. 2008, 17, 597–603. [Google Scholar]
- Shah, S.L.; Altindag, A. Hematological Parameters of Tench (Tinca tincaL.) after Acute and Chronic Exposure to Lethal and Sublethal Mercury Treatments. Bull. Environ. Contam. Toxicol. 2004, 73, 911–918. [Google Scholar] [CrossRef]
- Shah, S.L.; Altindağ, A. Effects of heavy metal accumulation on the 96-h LC_50 values in Tench Tinca tinca L., 1758. Turk. J. Vet. Anim. Sci. 2005, 29, 139–144. [Google Scholar]
- Aktumsek, A.; Gezgin, S. Seasonal variations of metal concentrations in muscle tissue of tench (Tinca tinca), water and sediment in Beysehir Lake (Turkey). Environ. Technol. 2011, 32, 1479–1485. [Google Scholar] [CrossRef]
- Vilizzi, L. The common carp, Cyprinus carpio, in the Mediterranean region: Origin, distribution, economic benefits, impacts and management. Fisher. Manage. Ecol. 2012, 19, 93–110. [Google Scholar] [CrossRef]
- Hupało, K.; Schmidt, S.; Macher, T.H.; Weiss, M.; Leese, F. Fresh insights into Mediterranean biodiversity: Environmental DNA reveals spatio-temporal patterns of stream invertebrate communities on Sicily. Hydrobiologia 2022, 849, 155–173. [Google Scholar] [CrossRef]
- Ottonello, D.; D’angelo, S.; Oneto, F.; Malavasi, S.; Zuffi, M.A.L.; Spadola, F. So close so dif-ferent: What makes the difference? Acta Herpetologica 2021, 16, 89–98. [Google Scholar] [CrossRef]
- Lorenzoni, M.; Corboli, M.; Doerr, A.J.M.; Giovinazzo, G.; Selvi, S.; Mearelli, M. Diets of Micropterus salmoides Lac. and Esox lucius L. in Lake Trasimeno (Umbria, Italy) and their diet overlap. Bull. Fr. Pêche Piscic. 2002, 365–366, 537–547. [Google Scholar] [CrossRef]
- Ventura, M.; Careddu, G.; Sporta Caputi, S.; Calizza, E.; Rossi, L.; Costantini, M.L. Role of body size and habitat complexity in the diet of the invasive Micropterus salmoides (Lacépède): Optimal foraging theory matters. Biol. Invasions 2025, 27, 1–15. [Google Scholar] [CrossRef]
- Pereira, F.W.; Vitule, J.R.S. The largemouth bass Micropterus salmoides (Lacepède, 1802): Impacts of a powerful freshwater fish predator outside of its native range. Rev. Fish Biol. Fisheries. 2019, 29, 639–652. [Google Scholar] [CrossRef]
- Han, J.H.; Paek, W.K.; An, K.G. Exotic species, Micropterus salmoides, as a key bioindicator influencing the reservoir health and fish community structure. J. Asia-Pac. Biodivers. 2016, 9, 403–411. [Google Scholar] [CrossRef]
- Orban, E.; Nevigato, T.; Masci, M.; Di Lena, G.; Casini, I.; Caproni, R.; Rampacci, M. Nutritional quality and safety of European perch (Perca fluviatilis) from three lakes of Central Italy. Food Chem. 2007, 100, 482–490. [Google Scholar] [CrossRef]
- Christensen, E.A.F.; Svendsen, M.B.S.; Steffensen, J.F. Population ecology, growth, and physico-chemical habitat of anadromous European perch Perca fluviatilis. Estuar. Coast. Shelf Sci. 2021, 249, 107091. [Google Scholar] [CrossRef]
- Froese, R.; Pauly, D. FishBase. World Wide Web electronic publication. 2022. Available online: www.fishbase.org (accessed on 26 September 2025).
- Walker, K.F.; Yang, H.Z. Fish and fisheries of western China. Fish Fish. High. Alt. Asia 1999, 385, 304. [Google Scholar]
- Ma, X.; Bangxi, X.; Yindong, W.; Mingxue, W. Intentionally introduced and transferred fishes in China’s inland waters. Asian Fish. Sci. 2003, 16, 279–290. [Google Scholar] [CrossRef]
- Allen, K.R. The food and migration of the perch (Perca fluviatilis) in Windermere. J. Animal. Ecol. 1935, 4, 264–273. [Google Scholar] [CrossRef]
- Banha, F.; Ilhéu, M.; Anastácio, P.M. Angling web forums as an additional tool for detection of new fish introductions: The first record of Perca fluviatilis in continental Portugal. Knowl. Manag. Aquat. Ecosyst. 2015, 3, 416. [Google Scholar] [CrossRef]
- Esposito, A.; Denys, G.P.; Haÿ, V.; Agostini, P.J.; Foata, J.; Quilichini, Y. Unregulated introduced fish (Perca fluviatilis Linnaeus, 1758) is host to zoonotic parasites in a small Mediterranean island. J. Parasitol. Res. 2024, 123, 247. [Google Scholar] [CrossRef] [PubMed]
- Bianco, P.G.; Ketmaier, V. Anthropogenic changes in the freshwater fish fauna of Italy, with reference to the central region and Barbus graellsii, a newly established alien species of Iberian origin. J. Fish Biol. 2001, 59, 190–208. [Google Scholar] [CrossRef]
- Persson, L.; Greenberg, L.A. Interspecific and intraspecific size class competition affecting resource use and growth of perch, Perca fluviatilis. Oikos 1990, 97–106. [Google Scholar] [CrossRef]
- Mehner, T.; Arlinghaus, R.; Berg, S.; Dörner, H.; Jacobsen, L.; Kasprzak, P.; Wysujack, K. How to link biomanipulation and sustainable fisheries management: A step-by-step guideline for lakes of the European temperate zone. Fish. Manage. Ecol. 2004, 11, 261–275. [Google Scholar] [CrossRef]
- Lorenzoni, M.; Ghetti, L.; Pedicillo, G.; Carosi, A. Analysis of the biological features of the goldfish Carassius auratus auratus in Lake Trasimeno (Umbria, Italy) with a view to drawing up plans for population control. Folia Zool. 2010, 59, 142–156. [Google Scholar] [CrossRef]
- Marrone, F.; Canale, D.E. Occurrence, distribution and bibliography of the medicinal leech Hirudo verbana Carena, 1820 (Hirudinea, Hirudinidae) in Sicily (Italy). Biogeogr. -J. Integr. Biogeogr. 2019, 34, 33–38. [Google Scholar] [CrossRef]
- Milana, V.; Franchini, P.; Sola, L.; Angiulli, E.; Rossi, A.R. Genetic structure in lagoons: The effects of habitat discontinuity and low dispersal ability on populations of Atherina boyeri. Mar. Biol. 2012, 159, 399–411. [Google Scholar] [CrossRef]
- Ibragimova, N.; Shalgimbayeva, S.; Popov, N.; Jumakhanova, G. The Study of the Heart of A. Boyeri caspia in Environmental Quality Assessment. EMCEI 2019, 2135–2140. [Google Scholar] [CrossRef]
- Bianco, P.G. An update on the status of native and exotic freshwater fishes of Italy. J. Appl. Ichthyol. 2014, 30, 62–77. [Google Scholar] [CrossRef]
- Caliani, I.; Rodríguez, L.P.; Casini, S.; Granata, A.; Zagami, G.; Pansera, M.; Querci, G.; Minutoli, R. Biochemical and genotoxic biomarkers in Atherina boyeri to evaluate the status of aquatic ecosystems. Reg. Stud. Mar. Sci. 2019, 28, 100566. [Google Scholar] [CrossRef]
- Evans, D.H.; Weingarten, K. The effect of cadmium and other metals on vascular smooth muscle of the dogfish shark, Squalus acanthias. Toxicology 1990, 61, 275–281. [Google Scholar] [CrossRef]
- Kottelat, M.; Freyhof, J. Handbook of European Freshwater Fishes; Kottelat, Cornol, Switzerland and Freyhof, Berlin, Germany. 2007; 646p. Available online: https://asih.kglmeridian.com/view/journals/cope/2008/3/article-p725.xml (accessed on 26 September 2025).
- Carapezza, A.; Faraci, F. CK2000–Carta ittica della Sicilia. In Checklist and Distribution of the Italian Fauna. CKmap, Memorie del Museo Civico di Storia Naturale di Verona; Ruffo, S., Stoch, F., Eds.; 2a serie, Sezione Scienze della Vita. 2006. Available online: https://www.google.com.hk/url?sa=t&source=web&rct=j&opi=89978449&url=https://faunaitalia.it/documents/CKmap_ENG.pdf&ved=2ahUKEwi4n6Hijo-RAxXqePUHHbhjBYYQFnoECCIQAQ&usg=AOvVaw2eP-K8NA2v724o6gApFiJJ (accessed on 26 September 2025).
- Öğlü, B.; Yorulmaz, B.; Genc, T.O.; Yılmaz, F. The Assessment of Heavy Metal Content by Using Bioaccumulation Indices in European Chub, Squalius cephalus (Linnaeus, 1758). Available online: https://www.researchgate.net/publication/274378970_The_assessment_of_heavy_metal_content_by_using_bioaccumulation_indices_in_European_chub_Squalius_cephalus_Linnaeus_1758 (accessed on 26 September 2025).
- Wang, R.; Wang, X.T.; Wu, L.; Mateescu, M.A. Toxic effects of cadmium and copper on the isolated heart of dogfish shark, Squalus acanthias. J. Toxicol. Environ. Health Part A 1999, 57, 507–519. [Google Scholar]
- Krasnići, N.; Dragun, Z.; Erk, M.; Raspor, B. Distribution of Co, Cu, Fe, Mn, Se, Zn, and Cd among cytosolic proteins of different molecular masses in gills of European chub (Squalius cephalus L.). Environ Sci Pollut Res Int. 2014, 21, 13512–13521. [Google Scholar] [CrossRef]
- Jacoby, D.M.; Casselman, J.M.; Crook, V.; DeLucia, M.B.; Ahn, H.; Kaifu, K.; Gollock, M.J. Synergistic patterns of threat and the challenges facing global anguillid eel conservation. Glob. Ecol. Conserv. 2015, 4, 321–333. [Google Scholar] [CrossRef]
- Barcala, E.; Romero, D.; Bulto, C.; Boza, C.; Peñalver, J.; María-Dolores, E.; Muñoz, P. An endangered species living in an endangered ecosystem: Population structure and growth of European eel Anguilla anguilla in a Mediterranean coastal lagoon. Reg. Stud. Mar. Sci. 2022, 50, 102163. [Google Scholar] [CrossRef]
- Canestrelli, D.; Nascetti, G. Phylogeography of the pool frog Rana (Pelophylax) lessonae in the Italian peninsula and Sicily: Multiple refugia, glacial expansions and nuclear–mitochondrial discordance. J. Biogeogr. 2008, 35, 1923–1936. [Google Scholar] [CrossRef]
- Dufresnes, C.; Golay, J.; Schuerch, J.; Dejean, T.; Dubey, S. Monitoring of the last stronghold of native pool frogs (Pelophylax lessonae) in Western Europe, with implications for their conservation. Eur. J. Wildl. Res. 2020, 66, 45. [Google Scholar] [CrossRef]
- Lillo, F.; Marrone, F.; Sicilia, A.; Castelli, G.; Zava, B. An invasive population of Xenopus laevis (Daudin, 1802) in Italy. Herpetozoa 2005, 18, 63–64. [Google Scholar]
- Lo Valvo, M.; Faraone, F.P.; Giacalone, G.; Lillo, F. Fauna di Sicilia 2017; Anfibi. Monografie Naturalistiche, 5; Edizioni Danaus: Palermo, Italy, 2017; p. 136. [Google Scholar]
- Lillo, F.; Faraone, F.P.; Lo Valvo, M. Can the introduction of Xenopus laevis affect native amphibian populations? Reduction of reproductive occurrence in presence of the invasive species. Biol. Invasions 2011, 13, 1533–1541. [Google Scholar] [CrossRef]
- Faraone, F.P.; Lillo, F.; Giacalone, G.; Valvo, M.L. The large invasive population of Xenopus laevis in Sicily, Italy. Amphibi. Reptil. 2008, 29, 405–412. [Google Scholar] [CrossRef]
- Wen, Y.; Schoups, G.; Van De Giesen, N. Organic pollution of rivers: Combined threats of urbanization, livestock farming and global climate change. Sci. Rep. 2017, 7, 43289. [Google Scholar] [CrossRef]
- Tullo, E.; Finzi, A.; Guarino, M. Environmental impact of livestock farming and Precision Livestock Farming as a mitigation strategy. Sci. Total Environ. 2019, 650, 2751–2760. [Google Scholar] [CrossRef] [PubMed]
- Patarón, E.R.O.; Guambo, V.M.V.; Orozco, E.G.; Casco, E.R.G. Anthropic activities and risks of contamination due to the emission of liquid waste in agriculture. Ann. For. Res. 2023, 66, 2936–2945. [Google Scholar]
- Cocchi, R.; Riga, F. Control of a coypu Myocastor coypus population in northern Italy and management implications. Ital. J. Zool. 2008, 75, 37–42. [Google Scholar] [CrossRef]
- Bertolino, S.; Genovesi, P. Semiaquatic mammals introduced into Italy: Case studies in biological invasion. In Biological Invaders in Inland Waters: Profiles, Distribution, and Threats; Springer: Dordrecht, The Netherlands, 2007; pp. 175–191. [Google Scholar]
- Petralia, E. Status della Nutria (Myocastor coypus Molina, 1782) nella Riserva Naturale Speciale Biologica “Macchia Foresta del Fiume Irminio” (Ragusa). Tec. Agric. 2003, 1–3, 87–97. [Google Scholar]
- Loy, A.; Bon, M.; De Febbraro, M.; Baisero, D.; Amori, G. (Eds.) Atlante dei Mammiferi in Italia/Atlas of Mammals in Italy; historia naturae (13); Associazione Teriologica Italiana Edizioni Belvedere: Latina, Italy, 2025; 520p. [Google Scholar]
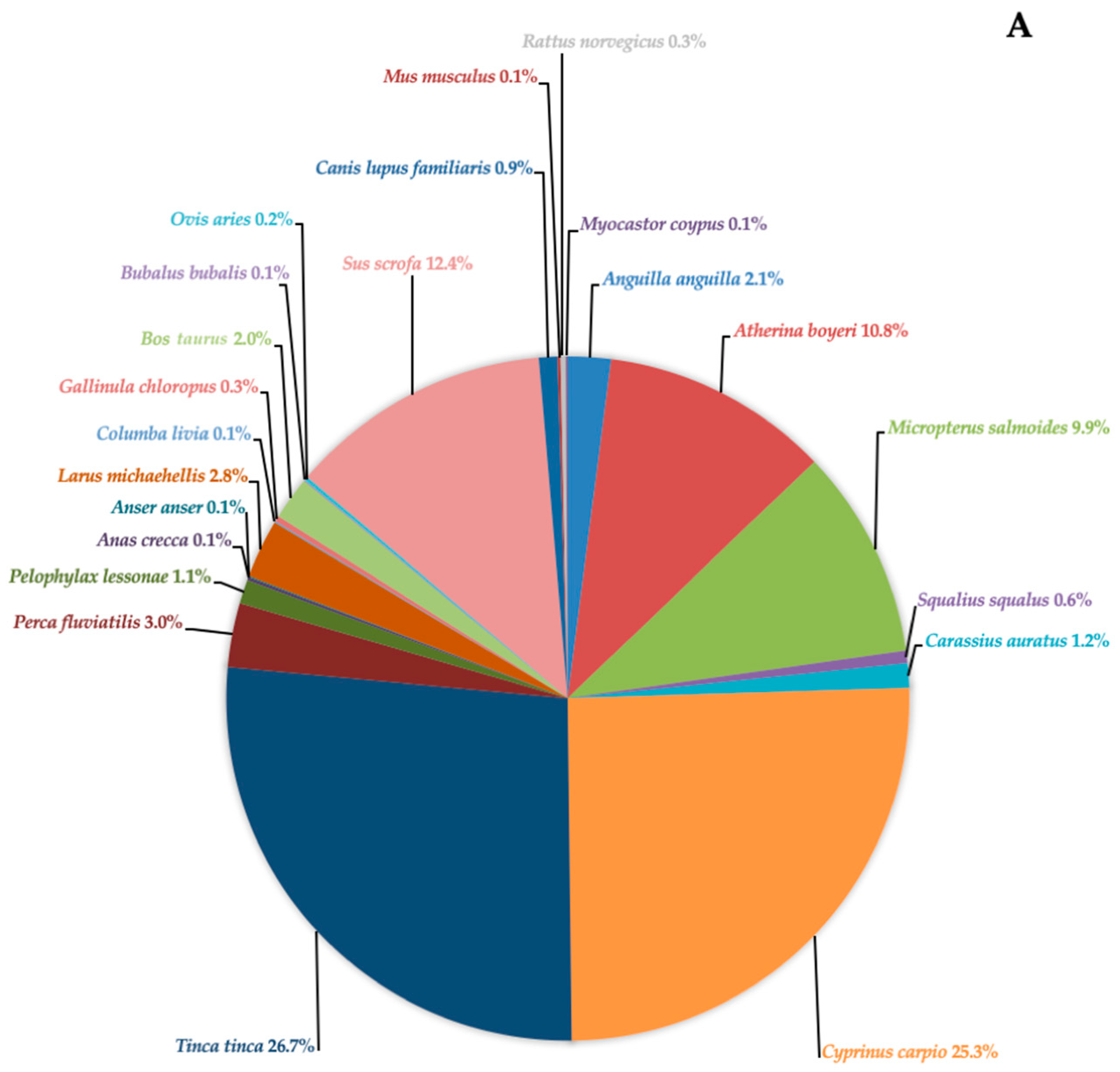

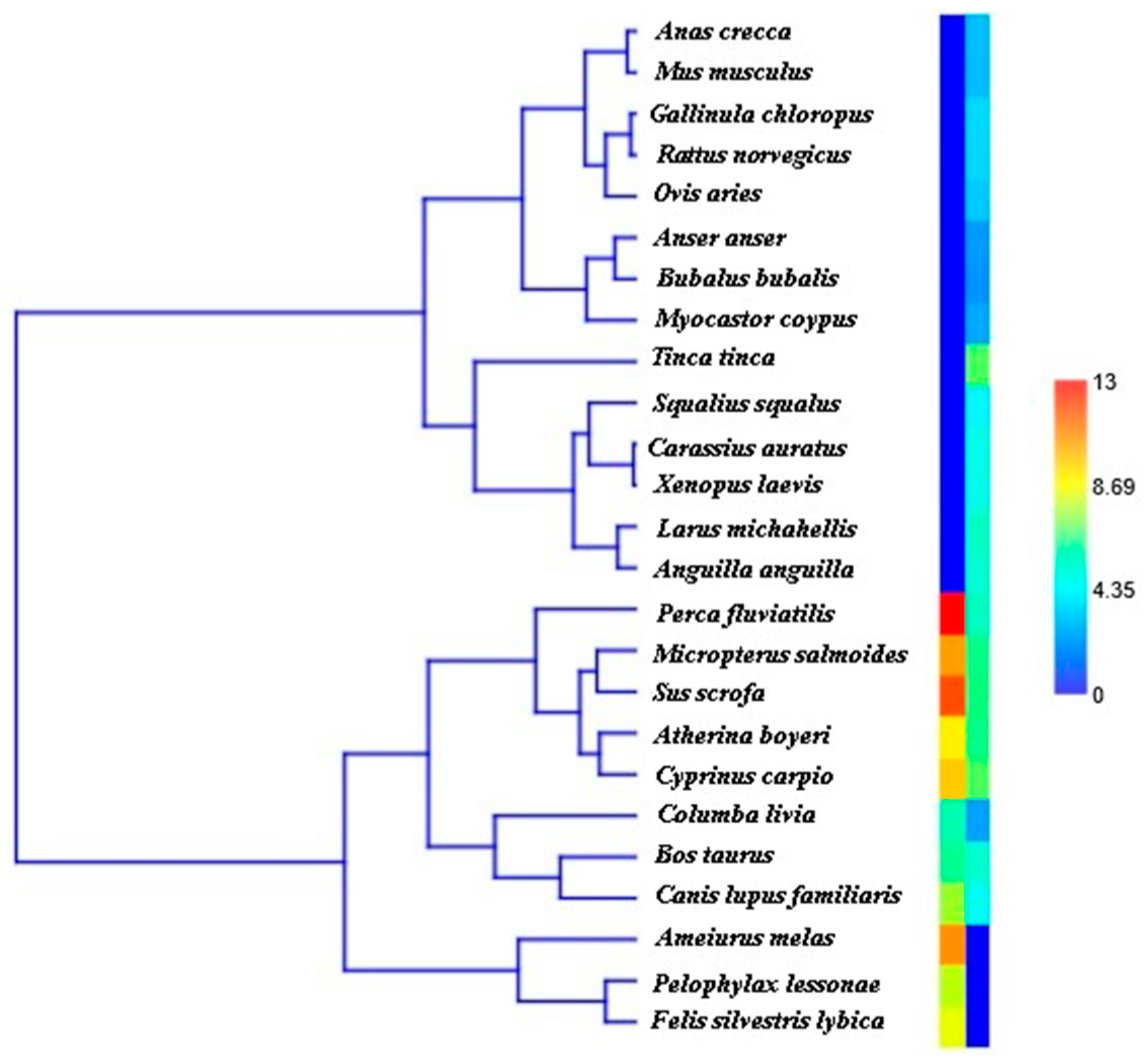
| Lake Rosamarina | Lake Garcia | |
|---|---|---|
| Taxa_S | 24 | 16 |
| Shannon_H | 3.148 | 2.419 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mauro, M.; Longo, F.; Vizzini, A.; Lo Valvo, M.; Radovic, S.; Orecchio, G.; De Luca, R.; Luparello, C.; Mauro, A.M.; Cuttitta, A.; et al. Environmental DNA as a Tool for the Preliminary Assessment of Vertebrate Biodiversity: A Case Study from Sicilian Freshwater Ecosystems. Biology 2025, 14, 1681. https://doi.org/10.3390/biology14121681
Mauro M, Longo F, Vizzini A, Lo Valvo M, Radovic S, Orecchio G, De Luca R, Luparello C, Mauro AM, Cuttitta A, et al. Environmental DNA as a Tool for the Preliminary Assessment of Vertebrate Biodiversity: A Case Study from Sicilian Freshwater Ecosystems. Biology. 2025; 14(12):1681. https://doi.org/10.3390/biology14121681
Chicago/Turabian StyleMauro, Manuela, Francesco Longo, Aiti Vizzini, Mario Lo Valvo, Slobodanka Radovic, Grazia Orecchio, Rosi De Luca, Claudio Luparello, Anna Maria Mauro, Angela Cuttitta, and et al. 2025. "Environmental DNA as a Tool for the Preliminary Assessment of Vertebrate Biodiversity: A Case Study from Sicilian Freshwater Ecosystems" Biology 14, no. 12: 1681. https://doi.org/10.3390/biology14121681
APA StyleMauro, M., Longo, F., Vizzini, A., Lo Valvo, M., Radovic, S., Orecchio, G., De Luca, R., Luparello, C., Mauro, A. M., Cuttitta, A., & Vazzana, M. (2025). Environmental DNA as a Tool for the Preliminary Assessment of Vertebrate Biodiversity: A Case Study from Sicilian Freshwater Ecosystems. Biology, 14(12), 1681. https://doi.org/10.3390/biology14121681










