Transcriptomic Analysis Provides New Insights into the Tolerance Mechanisms of Green Macroalgae Ulva prolifera to High Temperature and Light Stress
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Algal Strain Collection and Culture Conditions
2.2. Temperature Treatment and Tissue Sampling
2.3. RNA Extraction
2.4. Transcriptome Sequencing and Quality Control
2.5. Assembly and Annotation
2.6. Differential Expression Analysis
2.7. Validation of the Genes
3. Results
3.1. Transcriptome Analysis
3.2. Analysis of DEGs and Identification of Temperature-Responsive Genes
3.3. Functional Classification
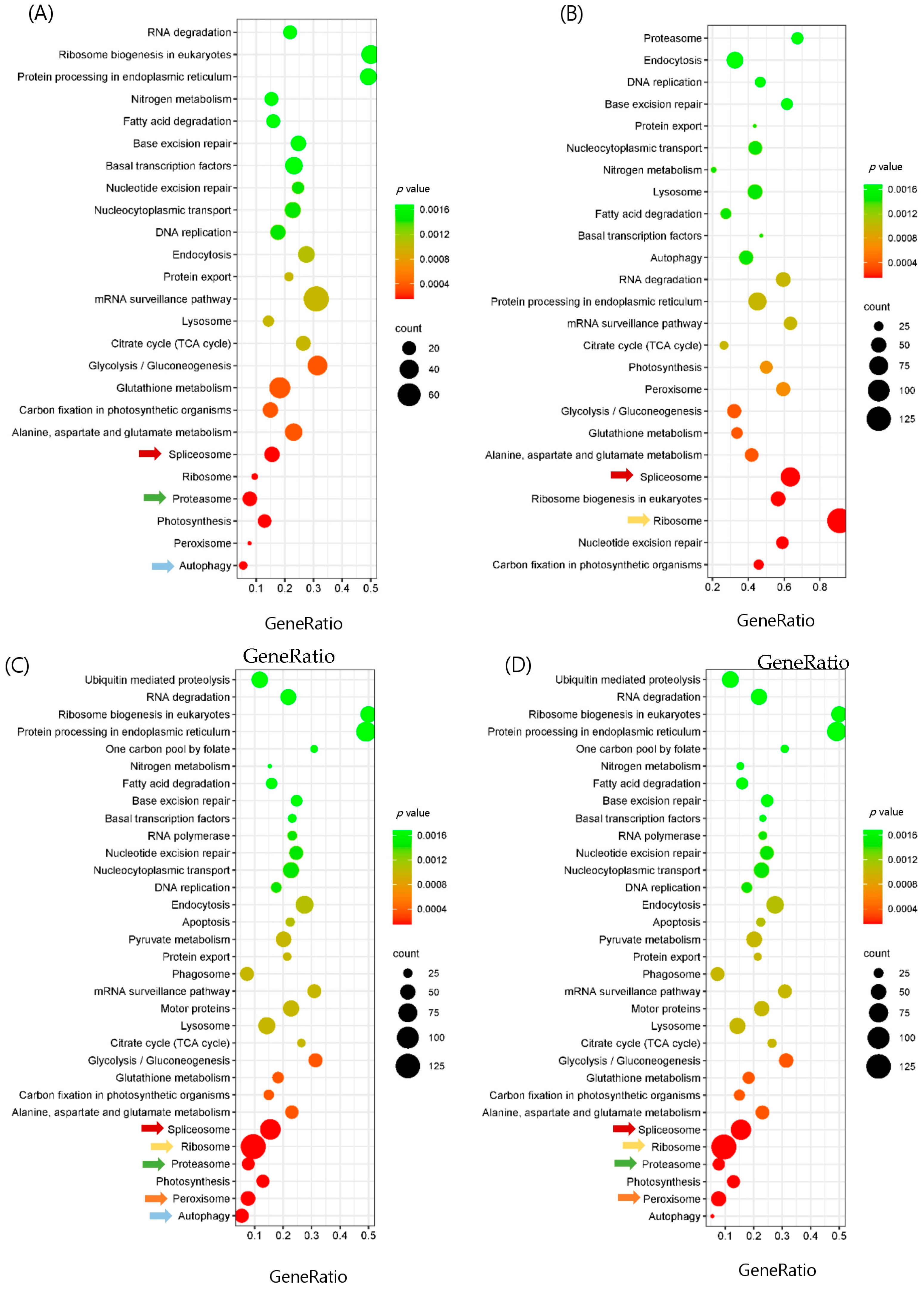
3.4. Enrichment and Pathway Analysis and Comparison of the Group
3.5. Analysis of Major Pathway-Related Differentially Express Genes (DEGs) in Different Comparison Groups
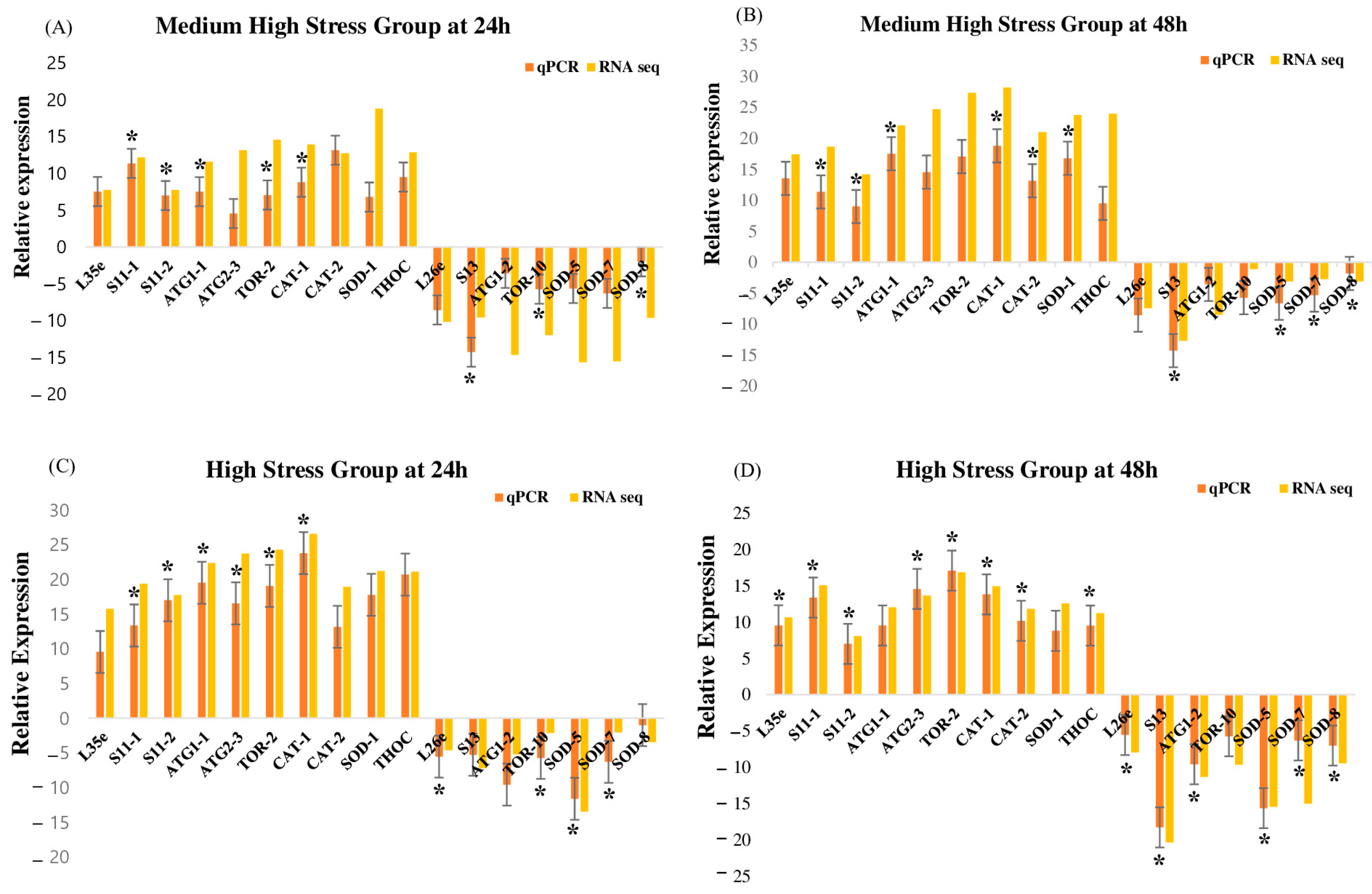
3.6. Validation of RNA-seq Results with RT-qPCR
4. Discussion
4.1. Ribosome Metabolism
4.2. Spliceosome Pathway
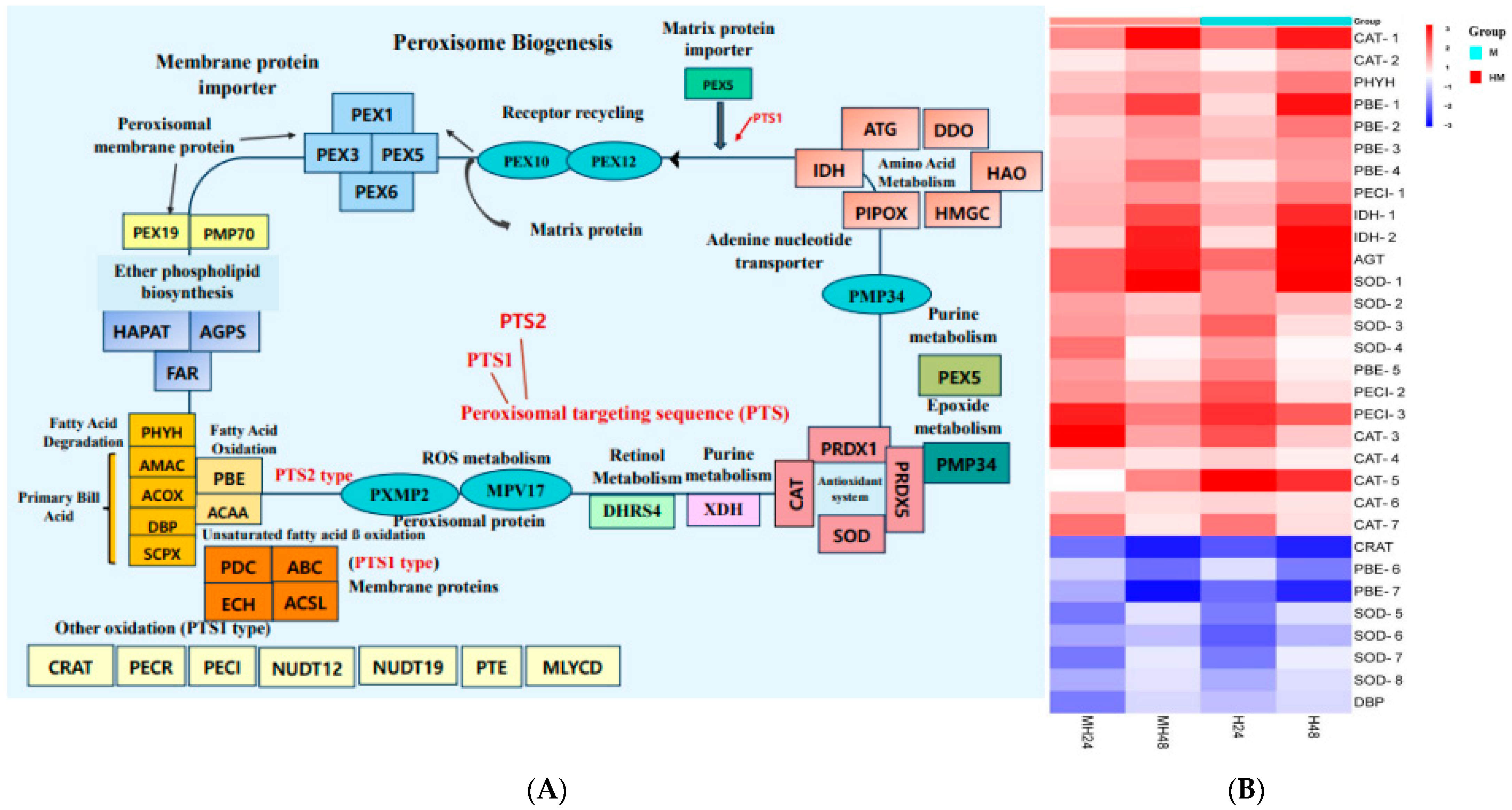
4.3. Peroxisome Metabolism
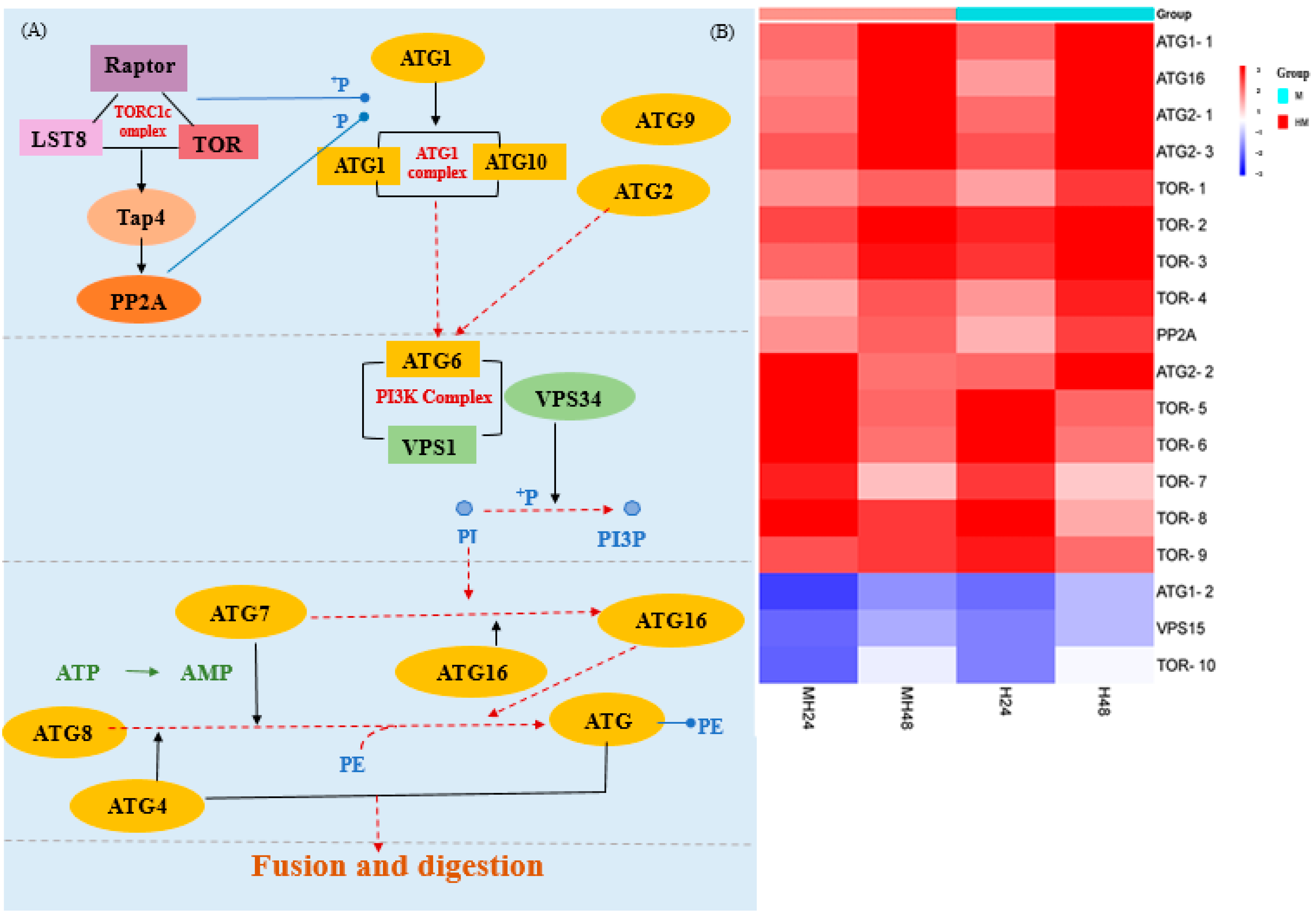
4.4. Autophagy Pathway
4.5. Energy Metabolism
4.6. Glutathione Pathway
4.7. Carbon Fixation in Photosynthetic Organisms
4.8. Alanine, Aspartate, and Glutamate Metabolism
4.9. Differentially Expressed Genes
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Müller, O.F. Flora Danica; Havniae: Copenhagen, Denmark, 1778; Volume 5, p. 8. [Google Scholar]
- Fan, M.H.; Sun, X.; Liao, Z.; Wang, J.X.; Li, Y.H.; Xu, N.J. Comparative proteomic analysis of Ulva prolifera response to high temperature stress. Proteome Sci. 2018, 16, 17–26. [Google Scholar] [CrossRef] [PubMed]
- Xue, M.; Wu, M.; Zheng, L.; Liu, J.; Liu, L.; Zhu, S.; Liu, S.; Liu, L. Multi-Factors Synthetically Contribute to Ulva prolifera Outbreaks in the South Yellow Sea of China. Remote Sens. 2023, 15, 5151. [Google Scholar] [CrossRef]
- Liu, X.Q.; Wang, Z.L.; Zhang, X.L. A review of the green tides in the Yellow Sea, China. Mar. Environ. Res. 2016, 119, 189–196. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.L.; Xiao, J.; Fan, S.; Li, Y.; Liu, X.; Liu, D. Who made the world’s largest green tide in China? An integrated study on the initiation and early development of the green tide in Yellow Sea. Limnol. Oceanogr. 2015, 60, 1105–1117. [Google Scholar] [CrossRef]
- Xu, Z.G.; Wu, H.Y.; Zhan, D.M.; Sun, F.; Sun, J.Z.; Wang, G. Combined effects of light intensity and NH4+-enrichment on growth, pigmentation, and photosynthetic performance of Ulva prolifera (Chlorophyta). Chin. J. Ocean Limnol. 2014, 32, 1016–1023. [Google Scholar] [CrossRef]
- Taylor, R.; Fletcher, R.L.; Raven, J.A. Preliminary studies on the growth of selected green tide algae in laboratory culture: Effects of irradiance, temperature, salinity and nutrients on growth rate. Bot. Mar. 2005, 44, 327–336. [Google Scholar] [CrossRef]
- Largo, D.B.; Sembrano, J.; Hiraoka, M.; Ohno, M. Taxonomic and ecological profile of ‘green tide’ species of Ulva (Ulvales, Chlorophyta) in Central Philippines. Hydrobiologia 2004, 512, 247–253. [Google Scholar] [CrossRef]
- He, Y.L.; Hu, C.Y.; Wang, Y.H.; Cui, D.D.; Sun, X.; Li, Y.H.; Xu, N.J. The metabolic survival strategy of marine macroalga Ulva prolifera under temperature stress. J. Appl. Phycol. 2018, 30, 3611–3621. [Google Scholar] [CrossRef]
- Bao, M.; Park, J.S.; Xing, Q.; He, P.; Zhang, J.; Yarish, C.; Yoo, H.I.; Kim, J.K. Comparative Analysis of Physiological Responses in Two Ulva prolifera Strains Revealed the Effect of Eutrophication on High Temperature and Copper Stress Tolerance. Front. Mar. Sci. 2022, 9, 863918. [Google Scholar] [CrossRef]
- Bao, M.; Xing, Q.; Park, J.S.; He, P.; Zhang, J.; Yarish, C.; Kim, J.K. Temperature and high nutrients enhance hypo-salinity tolerance of the bloom forming green alga, Ulva prolifera. Harmful Algae 2023, 123, 102402. [Google Scholar] [CrossRef]
- Zhang, J.; Kim, J.K.; Yarish, C.; He, P. The expansion of Ulva prolifera O.F. Müller macroalgal blooms in the Yellow Sea, PR China, through asexual reproduction. Mar. Pollut. Bull. 2016, 104, 101–106. [Google Scholar] [PubMed]
- Zhang, Y.; He, P.; Li, H.; Li, G.; Liu, J.; Jiao, F.; Zhang, J.; Huo, Y.; Shi, X.; Su, R.; et al. Ulva prolifera green-tide outbreaks and their environmental impact in the Yellow Sea, China. Natl. Sci. Rev. 2019, 6, 825–838. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Yan, B.; Lin, A.; Hu, J.; Shen, S. Ecological factor research on the growth and induction of spores release in Enteromorpha prolifera (Chlorophyta). Mar. Sci. Bull. 2007, 26, 60–65. [Google Scholar]
- Gu, K.; Liu, Y.; Jiang, T.; Cai, C.; Zhao, H.; Liu, X.; He, P. Molecular Response of Ulva prolifera to Short-Term High Light Stress Revealed by a Multi-Omics Approach. Biology 2022, 11, 1563. [Google Scholar] [CrossRef]
- Chen, Y.; Zheng, M.; Jiang, J.; Hu, W.; Xu, N.; Li, Y. Enhancement of growth in Ulva prolifera by diurnal temperature difference combined with nitrogen enrichment. Mar. Environ. Res. 2023, 186, 105905. [Google Scholar] [CrossRef]
- Xu, L.; Luo, L.; Zuo, X.; Cao, C.; Lin, L.; Zheng, H.; Ma, Z.; Chen, B.; Wu, M. Effects of temperature and irradiance on the regeneration of juveniles from the holdfasts of Sargassum fusiforme, a commercial seaweed. Aquaculture 2022, 557, 738317. [Google Scholar] [CrossRef]
- Meihua, F.; Xiaowen, T.; Zongxin, Y.; Jianxin, W.; Xiaolin, Z.; Xiaojun, Y.; Peng, L.; Nianjun, X.; Zhi, L. Integration of the transcriptome and proteome provides insights into the mechanism calcium regulated of Ulva prolifera in response to high-temperature stress. Aquaculture 2022, 557, 738344. [Google Scholar]
- He, H.; Yang, J.; He, Y.; Li, Z.; Fu, C.; Zhang, D.; Li, M.; Lu, A.; Dong, J.; Liu, J.; et al. Proliferating cell nuclear antigen involves in temperature stress tolerance of Ulva prolifera. bioRxiv 2023. [Google Scholar] [CrossRef]
- Yang, J.J.; Yu, D.C.; Ma, Y.F.; Yin, Y.; Shen, S.D. Antioxidative defense response of Ulva prolifera under high or low-temperature stimulus. Algal Res. 2019, 44, 101703. [Google Scholar] [CrossRef]
- Chandhini, S.; Kumar, V.J.R. Transcriptomics in aquaculture: Current status and applications. Aquaculture 2019, 11, 1379–1397. [Google Scholar] [CrossRef]
- Li, R.; Xu, Y.; Wu, F.; Peng, Z.; Chan, J.; Zhang, L. Easy-to-Use CRISPER-Cas9 Genome Editing in the Cultured Pacific Abalone (Haliotis discus hannai). CRISPR J. 2024, 7, 41–52. [Google Scholar] [CrossRef] [PubMed]
- Alevizos, E.; Barille, L. Global ocean spatial suitability for macroalgae offshore cultivation and sinking. Front. Mar. Sci. 2023, 10, 1320642. [Google Scholar] [CrossRef]
- Xing, Q.; Han, S.; Park, J.S.; Yarish, C.; Kim, J.K. Comparative transcriptome analysis reveals the molecular mechanism of heat-tolerance in Neopyropia yezoensis induced by Sargassum horneri extract. Mar. Mol. Biol. Ecol. 2023, 10, 1142483. [Google Scholar] [CrossRef]
- Han, I.S.; Lee, J.S.; Jung, H.K. Long-term pattern changes of sea surface temperature during summer and winter due to climate change in the Korea Waters. Fish. Aquat. Sci. 2023, 26, 639–648. [Google Scholar] [CrossRef]
- Hiraoka, M.; Kinoshita, Y.; Higa, M.; Tsubaki, S.; Monotilla, A.P.; Onda, A.; Dan, A. Fourfold daily growth rate in multicellular marine alga Ulva meridionalis. Sci. Rep. 2020, 10, 12606. [Google Scholar] [CrossRef]
- Worm, B.; Heike, K.; Lotze, H.K. Marine biodiversity and climate change. Clim. Chang. 2021, 21, 445–464. [Google Scholar]
- Pentz, B.; Klenk, N. Will climate change degrade the efficacy of marine resource management policies? Mar. Policy 2023, 148, 105462. [Google Scholar] [CrossRef]
- Thien, V.Y.; Rodrigues, K.F.; Voo, C.L.Y.; Wong, C.M.V.L.; Yong, W.T.L. Comparative Transcriptome Profiling of Kappaphycus alvarezii (Rhodophyta, Solieriaceae) in Response to Light of Different Wavelengths and Carbon Dioxide Enrichment. Plants 2021, 10, 1236. [Google Scholar] [CrossRef]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Trinity: Reconstructing a full-length transcriptome without a genome from RNA-Seq data. Nat. Biotechnol. 2011, 29, 644. [Google Scholar] [CrossRef]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using realtime quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Jahan, K.; Nie, H.; Yin, Z.; Zhang, Y.; Li, N.; Yan, X. Comparative transcriptome analysis to reveal the genes and pathways associated with adaptation strategies in two different populations of Manila clam (Ruditapes philippinarum) under acute temperature challenge. Aquaculture 2022, 552, 737999. [Google Scholar] [CrossRef]
- Handayani, T.; Zulpikar, F.; Kusnadi, A. The roles of macroalgae in climate change mitigation: Opportunities and challenges for marine -based carbon donor. Conf. Ser. Earth Environ. Sci. 2022, 1119, 01201. [Google Scholar] [CrossRef]
- Sharifian, S.; Homaei, A.; Hemmati, R.; Khajeh, K. Light emission miracle in the sea and preeminent applications of bioluminescence in recent new biotechnology. J. Photochem. Photobiol. 2017, 172, 115–128. [Google Scholar] [CrossRef]
- Sharifian, S.; Homaei, A.; Hemmati, R.; Luwor, R.B.; Khajeh, K. The emerging use of bioluminescence in medical research. Biomed. Pharmacother. 2018, 101, 74–86. [Google Scholar] [CrossRef]
- Sharifian, S.; Homaei, A.; Kim, S.K.; Satari, M. Production of newfound alkaline phosphatases from marine organisms with potential functions and industrial applications. Process Biochem. 2018, 64, 103–115. [Google Scholar] [CrossRef]
- Diyanat, S.; Homaei, A.; Mosaddegh, E. Immobilization of Penaeus vannamei protease on ZnO nanoparticles for long-term use. Int. J. Biol. Macromol. 2018, 118, 92–98. [Google Scholar] [CrossRef]
- Navvabi, A.; Razzaghi, M.; Fernandes, P.; Karami, L.; Homaei, A. Novel lipases discovery specifically from marine organisms for industrial production and practical applications. Process Biochem. 2018, 70, 61–70. [Google Scholar] [CrossRef]
- Razzaghi, M.; Homaei, A.; Mosaddegh, E. Penaeus vannamei protease stabilizing process of ZnS nanoparticles. Int. J. Biol. Macromol. 2018, 112, 509–515. [Google Scholar] [CrossRef]
- Airoldi, L.; Balata, D.; Beck, M.W. The gray zone: Relationships between habitat loss and marine diversity and their applications in conservation. J. Exp. Mar. Biol. Ecol. 2008, 366, 8–15. [Google Scholar] [CrossRef]
- Connor, K.; Gracey, A.Y. Cycles of heat and aerial-exposure induce changes in the transcriptome related to cell regulation and metabolism in Mytilus californianus. Mar. Biol. 2020, 167, 132. [Google Scholar]
- Dong, C.; Seokwoo, H.; Kwon, Y.; Young Wook, K.; Jeong Ha, K. Population genetic diversity and connectivity of the kelp species Ecklonia cava from the Korean coast. J. Appl. Phycology. J. Appl. Phycol. 2024, 36, 1035–1046. [Google Scholar]
- Sandra, H.; Ana, G.; Francisco, A.; Escribano, M.; Pilar, E.M.; Alexander, J.; Olivier, D.C.; Christine, A.M.; João, N.F.; Brezo, D.C.M. Range-edge populations of seaweeds show niche unfilling and poor adaptation to increased temperatures. J. Biogeogr. 2023, 50, 780–791. [Google Scholar]
- Ma, L.H.; Tian, L.; Wang, Y.Q.; Xie, C.Y.; Du, G.Y. Physiological and transcriptome analysis of acclimatory response to cold stress in marine red alga Pyropia yezoensis. Algae 2024, 39, 17–30. [Google Scholar] [CrossRef]
- Mikami, K.; Takio, S.; Hiwatashi, Y.; Kumar, M. Editorial: Environmental Stress-Promoting Responses in Algae. Front. Mar. Sci. 2021, 8, 797613. [Google Scholar] [CrossRef]
- Fu, J.; Momčilović, I.; Prasad, V. Molecular bases and improvement of heat tolerance in crop plants. In Heat Stress: Causes, Prevention and Treatments; Nova Publishers: Hauppauge, NY, USA, 2012; pp. 185–214. [Google Scholar]
- Chen, H.C.; Cheng, S.C. Functional roles of protein splicing factors. Biosci. Rep. 2012, 32, 345–359. [Google Scholar]
- Zhao, X.Y.; Tang, X.X.; Zhang, H.X.; Qu, T.F.; Wang, Y. Photosynthetic adaptation strategy of Ulva prolifera floating on the sea surface to environmental changes. Plant Physiol. Biochem. 2016, 107, 116–125. [Google Scholar] [CrossRef]
- Jianming, F.; Scott, H.; Ivana, M.; Prasad, P.V.V. Roles of Protein Synthesis Elongation Factor EF-Tu in Heat Tolerance in Plants. J. Bot. 2012, 2012, 835836. [Google Scholar]
- Kameyama, R.; Nishihara, G.N.; Kawagoe, C.; Terada, R. The effects of four stressors, irradiance, temperature, desiccation, and salinity on the photosynthesis of a red alga, Agarophyton vermiculophyllum (Gracilariales) from a native distributional range in Japan. J. Appl. Phycol. 2021, 33, 2561–2575. [Google Scholar] [CrossRef]
- Kistler, A.L.; Guthrie, C. Deletion of MUD2, the yeast homolog of U2AF65, can bypass the requirement for sub2, an essential spliceosomal ATPase. Genes Dev. 2001, 15, 42–49. [Google Scholar] [CrossRef] [PubMed]
- Perriman, R.; Ares, M., Jr. ATP can be dispensable for prespliceosome formation in yeast. Genes Dev. 2000, 14, 97–107. [Google Scholar] [CrossRef] [PubMed]
- Perriman, R.; Barta, I.; Voeltz, G.K.; Abelson, J.; Ares, M., Jr. ATP requirement for Prp5p function is determined by Cus2p and the structure of U2 small nuclear RNA. Proc. Natl. Acad. Sci. USA 2003, 100, 13857–13862. [Google Scholar] [CrossRef] [PubMed]
- Raghunathan, P.L.; Guthrie, C. RNA unwinding in U4/U6 snRNPs requires ATP hydrolysis and the DEIH-box splicing factor Brr2. Curr. Biol. 1998, 8, 847–855. [Google Scholar] [CrossRef]
- Laggerbauer, B.; Achsel, T.; Lührmann, R. The human U5-200kD DEXH-box protein unwinds U4/U6 RNA duplices in vitro. Proc. Natl. Acad. Sci. USA 1998, 95, 4188–4192. [Google Scholar] [CrossRef]
- Warkocki, Z.; Odenwalder, P.; Schmitzova, J.; Platzmann, F.; Stark, H.; Urlaub, H.; Ficner, R.; Fabrizio, P.; Luhrmann, R. Reconstitution of both steps of Saccharomyces cerevisiae splicing with purified spliceosomal components. Nat. Struct. Mol. Biol. 2009, 16, 1237–1243. [Google Scholar] [CrossRef]
- Fabrizio, P.; Laggerbauer, B.; Lauber, J.; Lane, W.S.; Luhrmann, R. An evolutionarily conserved U5 ¨ snRNP-specific protein is a GTP-binding factor closely related to the ribosomal translocase EF-2. EMBO J. 1997, 16, 4092–4106. [Google Scholar] [CrossRef]
- Brenner, T.J.; Guthrie, C. Genetic analysis reveals a role for the C terminus of the Saccharomyces cerevisiae GTPase Snu114 during spliceosome activation. Genetics 2005, 170, 1063–1080. [Google Scholar] [CrossRef]
- Bartels, C.; Klatt, C.; Luhrmann, R.; Fabrizio, P. The ribosomal translocase homologue Snu114p is involved in unwinding U4/U6 RNA during activation of the spliceosome. EMBO Rep. 2002, 3, 875–880. [Google Scholar] [CrossRef]
- Kim, S.H.; Lin, R.J. Pre -mRNA splicing within an assembled yeast spliceosome requires an RNA-dependent ATPase and ATP hydrolysis. Proc. Natl. Acad. Sci. USA 1993, 90, 888–892. [Google Scholar] [CrossRef]
- Schwer, B.; Guthrie, C. PRP16 is an RNA-dependent ATPase that interacts transiently with the spliceosome. Nature 1991, 349, 494–499. [Google Scholar] [CrossRef] [PubMed]
- Lardelli, R.M.; Thompson, J.X.; Yates, J.R., III; Stevens, S.W. Release of SF3 from the intron branchpoint activates the first step of pre-mRNA splicing. RNA 2010, 16, 516–528. [Google Scholar] [CrossRef] [PubMed]
- Tseng, C.K.; Liu, H.L.; Cheng, S.C. DEAH-box ATPase Prp16 has dual roles in remodeling of the spliceosome in catalytic steps. RNA. 2011, 17, 145–154. [Google Scholar] [CrossRef] [PubMed]
- Arenas, J.E.; Abelson, J.N. Prp43: An RNA helicase-like factor involved in spliceosome disassembly. Proc. Natl. Acad. Sci. USA 1997, 94, 11798–11802. [Google Scholar] [CrossRef]
- Mandadi, K.K.; Petrillo, E.; Dubrovina, A.S.; Kiselev, K.V. Editorial: Regulation of alternative splicing in plant stress responses. Front. Plant Sci. 2023, 13, 1120961. [Google Scholar] [CrossRef]
- Suhorukova, A.V.; Denis, S.S.; Milovskaya, I.G.; Fadeev, V.S.; Goldenkova-Pavlova, I.V.; Tyurin, A.A. A Molecular Orchestration of Plant Translation under Abiotic Stress. Cells 2023, 20, 2445. [Google Scholar] [CrossRef]
- Kao, Y.; Gonzalez, K.L.; Bartel, B. Peroxisome function, biogenesis, and dynamics in plants. Plant Physiol. 2018, 176, 162–177. [Google Scholar] [CrossRef]
- Pan, R.; Liu, J.; Wang, S.; Hu, J. Peroxisomes: Versatile organelles with diverse roles in plants. New Phytol. 2020, 225, 1410–1427. [Google Scholar] [CrossRef]
- Reumann, S.; Chowdhary, G. Prediction of peroxisomal matrix proteins in plants. In Proteomics of Peroxisomes; Subcellular Biochemistry Volume 89; Springer: Singapore, 2018; pp. 125–138. [Google Scholar]
- Baker, A.; Hogg, T.L.; Warriner, S.L. Peroxisome protein import: A complex journey. Biochem. Soc. Trans. 2016, 44, 783–789. [Google Scholar] [CrossRef][Green Version]
- Sandalio, L.M.; Peláez-Vico, M.A.; Molina-Moya, E.; Romero-Puertas, M.C. Peroxisomes as redox-signaling nodes in intracellular communication and stress responses. Plant Physiol. 2021, 186, 22–35. [Google Scholar]
- Hasanuzzaman, M.; Bhuyan, M.H.M.B.; Zulfiqar, F.; Raza, A.; Mohsin, S.M.; Mahmud, J.A.; Fujita, M.; Fotopoulos, V. Reactive Oxygen Species and Antioxidant Defense in Plants under Abiotic Stress: Revisiting the Crucial Role of a Universal Defense Regulator. Antioxidants 2020, 9, 681. [Google Scholar] [CrossRef] [PubMed]
- Su, T.; Li, W.; Wang, P.; Ma, C. Dynamics of Peroxisome Homeostasis and Its Role in Stress Response and Signaling in Plants. Front. Plant Sci. 2019, 10, 705. [Google Scholar] [CrossRef] [PubMed]
- Han, H.; Niu, X.; Liang, W.; Liu, M. An increase in the number of peroxisomes is coupled to the initial infection stage and stress response of Botrytis cinerea. Phytopathol. Res. 2022, 4, 25. [Google Scholar] [CrossRef]
- Zhang, W.Y.; Storey, K.B.; Dong, Y.W. Adaptations to the mudflat: Insights from physiological and transcriptional responses to thermal stress in a burrowing bivalve Sinonovacula constricta. Sci. Total Environ. 2020, 710, 136280. [Google Scholar] [CrossRef]
- Zhang, Y.Y.; Yu, S.E.; Wang, W.L.; Yang, L.E.; Lu, Q.Q.; Xie, C.T.; Liu, T.; Dong, Y.W. Temperature sensitivity of marine macroalgae for aquaculture in China. Aquaculture 2023, 567, 739262. [Google Scholar] [CrossRef]
- Murano, C.; Gallo, A.; Nocerino, A.; Macina, A.; Cecchini Gualandi, S.; Boni, R. Short-Term Thermal Stress Affects Immune Cell Features in the Sea Urchin Paracentrotus lividus. Animals 2023, 13, 1954. [Google Scholar] [CrossRef]
- Case, R.J.; Longford, S.R.; Campbell, A.H.; Low, A.; Tujula, N.; Steinberg, P.D.; Kjelleberg, S. Temperature induced bacterial virulence and bleaching disease in a chemically defended marine macroalga. Environ. Microbiol. 2010, 13, 529–537. [Google Scholar] [CrossRef]
- Lu, D.C.; Wang, F.Q.; Amann, R.I.; Teeling, H.; Du, Z.J. Epiphytic common core bacteria in the microbiomes of co -located green (Ulva), brown (Saccharina) and red (Grateloupia, Gelidium) macroalgae. Microbiome 2023, 11, 126. [Google Scholar] [CrossRef]
- Hudson, J.; Deshpande, N.; Leblanc, C.; Egan, S. Pathogen exposure leads to a transcriptional downregulation of core cellular functions that may dampen the immune response in a macroalga. Mol. Ecol. 2022, 31, 3468–3480. [Google Scholar] [CrossRef]
- Morrissey, K.L.; Iveša, L.; Delva, S.; D’Hondt, S.; Willems, A.; De Clerck, O. Impacts of environmental stress on resistance and resilience of algal-associated bacterial communities. Ecol. Evol. 2021, 11, 15004–15019. [Google Scholar] [CrossRef]
- Monserrat, M.; Comeau, S.; Verdura, J.; Alliouane, S.; Spennato, G.; Priouzeau, F.; Romero, G.; Mangialajo, L. Climate change and species facilitation affect the recruitment of macroalgal marine forests. Sci. Rep. 2022, 12, 18103. [Google Scholar] [CrossRef] [PubMed]
- Ismaiel, M.M.S.; Piercey-Normore, M.D. Antioxidant enzymes of Pseudochlorella pringsheimii under two stressors: Variation of SOD Isoforms activity. J. Plant Res. 2023, 136, 755–767. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Cai, Y.; Wakisaka, M.; Yang, Z.; Yin, Y.; Fang, W.; Xu, Y.; Omura, T.; Yu, R.; Zheng, A.L.T. Mitigation of oxidative stress damage caused by abiotic stress to improve biomass yield of microalgae: A review. Sci. Total Environ. 2023, 896, 165200. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.B.; Peng, L.N.; Yan, X.H. Multi-omics responses of red algae Pyropia haitanensis to intertidal desiccation during low tides. Algal Res. 2021, 58, 102376. [Google Scholar] [CrossRef]
- Huang, Y.; Cui, J.; Wang, S.; Chen, X.; Liao, J.; Guo, Y.; Xin, R.; Huang, B.; Xie, E. Transcriptome analysis reveals the molecular mechanisms of adaptation to high temperatures in Gracilaria bailinae. Front. Plant Sci. 2023, 14, 1125324. [Google Scholar] [CrossRef]
- Li, Y.; Xu, X.; Qi, G.; Cui, D.; Huang, C.; Sui, X.; Li, G.; Fan, Q. Mechanisms of autophagy function and regulation in plant growth, development, and response to abiotic stress. Crop J. 2023, 11, 1611–1625. [Google Scholar] [CrossRef]
- Petricka, J.J.; Winter, C.M.; Benfey, P.N. Control of Arabidopsis root development. Annu. Rev. Plant Biol. 2012, 63, 563–590. [Google Scholar] [CrossRef]
- Deb, S.; Sankaranarayanan, S.; Wewala, G.; Widdup, E.; Samuel, M.A. The S-Domain receptor kinase Arabidopsis receptor kinase2 and the U Box/ Armadillo repeat-containing E3 ubiquitin ligase9 module mediates la teral root development under phosphate starvation in Arabidopsis. Plant Physiol. 2014, 165, 1647–1656. [Google Scholar] [CrossRef]
- Liao, C.Y.; Bassham, D.C. Combating stress: The interplay between hormone signaling and autophagy in plants. J. Exp. Bot. 2020, 71, 1723–1733. [Google Scholar] [CrossRef]
- Su, T.; Li, X.; Yang, M.; Shao, Q.; Zhao, Y.; Ma, C.; Wang, P. Autophagy: An Intracellular Degradation Pathway Regulating Plant Survival and Stress Response. Front. Plant Sci. 2020, 11, 164. [Google Scholar] [CrossRef]
- Sankaranarayanan, S.; Samuel, M.A. A proposed role for selective autophagy in regulating auxin-dependent lateral root development under phosphate starvation in Arabidopsis. Plant Signal. Behav. 2015, 10, e989749. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Wang, W.; Teng, F.; Lin, Y.; Ji, D.; Xu, Y.; Chen, C.; Xie, C. Transcriptomic study to understand thermal adaptation in a high temperature-tolerant strain of Pyropia haitanensis. PLoS ONE 2018, 13, e0195842. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Yu, X.; Xu, K.; Bi, G.; Cao, M.; Zelzion, E.; Fu, C.; Sun, P.; Liu, Y.; Kong, F.; et al. Pyropia yezoensis genome reveals diverse mechanisms of carbon acquisition in the intertidal environment. Nat. Commun. 2020, 11, 4028. [Google Scholar] [CrossRef] [PubMed]
- Obata, T.; Fernie, A.R. The use of metabolomics to dissect plant responses to abiotic stresses. Cell. Mol. Life Sci. 2012, 69, 3225–3243. [Google Scholar] [CrossRef]
- Vašková, J.; Kočan, L.; Vaško, L.; Perjési, P. Glutathione-Related Enzymes and Proteins: A Review. Molecules 2023, 28, 1447. [Google Scholar] [CrossRef]
- Hayes, J.D.; Pulford, D.J. The glutathione S-transferase gene family: Regulation of GST and the contribution of the isoenzymes to cancer chemoprotection and drug resistance. Crit. Rev. Biochem. Mol. Biol. 1995, 30, 445–600. [Google Scholar] [CrossRef]
- Yang, B.; Liu, J.; Ma, X.; Guo, B.; Liu, B.; Wu, T.; Jiang, Y.; Chen, F. Genetic engineering of the Calvin cycle toward enhanced photosynthetic CO2 fixation in microalgae. Biotechnol. Biofuels 2017, 10, 229. [Google Scholar] [CrossRef]
- Mughunth, R.J.; Velmurugan, S.; Mohanalakshmi, M.; Vanitha, K. A review of seaweed extract’s potential as a biostimulant to enhance growth and mitigate stress in horticulture crops. Sci. Hortic. 2024, 334, 113312. [Google Scholar] [CrossRef]
- Supplitt, S.; Karpinski, P.; Sasiadek, M.; Laczmanska, I. Current Achievements and Applications of Transcriptomics in Personalized Cancer Medicine. Int. J. Mol. Sci. 2021, 22, 1422. [Google Scholar] [CrossRef]
- Tyagi, P.; Singh, D.; Mathur, S.; Singh, A.; Ranjan, R. Upcoming progress of transcriptomics studies on plants: An ove rview. Front. Plant Sci. 2022, 15, 1030890. [Google Scholar]
- Heras, J. Fish Transcriptomics: Applied to our Understanding of Aquaculture. Preprints 2020, 2020010332. [Google Scholar] [CrossRef]
- Jiang, J.; Yu, Y.; Zheng, M.; Liu, N. High light might alleviate inhibitory effects of high temperature on growth and physiological parameters of Ulva prolifera. Aquac. Res. 2020, 51, 2062–2070. [Google Scholar] [CrossRef]



| Group | Average Logcpm | Mean Fold Change Differentially Expressed Genes | Differentially Expressed Genes DEGs p-Value (Average) | DEG | Up-Regulated DEGs | Down-Regulated DEGs |
|---|---|---|---|---|---|---|
| H24 vs. C | 1.177248 | −6.896698 | 0.017459 | 49,796 | 26,253 | 23,543 |
| H48 vs. C | 0.983384 | −14.470383 | 0.22426397 | 47,066 | 25,250 | 21,816 |
| MH24 vs. C | 1.276447 | −20.981178 | 0.004089 | 44,468 | 23,210 | 21,258 |
| MH48 vs. C | 1.287536 | −16.937280 | 0.001698 | 44,169 | 23,180 | 20,989 |
| GO Term | % Unigenes | Number of Unigenes |
|---|---|---|
| Molecular functions | 30.84% | 19,214 |
| Biological processes | 28.84% | 17,967 |
| Cellular components | 14.91% | 9287 |
| Total unigenes | 46,468 |
| H24 vs. C | H48 vs. C | MH24 vs. C | MH48 vs. C | |
|---|---|---|---|---|
| Total enriched pathway | 434 | 434 | 432 | 431 |
| Total enriched genes | 1207 | 1166 | 1115 | 1108 |
| Significantly up-regulated or down-regulated genes in ribosome pathway | 131 | 130 | 129 | 130 |
| Significantly up-regulated or down-regulated Genes in the peroxisome pathway | 47 | 48 | 41 | 44 |
| Significantly up-regulated or down-regulated genes in the autophagy pathway | 17 | 16 | 15 | 16 |
| Significantly up-regulated or down-regulated genes in spliceosome pathway | 89 | 85 | 78 | 78 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jahan, K.; Supty, M.S.A.; Lee, J.-S.; Choi, K.-H. Transcriptomic Analysis Provides New Insights into the Tolerance Mechanisms of Green Macroalgae Ulva prolifera to High Temperature and Light Stress. Biology 2024, 13, 725. https://doi.org/10.3390/biology13090725
Jahan K, Supty MSA, Lee J-S, Choi K-H. Transcriptomic Analysis Provides New Insights into the Tolerance Mechanisms of Green Macroalgae Ulva prolifera to High Temperature and Light Stress. Biology. 2024; 13(9):725. https://doi.org/10.3390/biology13090725
Chicago/Turabian StyleJahan, Kifat, Mst Shamim Ara Supty, Jun-Seok Lee, and Keun-Hyung Choi. 2024. "Transcriptomic Analysis Provides New Insights into the Tolerance Mechanisms of Green Macroalgae Ulva prolifera to High Temperature and Light Stress" Biology 13, no. 9: 725. https://doi.org/10.3390/biology13090725
APA StyleJahan, K., Supty, M. S. A., Lee, J.-S., & Choi, K.-H. (2024). Transcriptomic Analysis Provides New Insights into the Tolerance Mechanisms of Green Macroalgae Ulva prolifera to High Temperature and Light Stress. Biology, 13(9), 725. https://doi.org/10.3390/biology13090725







