Evaluation of Different Few-Shot Learning Methods in the Plant Disease Classification Domain
Simple Summary
Abstract
1. Introduction
2. Dataset
3. Methodology
x = x∗/||x∗||,
cos(θj,i) = WjTxi,
4. Results and Discussion
4.1. First Stage—Transfer Learning
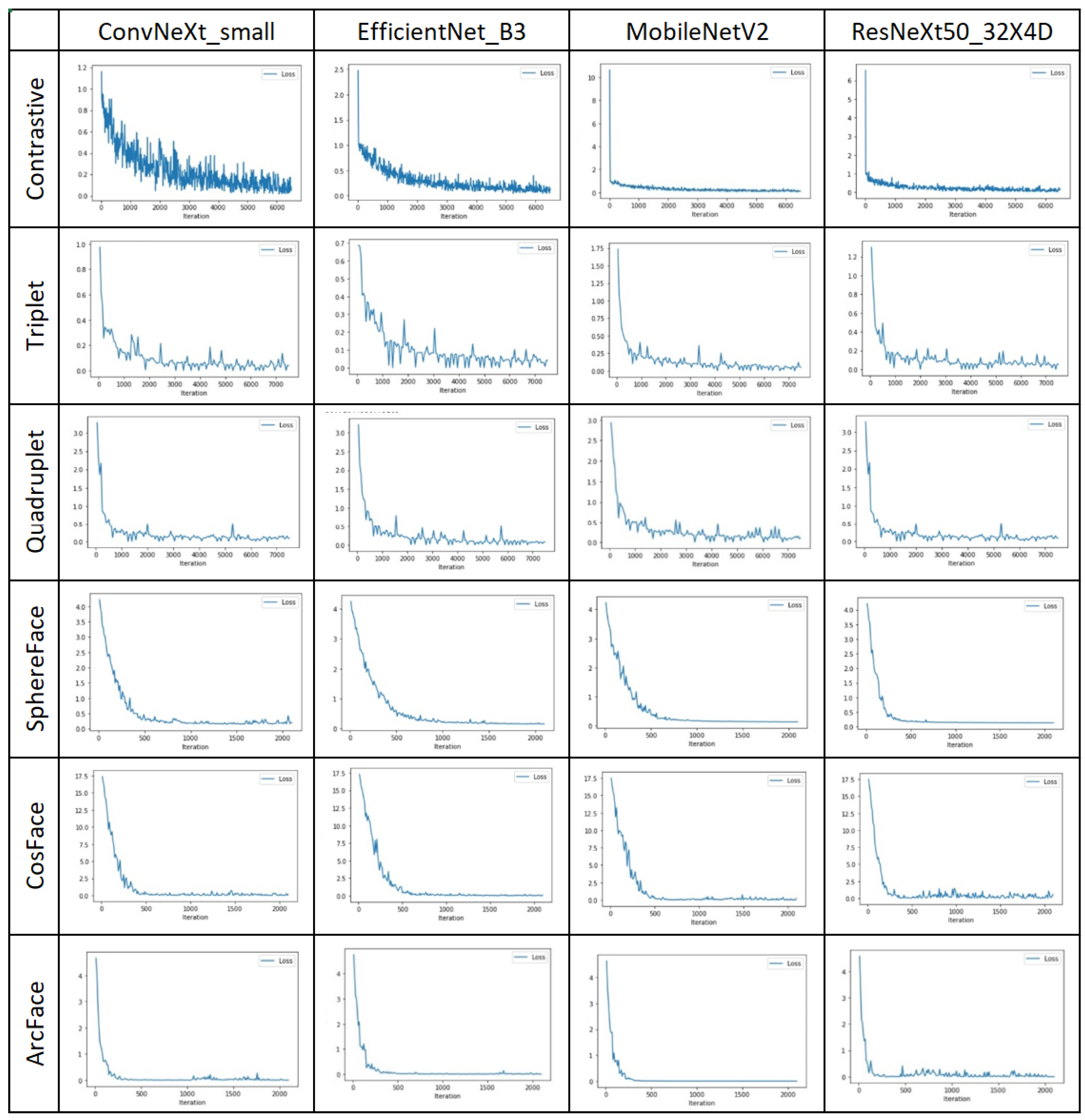
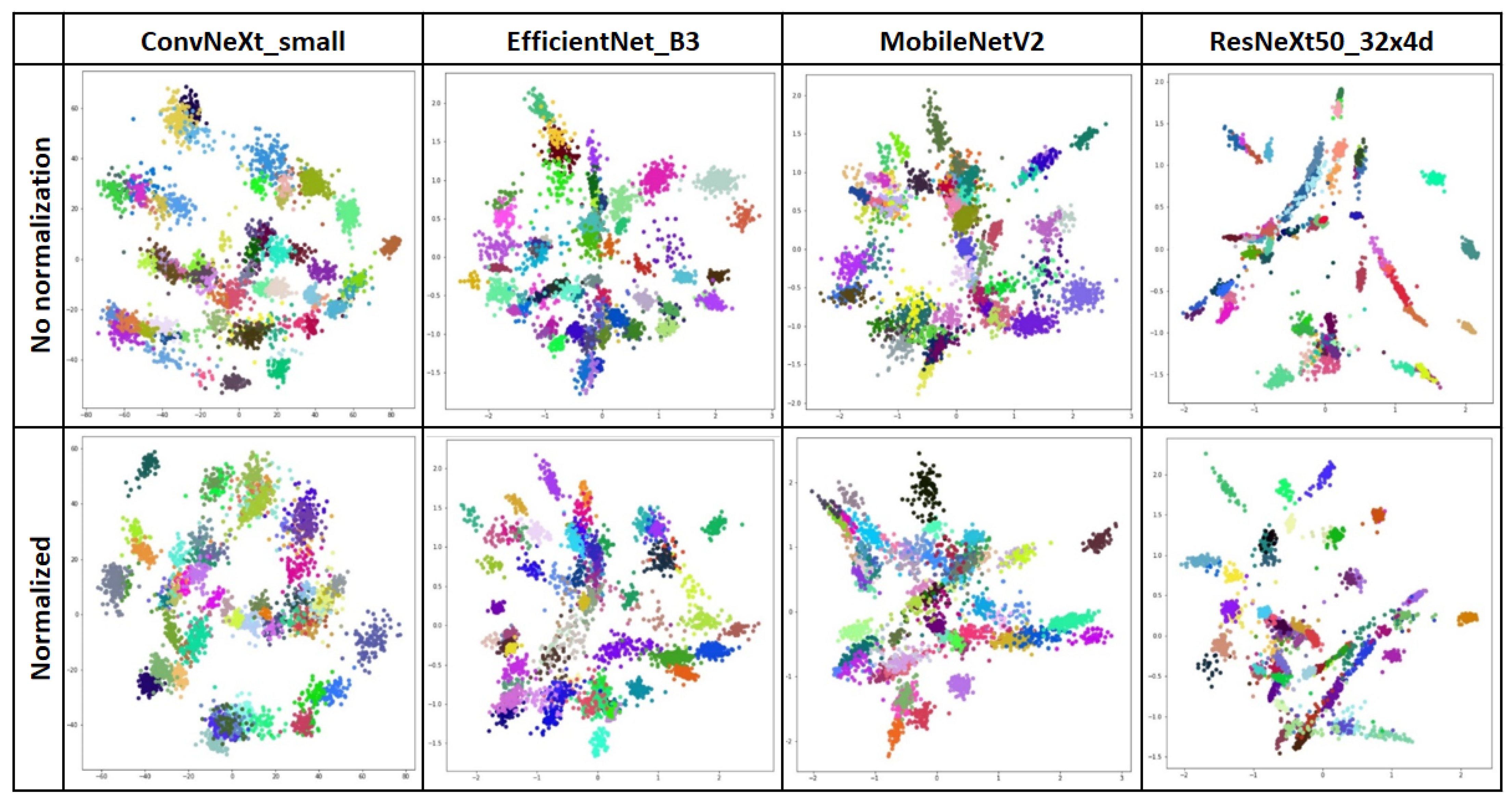
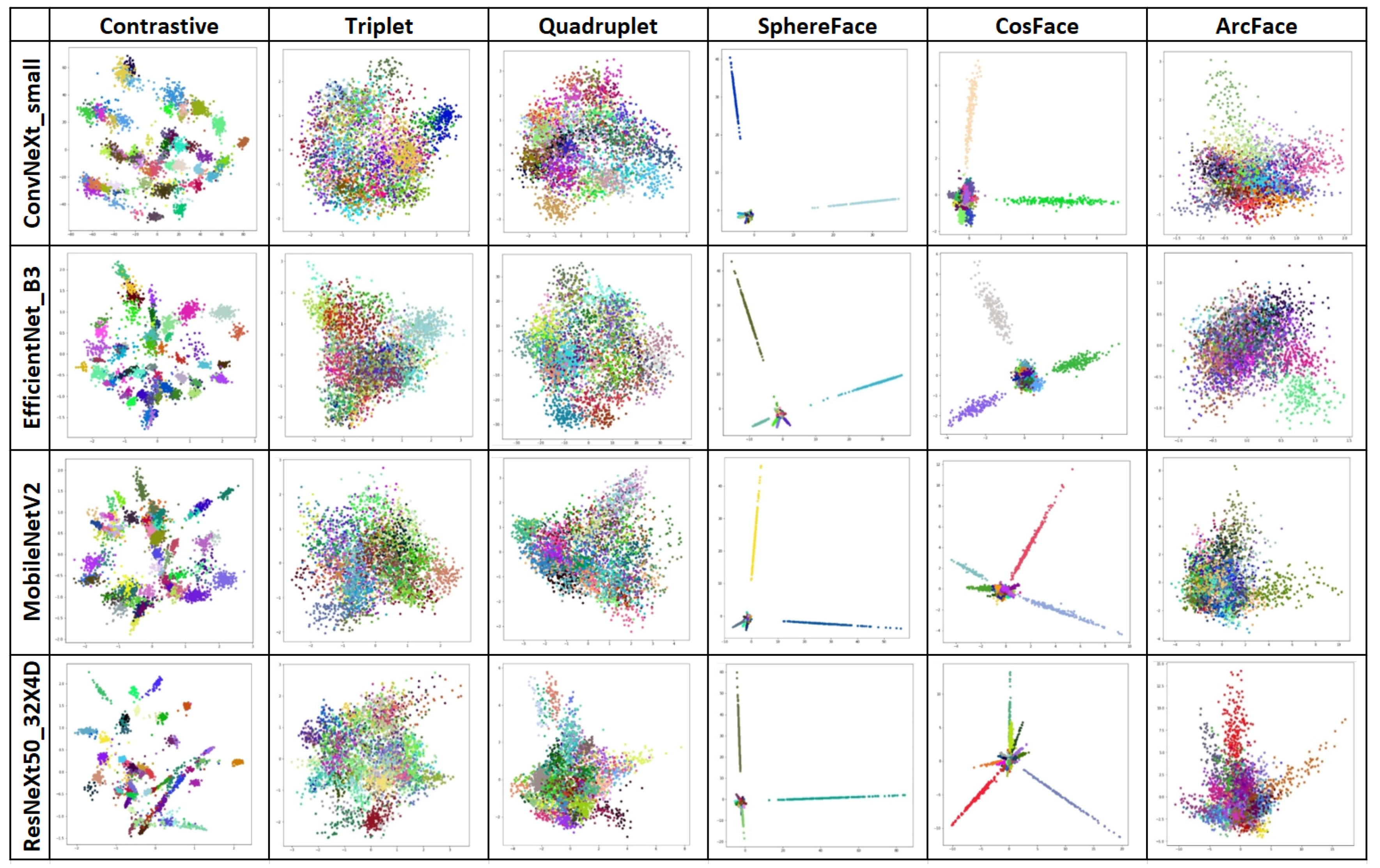
4.2. Second Stage—Similarity Learning
5. Conclusions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ramanjot; Mittal, U.; Wadhawan, A.; Singla, J.; Jhanjhi, N.Z.; Ghoniem, R.M.; Ray, S.K.; Abdelmaboud, A. Plant Disease Detection and Classification: A Systematic Literature Review. Sensors 2023, 23, 4769. [Google Scholar] [CrossRef] [PubMed]
- Shafik, W.; Tufail, A.; Namoun, A.; De Silva, L.C.; Apong, R.A.A.H.M. A systematic literature review on plant disease detection: Motivations, classification techniques, datasets, challenges, and future trends. IEEE Access 2023, 11, 59174–59203. [Google Scholar] [CrossRef]
- Demilie, W.B. Plant disease detection and classification techniques: A comparative study of the performances. J. Big Data 2024, 11, 5. [Google Scholar] [CrossRef]
- Muhammad, S.; Babar, S.; Shaker, E.; Akhtar, A.; Asad, U.; Fayadh, A.; Tsanko, G.; Tariq, H.; Farman, A. An advanced deep learning models-based plant disease detection: A review of recent research. Front. Plant Sci. 2023, 14, 1158933. [Google Scholar] [CrossRef]
- Porto, J.V.d.A.; Dorsa, A.C.; Weber, V.A.d.M.; Porto, K.R.d.A.; Pistori, H. Usage of few-shot learning and meta-learning in agriculture: A literature review. Smart Agric. Technol. 2023, 5, 100307. [Google Scholar] [CrossRef]
- Dyrmann, M.; Karstoft, H.; Midtiby, H.S. Plant species classification using deep convolutional neural network. Biosyst. Eng. 2016, 151, 72–80. [Google Scholar] [CrossRef]
- Chen, J.; Liu, Q.; Gao, L. Visual tea leaf disease recognition using a convolutional neural network model. Symmetry 2019, 11, 343. [Google Scholar] [CrossRef]
- Abasi, A.K.; Makhadmeh, S.N.; Alomari, O.A.; Tubishat, M.; Mohammed, H.J. Enhancing Rice Leaf Disease Classification: A Customized Convolutional Neural Network Approach. Sustainability 2023, 15, 15039. [Google Scholar] [CrossRef]
- Hughes, D.P.; Salathe, M. An Open Access Repository of Images on Plant Health to Enable the Development of Mobile Disease Diagnostics. arXiv 2015, arXiv:1511.08060. Available online: http://arxiv.org/abs/1511.08060 (accessed on 12 December 2024).
- Sladojevic, S.; Arsenovic, M.; Anderla, A.; Culibrk, D.; Stefanovic, D. Deep neural networks-based recognition of plant diseases by leaf image classification. Comput. Intell. Neurosci. 2016, 2016, 3289801. [Google Scholar] [CrossRef]
- Mohanty, S.P.; Hughes, D.P.; Salathé, M. Using deep learning for image-based plant disease detection. Front. Plant Sci. 2016, 7, 1419. [Google Scholar] [CrossRef] [PubMed]
- Ferentinos, K.P. Deep learning models for plant disease detection and diagnosis. Comput. Electron. Agric. 2018, 145, 311–318. [Google Scholar] [CrossRef]
- Picon, A.; Seitz, M.; Alvarez-Gila, A.; Mohnke, P.; Ortiz-Barredo, A.; Echazarra, J. Crop conditional convolutional neural networks for massive multi-crop plant disease classification over cell phone acquired images taken on real field conditions. Comput. Electron. Agric. 2019, 167, 105093. [Google Scholar] [CrossRef]
- Naik, B.N.; Ramanathan, M.; Palanisamy, P. Detection and classification of chilli leaf disease using a squeeze-and-excitation-based CNN model. Ecol. Inform. 2022, 69, 101663. [Google Scholar] [CrossRef]
- Lin, H.; Tse, R.; Tang, S.K.; Qiang, Z.P.; Pau, G. Few-shot learning approach with multi-scale feature fusion and attention for plant disease recognition. Front. Plant Sci. 2022, 13, 907916. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.; Tse, R.; Tang, S.K.; Qiang, Z.P.; Pau, G. The Positive Effect of Attention Module in Few-Shot Learning for Plant Disease Recognition. In Proceedings of the 5th International Conference on Pattern Recognition and Artificial Intelligence (PRAI), Chengdu, China, 19–21 August 2022; pp. 114–120. [Google Scholar]
- Nuthalapati, S.V.; Tunga, A. Multi-Domain Few-Shot Learning and Dataset for Agricultural Applications. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Montreal, QC, Canada, 11–17 October 2021; pp. 1399–1408. [Google Scholar]
- Wang, Y.; Yin, Y.; Li, Y.; Qu, T.; Guo, Z.; Peng, M.; Jia, S.; Wang, Q.; Zhang, W.; Li, F. Classification of Plant Leaf Disease Recognition Based on Self-Supervised Learning. Agronomy 2024, 14, 500. [Google Scholar] [CrossRef]
- Goodfellow, I.J.; Pouget-Abadie, J.; Mirza, M.; Xu, B.; Warde-Farley, D.; Ozair, S.; Courville, A.; Bengio, Y. Generative Adversarial Networks. arXiv 2014, arXiv:1406.2661. [Google Scholar] [CrossRef]
- Hu, G.; Wu, H.; Zhang, Y.; Wan, M. A low shot learning method for tea leaf’s disease identification. Comput. Electron. Agric. 2019, 163, 104852. [Google Scholar] [CrossRef]
- Nazki, H.; Yoon, S.; Fuentes, A.; Park, D.S. Unsupervised image translation using adversarial networks for improved plant disease recognition. Comput. Electron. Agric. 2020, 168, 105117. [Google Scholar] [CrossRef]
- Lu, Y.; Tao, X.; Zeng, N.; Du, J.; Shang, R. Enhanced CNN Classification Capability for Small Rice Disease Datasets Using Progressive WGAN-GP: Algorithms and Applications. Remote Sens. 2023, 15, 1789. [Google Scholar] [CrossRef]
- Alshammari, K.; Alshammari, R.; Alshammari, A.; TAlkhudaydi, T. An improved pear disease classification approach using cycle genera-tive adversarial network. Sci. Rep. 2024, 14, 6680. [Google Scholar] [CrossRef] [PubMed]
- Koch, G.; Zemel, R.; Salakhutdinov, R. Siamese neural networks for one-shot image recognition. In Proceedings of the ICML Deep Learning Workshop, Lille, France, 6–11 July 2015; Volume 2. [Google Scholar]
- Argüeso, D.; Picon, A.; Irusta, U.; Medela, A.; San-Emeterio, M.G.; Bereciartua, A.; Alvarez-Gila, A. Few-shot learning approach for plant disease classification using images taken in the field, Comput. Electron. Agric. 2020, 175, 105542. [Google Scholar] [CrossRef]
- Chopra, S.; Hadsell, R.; LeCun, Y. Learning a similarity metric discriminatively, with application to face verification. In Proceedings of the 2005 IEEE Computer Society Conference on Computer Vision and Pattern Recognition (CVPR’05), San Diego, CA, USA, 20–26 June 2005; Volume 1, pp. 539–546. [Google Scholar]
- Schroff, F.; Kalenichenko, D.; Philbin, J. FaceNet: A unified embedding for face recognition and clustering. In Proceedings of the 2015 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Boston, MA, USA, 7–12 June 2015; pp. 815–823. [Google Scholar] [CrossRef]
- Egusquiza, I.; Picon, A.; Irusta, U.; Bereciartua-Perez, A.; Eggers, T.; Klukas, C.; Aramendi, E.; Navarra-Mestre, R. Analysis of few-shot techniques for fungal plant disease classification and evaluation of clustering capabilities over real datasets. Front. Plant Sci. 2022, 13, 295. [Google Scholar] [CrossRef]
- Saad, M.H.; Salman, A.E. A plant disease classification using one-shot learning technique with field images. Multimed Tools Appl. 2023, 83, 58935–58960. [Google Scholar] [CrossRef]
- Tassis, L.M.; Krohling, R.A. Few-shot learning for biotic stress classification of coffee leaves. Artif. Intell. Agric. 2022, 6, 55–67. [Google Scholar] [CrossRef]
- Chen, W.; Chen, X.; Zhang, J.; Huang, K. Beyond Triplet Loss: A Deep Quadruplet Network for Person Re-identification. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017; pp. 1320–1329. [Google Scholar] [CrossRef]
- Liu, W.; Wen, Y.; Yu, Z.; Li, M.; Raj, S.; Song, L. SphereFace: Deep Hypersphere Embedding for Face Recognition. arXiv 2017, arXiv:1704.08063. Available online: https://arxiv.org/abs/1704.08063 (accessed on 12 December 2024).
- Deng, J.; Guo, J.; Yang, J.; Xue, N.; Kotsia, I.; Zafeiriou, S. ArcFace: Additive Angular Margin Loss for Deep Face Recognition. arXiv 2018, arXiv:1801.07698. Available online: https://arxiv.org/abs/1801.07698 (accessed on 12 December 2024).
- Pan, S.-Q.; Qiao, J.-F.; Wang, R.; Yu, H.-L.; Wang, C.; Taylor, K.; Pan, H.-Y. Intelligent diagnosis of northern corn leaf blight with deep learning model. J. Integr. Agricult. 2022, 21, 1094–1105. [Google Scholar] [CrossRef]
- Wang, H.; Wang, Y.; Zhou, Z.; Ji, X.; Gong, D.; Zhou, J.; Li, Z.; Liu, W. CosFace: Large Margin Cosine Loss for Deep Face Recognition. arXiv 2018, arXiv:1801.09414. Available online: https://arxiv.org/abs/1801.09414 (accessed on 12 December 2024).
- Dai, Q.; Guo, Y.; Li, Z.; Song, S.; Lyu, S.; Sun, D.; Wang, Y.; Chen, Z. Citrus Disease Image Generation and Classification Based on Improved FastGAN and EfficientNet-B5. Agronomy 2023, 13, 988. [Google Scholar] [CrossRef]
- Rezaei, M.; Diepeveen, D.; Laga, H.; Jones, M.G.K.; Sohel, F. Plant disease recognition in a low data scenario using few-shot learning. Comput. Electron. Agric. 2024, 219, 108812. [Google Scholar] [CrossRef]
- Jiawei, M.; Feng, Q.; Yang, J.; Zhang, J.; Yang, S. Few-shot disease recognition algorithm based on supervised contrastive learning. Front. Plant Sci. 2024, 15, 1341831. [Google Scholar] [CrossRef]
- Yan, W.; Feng, Q.; Yang, S.; Zhang, J.; Yang, W. HMFN-FSL: Heterogeneous Metric Fusion Network-Based Few-Shot Learning for Crop Disease Recognition. Agronomy 2023, 13, 2876. [Google Scholar] [CrossRef]
- Ososkov, G.; Nechaevskiy, A.; Uzhinskiy, A.; Goncharov, P. Architecture and basic principles of the multifunctional platform for plant disease detection CEUR Workshop Proceedings. In Proceedings of the Selected Papers of the 8th International Conference “Distributed Computing and Grid-Technologies in Science and Education, Dubna, Russia, 10–14 September 2018; pp. 200–206. [Google Scholar]
- Sandler, M.; Howard, A.; Zhu, Z.; Zhmoginov, A.; Chen, L.-C. MobileNetV2: Inverted Residuals and Linear Bottlenecks. arXiv 2018, arXiv:1801.04381. Available online: https://arxiv.org/abs/1801.04381 (accessed on 12 December 2024).
- Goncharov, P.; Nestsiarenia, I.; Ososkov, G.; Nechaevskiy, A.; Uzhinskiy, A. Disease detection on the plant leaves by deep learning. In Advances in Neural Computation, Machine Learning, and Cognitive Research II; Springer: Berlin/Heidelberg, Germany, 2018; pp. 151–159. [Google Scholar] [CrossRef]
- Uzhinskiy, A.; Ososkov, G.; Goncharov, P.; Nechaevskiy, A.; Smetanin, A. Oneshot learning with triplet loss for vegetation classification tasks. Comput. Opt. 2021, 45, 608–614. [Google Scholar] [CrossRef]
- Tan, M.; Le, Q.V. EfficientNet: Rethinking Model Scaling for Convolutional Neural Networks. arXiv 2019, arXiv:1905.11946. Available online: https://arxiv.org/abs/1905.11946 (accessed on 12 December 2024).
- Tan, M.; Chen, B.; Pang, R.; Vasudevan, V.; Sandler, M.; Howard, A.; Le, Q.V. MnasNet: Platform-Aware Neural Architecture Search for Mobile. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Long Beach, CA, USA, 15–20 June 2019. [Google Scholar]
- Radosavovic, I.; Kosaraju, R.P.; Girshick, R.; He, K.; Dollár, P. Designing Network Design Spaces. arXiv 2020, arXiv:2003.13678. Available online: https://arxiv.org/abs/2003.13678 (accessed on 12 December 2024).
- Liu, Z.; Mao, H.; Wu, C.-Y.; Feichtenhofer, C.; Darrell, T.; Xie, S. A ConvNet for the 2020s. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), New Orleans, LA, USA, 18–24 June 2022. [Google Scholar]
- Szegedy, C.; Vanhoucke, V.; Ioffe, S.; Shlens, J.; Wojna, Z. Rethinking the Inception Architecture for Computer Vision. arXiv 2015, arXiv:1512.00567. Available online: https://arxiv.org/abs/1512.00567 (accessed on 12 December 2024).
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. arXiv 2015, arXiv:1512.03385. Available online: https://arxiv.org/abs/1512.03385 (accessed on 12 December 2024).
- Xie, S.; Girshick, R.; Dollár, P.; Tu, Z.; He, K. Aggregated Residual Transformations for Deep Neural Networks. arXiv 2016, arXiv:1611.05431. Available online: https://arxiv.org/abs/1611.05431 (accessed on 12 December 2024).
- Deng, J.; Dong, W.; Socher, R.; Li, L.J.; Li, K.; Fei-Fei, L. ImageNet: A large-scale hierarchical image database. In Proceedings of the 2009 IEEE Conference on Computer Vision and Pattern Recognition, Miami, FL, USA, 20–25 June 2009; pp. 248–255. [Google Scholar] [CrossRef]
- Howard, A.; Sandler, M.; Chu, G.; Chen, L.-C.; Chen, B.; Tan, M.; Wang, W.; Zhu, Y.; Pang, R.; Vasudevan, V.; et al. Searching for MobileNetV3. arXiv 2019, arXiv:1905.02244. Available online: https://arxiv.org/abs/1905.02244 (accessed on 12 December 2024).
- Zuev, M.; Butenko, Y.; Ćosić, M.; Nechaevskiy, A.; Podgainy, D.; Rahmonov, I.; Stadnik, A.; Streltsova, O. ML/DL/HPC Ecosystem of the HybriLIT Heterogeneous Platform (MLIT JINR): New Opportunities for Applied Research. In Proceedings of the 6th International Workshop on Deep Learning in Computational Physics (DLCP2022), Dubna, Russia, 6–8 July 2022. [Google Scholar]
- Maaten, L.J.P.; Hinton, G.E. Visualizing High-Dimensional Data Using t-SNE. J. Mach. Learn. Res. 2008, 9, 2579–2605. [Google Scholar]




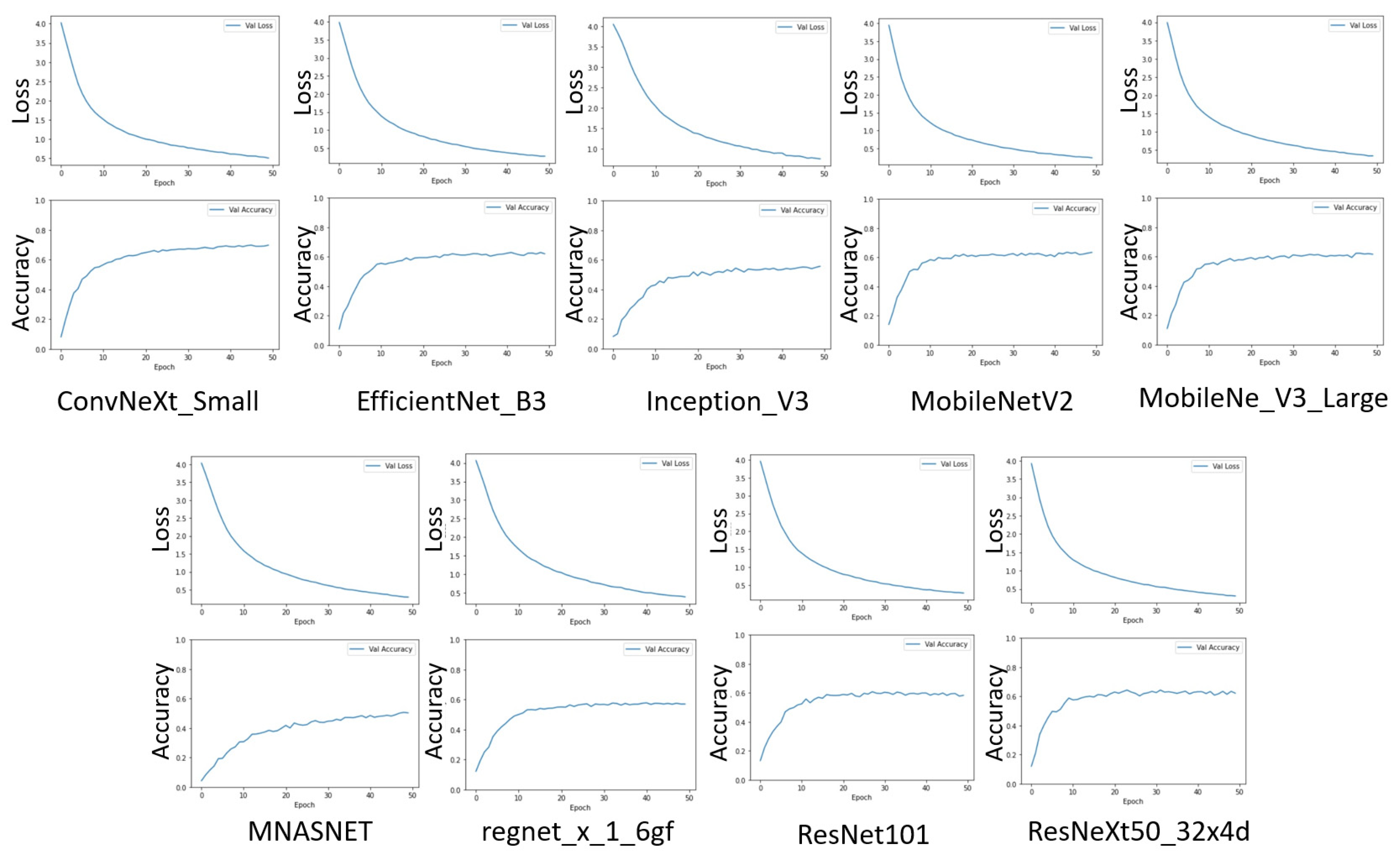
| Acc@1 (on ImageNet-1K) (%) | Model File Size (Mb) | Num. of Parameters | |
|---|---|---|---|
| ConvNeXt_Small [47] | 82.52 | 191.7 | 50,223,688 |
| EfficientNet_B3 [44] | 82.008 | 47.2 | 12,233,232 |
| Inception_V3 [48] | 77.294 | 103.9 | 27,161,264 |
| MNASNet1_3 [45] | 76.506 | 24.2 | 6,282,256 |
| MobileNetV2 [41] | 71.878 | 13.6 | 3,504,872 |
| MobileNet_V3_Large [52] | 74.042 | 21.1 | 5,483,032 |
| RegNet_X_1_6GF [46] | 77.04 | 35.3 | 9,190,136 |
| ResNet101 [49] | 77.374 | 170.5 | 44,549,160 |
| ResNeXt50_32X4D [50] | 77.618 | 95.8 | 25,028,904 |
| Accuracy (%) | MEPT (Sec.) | Wa Precision | Wa Recall | Wa F1 Score | |
|---|---|---|---|---|---|
| ConvNeXt_small | 68.41 | 17.84 | 0.71 | 0.70 | 0.69 |
| EfficientNet_B3 | 62.87 | 5.36 | 0.64 | 0.62 | 0.61 |
| Inception_V3 | 55.68 | 6.86 | 0.58 | 0.56 | 0.54 |
| MNASNet1_3 | 54.09 | 3.25 | 0.56 | 0.54 | 0.53 |
| MobileNetV2 | 62.79 | 2.81 | 0.64 | 0.63 | 0.62 |
| MobileNet_V3_Large | 61.87 | 2.48 | 0.62 | 0.62 | 0.61 |
| RegNet_X_1_6GF | 62.25 | 4.78 | 0.63 | 0.61 | 0.61 |
| ResNet101 | 62.62 | 9.02 | 0.65 | 0.64 | 0.62 |
| ResNeXt50_32X4D | 62.73 | 8.02 | 0.66 | 0.64 | 0.63 |
| Accuracy (%) | MEPT (Sec.) | Wa Precision | Wa Recall | Wa F1 Score | |
|---|---|---|---|---|---|
| ConvNeXt_small (CL) | 87.89 | 150.51 | 0.87 | 0.86 | 0.85 |
| ConvNeXt_small (CL, norm) | 89.53 | 148.43 | 0.89 | 0.90 | 0.89 |
| ConvNeXt_small (TL) | 92.88 | 213.62 | 0.94 | 0.94 | 0.93 |
| ConvNeXt_small (TL, norm) | 92.56 | 214.31 | 0.93 | 0.93 | 0.93 |
| ConvNeXt_small (QL) | 96.12 | 278.98 | 0.97 | 0.97 | 0.96 |
| ConvNeXt_small (QL, norm) | 96.85 | 279.41 | 0.97 | 0.97 | 0.97 |
| EfficientNet_B3 (CL) | 94.62 | 45.54 | 0.94 | 0.94 | 0.94 |
| EfficientNet_B3 (CL, norm) | 95.15 | 45.52 | 0.96 | 0.95 | 0.95 |
| EfficientNet_B3 (TL) | 95.73 | 67.13 | 0.96 | 0.96 | 0.96 |
| EfficientNet_B3 (TL, norm) | 95.13 | 67.56 | 0.95 | 0.95 | 0.95 |
| EfficientNet_B3 (QL) | 97.33 | 86.85 | 0.98 | 0.98 | 0.98 |
| EfficientNet_B3 (QL, norm) | 96.98 | 86.87 | 0.97 | 0.97 | 0.97 |
| MobileNetV2 (CL) | 90.94 | 22.42 | 0.92 | 0.91 | 0.90 |
| MobileNetV2 (CL, norm) | 92.23 | 22.37 | 0.93 | 0.92 | 0.92 |
| MobileNetV2 (TL) | 90.21 | 50.79 | 0.91 | 0.90 | 0.89 |
| MobileNetV2 (TL, norm) | 90.89 | 52.93 | 0.92 | 0.92 | 0.92 |
| MobileNetV2 (QL) | 92.87 | 68.91 | 0.94 | 0.93 | 0.93 |
| MobileNetV2 (Ql, norm) | 93.79 | 68.65 | 0.95 | 0.94 | 0.94 |
| ResNeXt50_32X4D (CL) | 88.67 | 83.34 | 0.89 | 0.88 | 0.88 |
| ResNeXt50_32X4D (CL, norm) | 89.56 | 82.63 | 0.89 | 0.89 | 0.88 |
| ResNeXt50_32X4D (TL) | 91.63 | 57.34 | 0.93 | 0.92 | 0.91 |
| ResNeXt50_32X4D (TL, norm) | 91.92 | 56.63 | 0.93 | 0.93 | 0.92 |
| ResNeXt50_32X4D (QL) | 92.51 | 107.34 | 0.94 | 0.94 | 0.94 |
| ResNeXt50_32X4D (QL, norm) | 93.79 | 107.16 | 0.95 | 0.94 | 0.94 |
| Accuracy (%) | MEPT (Sec.) | Wa Precision | Wa Recall | Wa F1 Score | |
|---|---|---|---|---|---|
| ConvNeXt_small (SF) | 100.0 | 86.05 | 1.00 | 1.00 | 1.00 |
| ConvNeXt_small (SF, norm) | 99.87 | 86.17 | 1.00 | 1.00 | 1.00 |
| ConvNeXt_small (CF) | 100.0 | 86.13 | 1.00 | 1.00 | 1.00 |
| ConvNeXt_small (CF, norm) | 100.0 | 86.19 | 1.00 | 1.00 | 1.00 |
| ConvNeXt_small (AF) | 100.0 | 86.13 | 1.00 | 1.00 | 1.00 |
| ConvNeXt_small (AF, norm) | 100.0 | 86.19 | 1.00 | 1.00 | 1.00 |
| EfficientNet_B3 (SF) | 99.95 | 26.03 | 1.00 | 1.00 | 1.00 |
| EfficientNet_B3 (SF, norm) | 100.0 | 26.00 | 1.00 | 1.00 | 1.00 |
| EfficientNet_B3 (CF) | 99.95 | 26.10 | 1.00 | 1.00 | 1.00 |
| EfficientNet_B3 (CF, norm) | 99.95 | 26.01 | 1.00 | 1.00 | 1.00 |
| EfficientNet_B3 (AF) | 97.87 | 26.10 | 0.98 | 0.98 | 0.98 |
| EfficientNet_B3 (AF, norm) | 98.22 | 26.01 | 0.98 | 0.98 | 0.98 |
| MobileNetV2 (SF) | 99.95 | 20.42 | 1.00 | 1.00 | 1.00 |
| MobileNetV2 (SF, norm) | 99.95 | 20.73 | 1.00 | 1.00 | 1.00 |
| MobileNetV2 (CF) | 99.87 | 19.42 | 1.00 | 1.00 | 1.00 |
| MobileNetV2 (CF, norm) | 99.95 | 19.73 | 1.00 | 1.00 | 1.00 |
| MobileNetV2 (AF) | 98.42 | 19.62 | 0.99 | 0.99 | 0.99 |
| MobileNetV2 (AF, norm) | 98.49 | 19.68 | 0.99 | 0.99 | 0.99 |
| ResNeXt50_32X4D (SF) | 100.0 | 31.19 | 1.00 | 1.00 | 1.00 |
| ResNeXt50_32X4D (SF, norm) | 100.0 | 31.12 | 1.00 | 1.00 | 1.00 |
| ResNeXt50_32X4D (CF) | 99.91 | 31.22 | 1.00 | 1.00 | 1.00 |
| ResNeXt50_32X4D (CF, norm) | 99.95 | 31.19 | 1.00 | 1.00 | 1.00 |
| ResNeXt50_32X4D (AF) | 99.91 | 31.28 | 1.00 | 1.00 | 1.00 |
| ResNeXt50_32X4D (AF, norm) | 100.0 | 31.26 | 1.00 | 1.00 | 1.00 |
| ConvNeXt_small | EfficientNet_B3 | MobileNetV2 | ResNeXt50_32X4D | |||||
|---|---|---|---|---|---|---|---|---|
| Accuracy | MEPT | Accuracy | MEPT | Accuracy | MEPT | Accuracy | MEPT | |
| Contrastive | 87.89 | 150.51 | 88.80 | 45.54 | 89.51 | 21.32 | 88.80 | 57.34 |
| Triplet | 92.88 | 213.62 | 95.33 | 67.13 | 90.21 | 50.79 | 88.67 | 83.34 |
| Quadruplet | 96.12 | 279.41 | 97.33 | 86.85 | 92.87 | 68.91 | 92.51 | 107.34 |
| SphereFace | 99.95 | 86.31 | 99.95 | 26.03 | 99.95 | 19.41 | 100 | 31.19 |
| CosFace | 100.0 | 86.19 | 99.95 | 26.10 | 99.87 | 18.40 | 99.91 | 31.22 |
| ArcFace | 100.0 | 86.26 | 97.87 | 27.02 | 98.45 | 18.51 | 99.91 | 31.28 |
| ConvNeXt_small (191.7 Mb) | EfficientNet_B3 (47.2 Mb) | MobileNetV2 (13.6 Mb) | ResNeXt50_32X4D (95.8 Mb) | |||||
|---|---|---|---|---|---|---|---|---|
| Vanilla (%) | Norm (%) | Vanilla (%) | Norm (%) | Vanilla (%) | Norm (%) | Vanilla (%) | Norm (%) | |
| Contrastive | 44 | 46.4 | 43.4 | 46.2 | 38.4 | 40 | 37.6 | 43 |
| Triplet | 58.2 | 58.6 | 54.4 | 53.4 | 39.4 | 40.2 | 47.4 | 51.8 |
| Quadruplet | 58.6 | 62.4 | 55 | 53.4 | 40.6 | 40.6 | 50.4 | 51.6 |
| SphereFace | 72.2 | 75.8 | 57.6 | 62 | 45.8 | 43.4 | 53.4 | 55.2 |
| CosFace | 71.2 | 73.6 | 56.8 | 59.8 | 43.6 | 43.8 | 52.6 | 49.6 |
| ArcFace | 70 | 67.8 | 55 | 57.2 | 40.6 | 41.2 | 51.6 | 49.6 |
| Vainilla TL | 43.2 | 37 | 32 | 38.6 | ||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Uzhinskiy, A. Evaluation of Different Few-Shot Learning Methods in the Plant Disease Classification Domain. Biology 2025, 14, 99. https://doi.org/10.3390/biology14010099
Uzhinskiy A. Evaluation of Different Few-Shot Learning Methods in the Plant Disease Classification Domain. Biology. 2025; 14(1):99. https://doi.org/10.3390/biology14010099
Chicago/Turabian StyleUzhinskiy, Alexander. 2025. "Evaluation of Different Few-Shot Learning Methods in the Plant Disease Classification Domain" Biology 14, no. 1: 99. https://doi.org/10.3390/biology14010099
APA StyleUzhinskiy, A. (2025). Evaluation of Different Few-Shot Learning Methods in the Plant Disease Classification Domain. Biology, 14(1), 99. https://doi.org/10.3390/biology14010099






