Imprinting as Basis for Complex Evolutionary Novelties in Eutherians
Abstract
Simple Summary
Abstract
1. Introduction
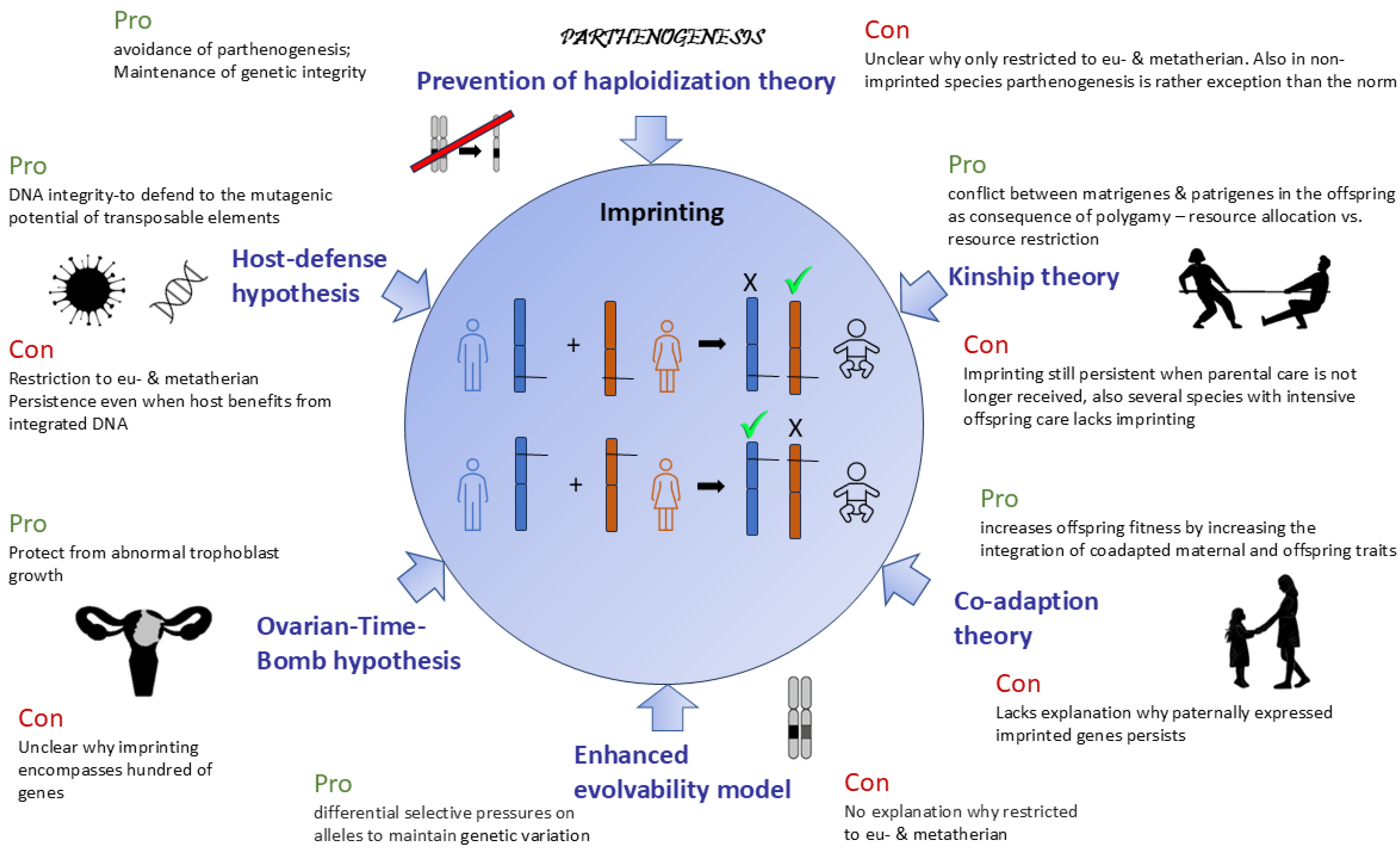
2. Theories of Evolution of Genomic Imprinting
3. Imprinting from the Vantage Point of Diploidy and Haploidy
4. The Importance of Dosage Compensation
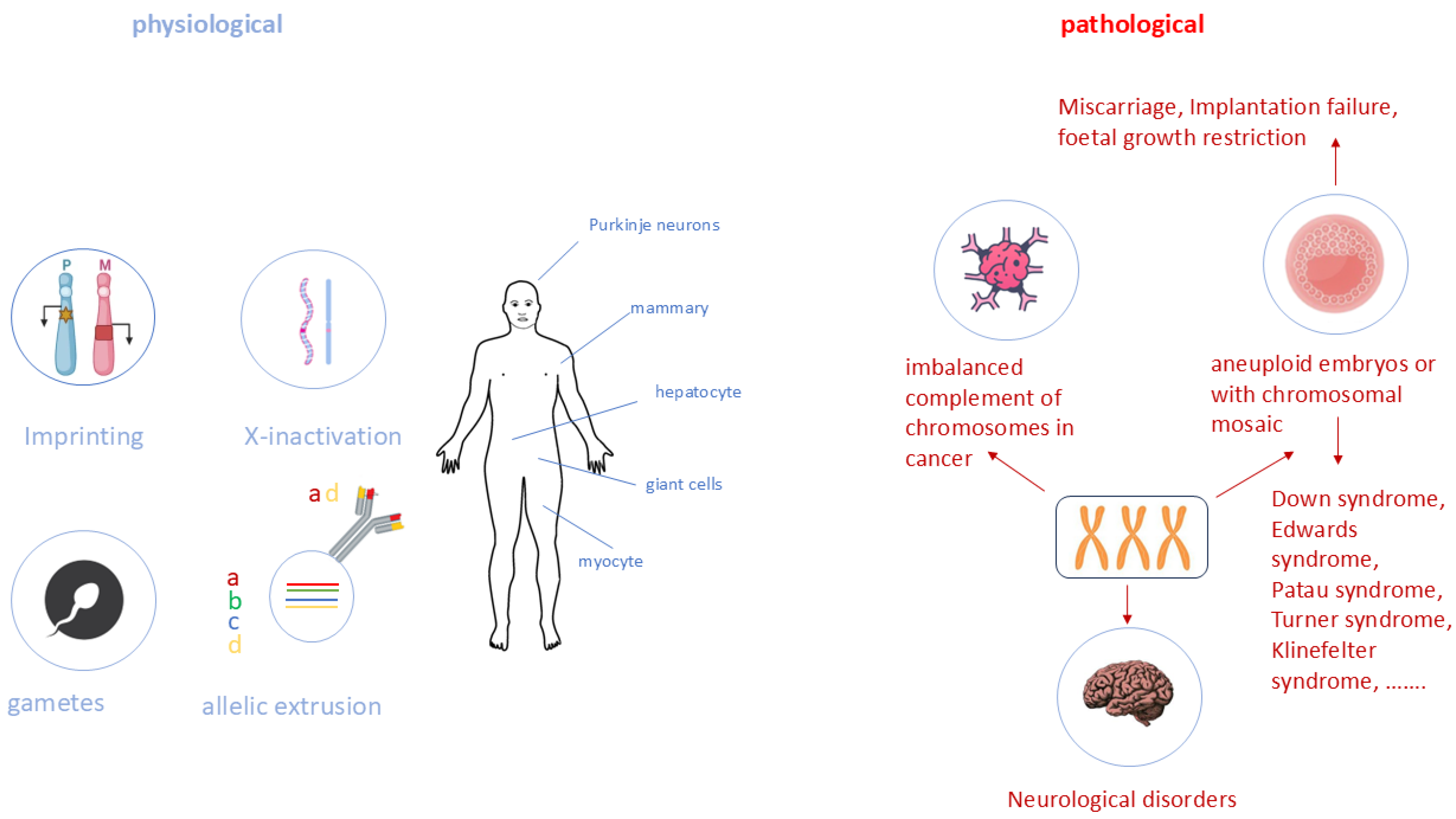
5. Monoallelic Expression in Development and Disease
6. The Broader Picture of Allele-Specific Expression
7. Evolution of Imprinting in Organs
- (1)
- Placenta
- (2)
- Brain
8. Conclusions
- (1)
- The comparison of theories regarding the origin of imprinting demonstrates that all current hypotheses explain certain, but not all, aspects of why such a complex and vulnerable regulatory process evolved with the origin of mammals.
- (2)
- Imprinting might represent a functional haploidy of some genes. Regarding an allele-specific expression, imprinting is not a unique feature in mammals. This aspect is also known in immune and neuronal adhesion molecules. Therefore, comparing effects of imprinting in several regions of the body could provide insights into the evolution of this mechanism.
- (3)
- Imprinting is important in gene dosage regulation, while the origin of expression (maternal or paternal) might be irrelevant in these instances. A monoallelic expression possibly allows a better fine-tuning of gene regulation. Over- and under-expression of distinct genes can cause a phenotypic effect, which are often contrary in their phenotypic aspects. Several imprinted genes are dysregulated in cancer and other severe pathological processes. Notably, XCI and imprinting share many common features, including gene dosage compensation. Research into the similarities and differences of XCI and imprinting (silencing) of single genes potentially enables not only the understanding of dosage effects, but also of threshold effects and roles in gene regulatory networks that depend on precise dosages.
- (4)
- Disturbed monoallelic expression is not only known for imprinting disorders. For example, an escape of XCI is regularly suggested as a reason for higher prevalence of auto-immune diseases women. LOI (loss of prominent imprinting) is associated with many congenital disorders and might be also involved in tumorigenesis, neurological disorders, and complications of pregnancy such as pre-eclampsia. For that reason, and following up on point 2 and 3 above, is it important to further investigate the mechanisms of gene regulations (dosage, imprinting), as this might lead to new treatments using, for example, CRISPR to regulate gene dosages, and therefore reduce the impact of XCI escape or LOI.
- (5)
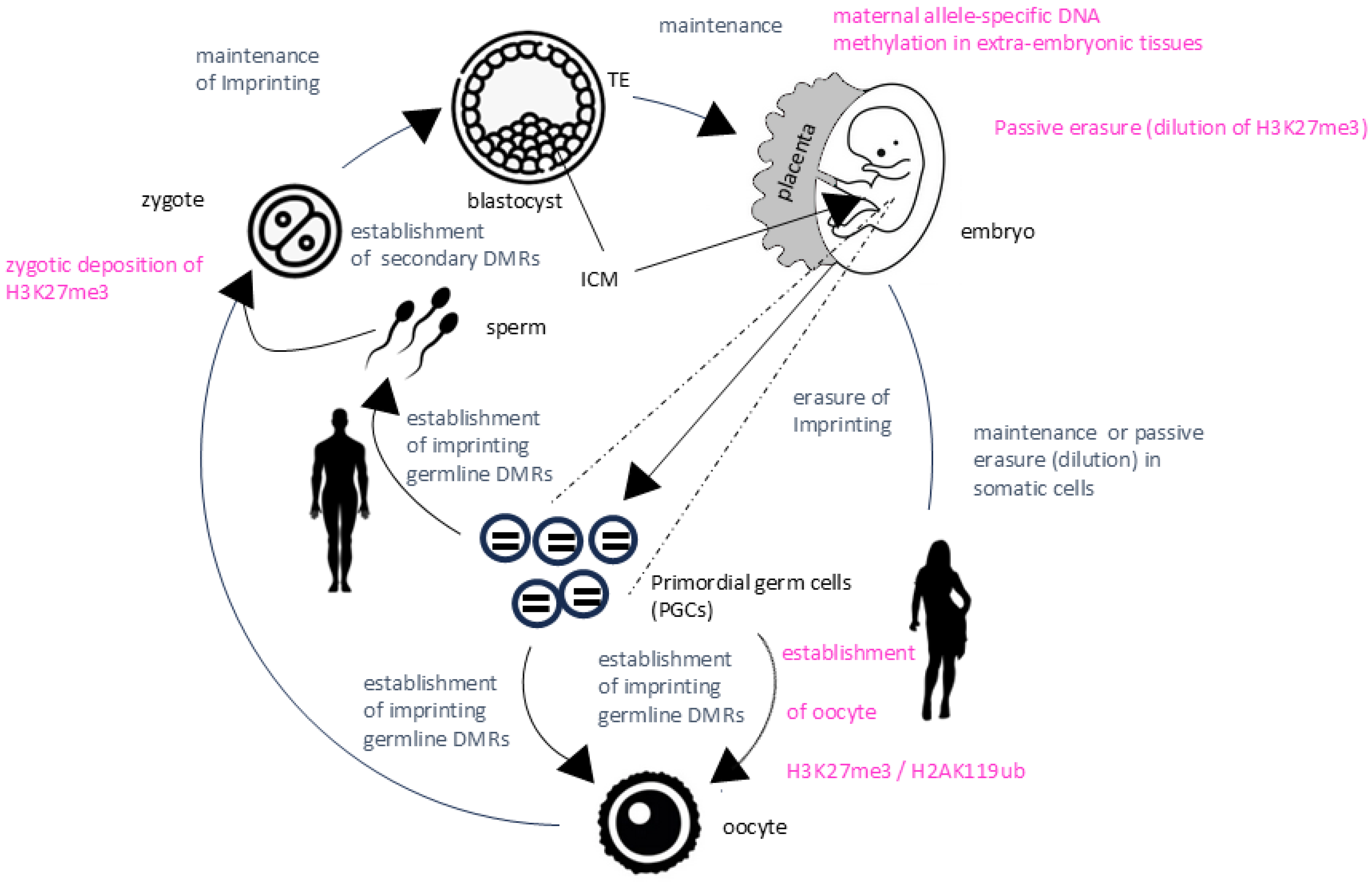
- Beside the classical canonical imprinting based on parent-specific DNA methylation, an additional mechanism of imprinting exists. The non-canonical imprinting is based primarily on histone modifications and was described in embryonic stages and extraembryonic tissues. However, this layer of epigenetic regulation is poorly conserved among species and the detailed role(s) of non-canonical imprinting remains to be elucidated. The description of developmental stage specific imprinted genes in mice suggests that imprinting is more complex than previously thought.
- (6)
- The biased allele-specific expression (ASE), erroneously also designated as non-canonical imprinting, stresses the concept of a uniform biallelic expression and the current hypothesis of the occurrence of imprinting. With the implementation of single-cell expression analysis and the investigation of epigenetic modifications, future research will not only focus on the spatiotemporal expression patterns but also ASE.
- (7)
- The placenta as an evolutionary novelty and the massive brain enlargement are the hallmarks of mammals. Both organs represent hotspots of imprinting, and several imprinting disorders are associated with neurological and placental anomalies. The placenta–brain axis signifies how the placenta modulates fetal brain development. Genomic imprinting allows a high plasticity on cells and organs, and both organs are characterized by high plasticity and their capability of fast adaptations to environmental changes. Genomic imprinting might not only contribute substantially to the plasticity of the placenta–brain axis but might be the key evolutionary driver of these organs.
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Surani, M.; Barton, S.C.; Norris, M. Development of reconstituted mouse eggs suggests imprinting of the genome during gametogenesis. Nature 1984, 308, 548–550. [Google Scholar] [CrossRef] [PubMed]
- Malnou, E.C.; Umlauf, D.; Mouysset, M.; Cavaille, J. Imprinted MicroRNA Gene Clusters in the Evolution, Development, and Functions of Mammalian Placenta. Front. Genet. 2019, 9, 706. [Google Scholar] [CrossRef]
- McGrath, J.; Solter, D. Completion of mouse embryogenesis requires both the maternal and paternal genomes. Cell 1984, 37, 179–183. [Google Scholar] [CrossRef] [PubMed]
- Gardner, D.K.; Lane, M. Ex vivo early embryo development and effects on gene expression and imprinting. Reprod. Fertil. Dev. 2005, 17, 361–370. [Google Scholar] [CrossRef] [PubMed]
- Consortium, I.H.G.S. Finishing the euchromatic sequence of the human genome. Nature 2004, 431, 931–945. [Google Scholar]
- Wilkins, J.F.; Úbeda, F.; Van Cleve, J. The evolving landscape of imprinted genes in humans and mice: Conflict among alleles, genes, tissues, and kin. Bioessays 2016, 38, 482–489. [Google Scholar] [CrossRef]
- Elbracht, M.; Mackay, D.; Begemann, M.; Kagan, K.O.; Eggermann, T. Disturbed genomic imprinting and its relevance for human reproduction: Causes and clinical consequences. Hum. Reprod. Update 2020, 26, 197–213. [Google Scholar] [CrossRef]
- Eggermann, T.; Davies, J.H.; Tauber, M.; van den Akker, E.; Hokken-Koelega, A.; Johansson, G.; Netchine, I. Growth Restriction and Genomic Imprinting-Overlapping Phenotypes Support the Concept of an Imprinting Network. Genes 2021, 12, 585. [Google Scholar] [CrossRef]
- Wang, T.; Li, J.; Yang, L.; Wu, M.; Ma, Q. The role of long non-coding RNAs in human imprinting disorders: Prospective therapeutic targets. Front. Cell Dev. Biol. 2021, 9, 730014. [Google Scholar] [CrossRef]
- Bartolomei, M.S. Genomic imprinting: Employing and avoiding epigenetic processes. Genes Dev. 2009, 23, 2124–2133. [Google Scholar] [CrossRef]
- Rakyan, V.K.; Down, T.A.; Balding, D.J.; Beck, S. Epigenome-wide association studies for common human diseases. Nat. Rev. Genet. 2011, 12, 529–541. [Google Scholar] [CrossRef]
- Nasrullah; Hussain, A.; Ahmed, S.; Rasool, M.; Shah, A.J. DNA methylation across the tree of life, from micro to macro-organism. Bioengineered 2022, 13, 1666–1685. [Google Scholar] [CrossRef]
- Aamidor, S.E.; Yagound, B.; Ronai, I.; Oldroyd, B.P. Sex mosaics in the honeybee: How haplodiploidy makes possible the evolution of novel forms of reproduction in social Hymenoptera. Biol. Lett. 2018, 14, 20180670. [Google Scholar] [CrossRef] [PubMed]
- Scott, R.J.; Spielman, M. Genomic imprinting in plants and mammals: How life history constrains convergence. Cytogenet. Genome Res. 2006, 113, 53–67. [Google Scholar] [CrossRef] [PubMed]
- Renfree, M.B.; Hore, T.A.; Shaw, G.; Marshall Graves, J.A.; Pask, A.J. Evolution of genomic imprinting: Insights from marsupials and monotremes. Annu. Rev. Genom. Hum. Genet. 2009, 10, 241–262. [Google Scholar] [CrossRef]
- Barlow, D.P.; Bartolomei, M.S. Genomic imprinting in mammals. Cold Spring Harb. Perspect. Biol. 2014, 6, a018382. [Google Scholar] [CrossRef] [PubMed]
- Feil, R.; Berger, F. Convergent evolution of genomic imprinting in plants and mammals. Trends Genet. 2007, 23, 192–199. [Google Scholar] [CrossRef]
- Harris, L.K.; Crocker, I.P.; Baker, P.N.; Aplin, J.D.; Westwood, M. IGF2 actions on trophoblast in human placenta are regulated by the insulin-like growth factor 2 receptor, which can function as both a signaling and clearance receptor. Biol. Reprod. 2011, 84, 440–446. [Google Scholar] [CrossRef]
- Sibley, C.; Coan, P.; Ferguson-Smith, A.; Dean, W.; Hughes, J.; Smith, P.; Reik, W.; Burton, G.; Fowden, A.; Constancia, M. Placental-specific insulin-like growth factor 2 (Igf2) regulates the diffusional exchange characteristics of the mouse placenta. Proc. Natl. Acad. Sci. USA 2004, 101, 8204–8208. [Google Scholar] [CrossRef]
- Killian, J.K.; Nolan, C.M.; Stewart, N.; Munday, B.L.; Andersen, N.A.; Nicol, S.; Jirtle, R.L. Monotreme IGF2 expression and ancestral origin of genomic imprinting. J. Exp. Zool. 2001, 291, 205–212. [Google Scholar] [CrossRef]
- O’Neill, M.J.; Ingram, R.S.; Vrana, P.B.; Tilghman, S.M. Allelic expression of IGF2 in marsupials and birds. Dev. Genes Evol. 2000, 210, 18–20. [Google Scholar] [CrossRef] [PubMed]
- Nolan, C.M.; Killian, J.K.; Petitte, J.N.; Jirtle, R.L. Imprint status of M6P/IGF2R and IGF2 in chickens. Dev. Genes Evol. 2001, 211, 179–183. [Google Scholar] [CrossRef]
- Hore, T.A.; Rapkins, R.W.; Graves, J.A.M. Construction and evolution of imprinted loci in mammals. Trends Genet. 2007, 23, 440–448. [Google Scholar] [CrossRef] [PubMed]
- Keverne, E.B. Genomic imprinting, maternal care, and brain evolution. Horm. Behav. 2001, 40, 146–155. [Google Scholar] [CrossRef]
- Leung, K.N.; Vallero, R.O.; DuBose, A.J.; Resnick, J.L.; LaSalle, J.M. Imprinting regulates mammalian snoRNA-encoding chromatin decondensation and neuronal nucleolar size. Hum. Mol. Genet. 2009, 18, 4227–4238. [Google Scholar] [CrossRef] [PubMed]
- Hunter, P. The silence of genes: Is genomic imprinting the software of evolution or just a battleground for gender conflict? EMBO Rep. 2007, 8, 441–443. [Google Scholar] [CrossRef] [PubMed]
- Vrana, P.B.; Fossella, J.A.; Matteson, P.; Del Rio, T.; O’Neill, M.J.; Tilghman, S.M. Genetic and epigenetic incompatibilities underlie hybrid dysgenesis in Peromyscus. Nat. Genet. 2000, 25, 120–124. [Google Scholar] [CrossRef]
- Zeyl, C.; Vanderford, T.; Carter, M. An evolutionary advantage of haploidy in large yeast populations. Science 2003, 299, 555–558. [Google Scholar] [CrossRef]
- Otto, S.; Goldstein, D. Recombination and the evolution of diploidy. Genetics 1992, 131, 745–751. [Google Scholar] [CrossRef]
- Haig, D. The kinship theory of genomic imprinting. Annu. Rev. Ecol. Syst. 2000, 31, 9–32. [Google Scholar] [CrossRef]
- Haig, D. Maternal-fetal conflict, genomic imprinting and mammalian vulnerabilities to cancer. Philos. Trans. R Soc. Lond. B Biol. Sci. 2015, 370, 20140178. [Google Scholar] [CrossRef]
- Pilvar, D.; Reiman, M.; Pilvar, A.; Laan, M. Parent-of-origin-specific allelic expression in the human placenta is limited to established imprinted loci and it is stably maintained across pregnancy. Clin Epigenetics 2019, 11, 94. [Google Scholar] [CrossRef] [PubMed]
- Cao, W.; Douglas, K.C.; Samollow, P.B.; VandeBerg, J.L.; Wang, X.; Clark, A.G. Origin and evolution of marsupial-specific imprinting clusters through lineage-specific gene duplications and acquisition of promoter differential methylation. Mol. Biol. Evol. 2023, 40, msad022. [Google Scholar] [CrossRef] [PubMed]
- Jirtle, R.L. Geneimprint. Available online: https://www.geneimprint.com/site/genes-by-species (accessed on 6 February 2024).
- Wolf, J.B.; Hager, R. A maternal–offspring coadaptation theory for the evolution of genomic imprinting. PLoS Biol. 2006, 4, e380. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, E.K.; Wolf, J.B. The coadaptation theory for genomic imprinting. Evol. Lett. 2017, 1, 49–59. [Google Scholar] [CrossRef] [PubMed]
- Beaudet, A.L.; Jiang, Y.-h. A rheostat model for a rapid and reversible form of imprinting-dependent evolution. Am. J. Hum. Genet. 2002, 70, 1389–1397. [Google Scholar] [CrossRef][Green Version]
- Thomas, J.H. Genomic imprinting proposed as a surveillance mechanism for chromosome loss. Proc. Natl. Acad. Sci. USA 1995, 92, 480–482. [Google Scholar] [CrossRef]
- Hoppe, P.C.; Illmensee, K. Full-term development after transplantation of parthenogenetic embryonic nuclei into fertilized mouse eggs. Proc. Natl. Acad. Sci. USA 1982, 79, 1912–1916. [Google Scholar] [CrossRef]
- Fujita, M.K.; Singhal, S.; Brunes, T.O.; Maldonado, J.A. Evolutionary dynamics and consequences of parthenogenesis in vertebrates. Annu. Rev. Ecol. Evol. Syst. 2020, 51, 191–214. [Google Scholar] [CrossRef]
- Varmuza, S.; Mann, M. Genomic imprinting—Defusing the ovarian time bomb. Trends Genet. 1994, 10, 118–123. [Google Scholar] [CrossRef]
- Weisstein, A.E.; Feldman, M.W.; Spencer, H.G. Evolutionary genetic models of the ovarian time bomb hypothesis for the evolution of genomic imprinting. Genetics 2002, 162, 425–439. [Google Scholar] [CrossRef] [PubMed]
- Kshitiz; Afzal, J.; Maziarz, J.D.; Hamidzadeh, A.; Liang, C.; Erkenbrack, E.M.; Kim, H.N.; Haeger, J.-D.; Pfarrer, C.; Hoang, T. Evolution of placental invasion and cancer metastasis are causally linked. Nat. Ecol. Evol. 2019, 3, 1743–1753. [Google Scholar] [CrossRef] [PubMed]
- Barlow, D.P. Methylation and imprinting: From host defense to gene regulation? Science 1993, 260, 309–310. [Google Scholar] [CrossRef] [PubMed]
- Jähner, D.; Stuhlmann, H.; Stewart, C.L.; Harbers, K.; Löhler, J.; Simon, I.; Jaenisch, R. De novo methylation and expression of retroviral genomes during mouse embryogenesis. Nature 1982, 298, 623–628. [Google Scholar] [CrossRef]
- Geis, F.K.; Goff, S.P. Silencing and transcriptional regulation of endogenous retroviruses: An overview. Viruses 2020, 12, 884. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Ito, M.; Zhou, F.; Youngson, N.; Zuo, X.; Leder, P.; Ferguson-Smith, A.C. A maternal-zygotic effect gene, Zfp57, maintains both maternal and paternal imprints. Dev. Cell 2008, 15, 547–557. [Google Scholar] [CrossRef] [PubMed]
- Kuramochi-Miyagawa, S.; Watanabe, T.; Gotoh, K.; Totoki, Y.; Toyoda, A.; Ikawa, M.; Asada, N.; Kojima, K.; Yamaguchi, Y.; Ijiri, T.W. DNA methylation of retrotransposon genes is regulated by Piwi family members MILI and MIWI2 in murine fetal testes. Genes Dev. 2008, 22, 908–917. [Google Scholar] [CrossRef] [PubMed]
- Hanna, C.W.; Perez-Palacios, R.; Gahurova, L.; Schubert, M.; Krueger, F.; Biggins, L.; Andrews, S.; Colome-Tatche, M.; Bourc’his, D.; Dean, W.; et al. Endogenous retroviral insertions drive non-canonical imprinting in extra-embryonic tissues. Genome Biol. 2019, 20, 225. [Google Scholar] [CrossRef] [PubMed]
- Shiura, H.; Kitazawa, M.; Ishino, F.; Kaneko-Ishino, T. Roles of retrovirus-derived PEG10 and PEG11/RTL1 in mammalian development and evolution and their involvement in human disease. Front. Cell Dev. Biol. 2023, 11, 1273638. [Google Scholar] [CrossRef] [PubMed]
- Kaneko-Ishino, T.; Ishino, F. The evolutionary advantage in mammals of the complementary monoallelic expression mechanism of genomic imprinting and its emergence from a defense against the insertion into the host genome. Front. Genet. 2022, 13, 832983. [Google Scholar] [CrossRef] [PubMed]
- Das, R.; Hampton, D.D.; Jirtle, R.L. Imprinting evolution and human health. Mamm. Genome 2009, 20, 563–572. [Google Scholar] [CrossRef] [PubMed]
- Sagi, I.; Benvenisty, N. Haploidy in Humans: An Evolutionary and Developmental Perspective. Dev. Cell 2017, 41, 581–589. [Google Scholar] [CrossRef] [PubMed]
- Comai, L. The advantages and disadvantages of being polyploid. Nat. Rev. Genet. 2005, 6, 836–846. [Google Scholar] [CrossRef]
- Tarkowski, A.K.; Witkowska, A.; Nowicka, J. Experimental parthenogenesis in the mouse. Nature 1970, 226, 162–165. [Google Scholar] [CrossRef] [PubMed]
- Graham, C. Parthenogenetic mouse blastocysts. Nature 1970, 226, 165–167. [Google Scholar] [CrossRef]
- Elling, U.; Taubenschmid, J.; Wirnsberger, G.; O’Malley, R.; Demers, S.-P.; Vanhaelen, Q.; Shukalyuk, A.I.; Schmauss, G.; Schramek, D.; Schnuetgen, F. Forward and reverse genetics through derivation of haploid mouse embryonic stem cells. Cell Stem Cell 2011, 9, 563–574. [Google Scholar] [CrossRef]
- Leeb, M.; Wutz, A. Derivation of haploid embryonic stem cells from mouse embryos. Nature 2011, 479, 131–134. [Google Scholar] [CrossRef]
- Xu, H.; Yue, C.; Zhang, T.; Li, Y.; Guo, A.; Liao, J.; Pei, G.; Li, J.; Jing, N. Derivation of haploid neurons from mouse androgenetic haploid embryonic stem cells. Neurosci. Bull. 2017, 33, 361–364. [Google Scholar] [CrossRef]
- Aguila, L.; Nociti, R.P.; Sampaio, R.V.; Therrien, J.; Meirelles, F.V.; Felmer, R.N.; Smith, L.C. Haploid androgenetic development of bovine embryos reveals imbalanced WNT signaling and impaired cell fate differentiation. Biol. Reprod. 2023, 109, 821–838. [Google Scholar] [CrossRef]
- Aguila, L.; Suzuki, J.; Hill, A.B.; García, M.; de Mattos, K.; Therrien, J.; Smith, L.C. Dysregulated gene expression of imprinted and X-linked genes: A link to poor development of bovine haploid androgenetic embryos. Front. Cell Dev. Biol. 2021, 9, 640712. [Google Scholar] [CrossRef]
- Leeb, M.; Wutz, A. Haploid genomes illustrate epigenetic constraints and gene dosage effects in mammals. Epigenet. Chromatin 2013, 6, 1–10. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Girirajan, S.; Campbell, C.D.; Eichler, E.E. Human copy number variation and complex genetic disease. Annu. Rev. Genet. 2011, 45, 203–226. [Google Scholar] [CrossRef] [PubMed]
- Marín, I.; Siegal, M.L.; Baker, B.S. The evolution of dosage-compensation mechanisms. Bioessays 2000, 22, 1106–1114. [Google Scholar] [CrossRef]
- Gorman, M.; Baker, B.S. How flies make one equal two: Dosage compensation in Drosophila. Trends Genet. 1994, 10, 376–380. [Google Scholar] [CrossRef] [PubMed]
- Nicoll, M.; Akerib, C.C.; Meyer, B.J. X-chromosome-counting mechanisms that determine nematode sex. Nature 1997, 388, 200–204. [Google Scholar] [CrossRef]
- Zeng, S.-M.; Yankowitz, J. X-inactivation patterns in human embryonic and extra-embryonic tissues. Placenta 2003, 24, 270–275. [Google Scholar] [CrossRef]
- Veitia, R.A.; Veyrunes, F.; Bottani, S.; Birchler, J.A. X chromosome inactivation and active X upregulation in therian mammals: Facts, questions, and hypotheses. J. Mol. Cell. Biol. 2015, 7, 2–11. [Google Scholar] [CrossRef]
- Meester, I.; Manilla-Muñoz, E.; León-Cachón, R.B.; Paniagua-Frausto, G.A.; Carrión-Alvarez, D.; Ruiz-Rodríguez, C.O.; Rodríguez-Rangel, X.; García-Martínez, J.M. SeXY chromosomes and the immune system: Reflections after a comparative study. Biol. Sex Differ. 2020, 11, 1–13. [Google Scholar] [CrossRef]
- Harris, V.M.; Sharma, R.; Cavett, J.; Kurien, B.T.; Liu, K.; Koelsch, K.A.; Rasmussen, A.; Radfar, L.; Lewis, D.; Stone, D.U. Klinefelter’s syndrome (47, XXY) is in excess among men with Sjögren’s syndrome. Clin. Immunol. 2016, 168, 25–29. [Google Scholar] [CrossRef]
- Zhao, Y.; Li, J.; Dai, L.; Ma, Y.; Bai, Y.; Guo, H. X chromosome inactivation pattern and pregnancy outcome of female carriers of pathogenic heterozygous X-linked deletions. Front. Genet. 2021, 12, 782629. [Google Scholar] [CrossRef]
- Amano, K.; Sago, H.; Uchikawa, C.; Suzuki, T.; Kotliarova, S.E.; Nukina, N.; Epstein, C.J.; Yamakawa, K. Dosage-dependent over-expression of genes in the trisomic region of Ts1Cje mouse model for Down syndrome. Hum. Mol. Genet. 2004, 13, 1333–1340. [Google Scholar] [CrossRef] [PubMed]
- Lana-Elola, E.; Aoidi, R.; Llorian, M.; Gibbins, D.; Buechsenschuetz, C.; Bussi, C.; Flynn, H.; Gilmore, T.; Watson-Scales, S.; Haugsten Hansen, M. Increased dosage of DYRK1A leads to congenital heart defects in a mouse model of Down syndrome. Sci. Transl. Med. 2024, 16, eadd6883. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.T. Molecular links between X-inactivation and autosomal imprinting: X-inactivation as a driving force for the evolution of imprinting? Curr. Biol. 2003, 13, R242–R254. [Google Scholar] [CrossRef]
- Okabe, H.; Satoh, S.; Furukawa, Y.; Kato, T.; Hasegawa, S.; Nakajima, Y.; Yamaoka, Y.; Nakamura, Y. Involvement of PEG10 in human hepatocellular carcinogenesis through interaction with SIAH1. Cancer Res. 2003, 63, 3043–3048. [Google Scholar] [PubMed]
- Chou, M.-Y.; Hu, M.-C.; Chen, P.-Y.; Hsu, C.-L.; Lin, T.-Y.; Tan, M.-J.; Lee, C.-Y.; Kuo, M.-F.; Huang, P.-H.; Wu, V.-C. RTL1/PEG11 imprinted in human and mouse brain mediates anxiety-like and social behaviors and regulates neuronal excitability in the locus coeruleus. Hum. Mol. Genet. 2022, 31, 3161–3180. [Google Scholar] [CrossRef]
- Kitazawa, M.; Sutani, A.; Kaneko-Ishino, T.; Ishino, F. The role of eutherian-specific RTL1 in the nervous system and its implications for the Kagami-Ogata and Temple syndromes. Genes Cells 2021, 26, 165–179. [Google Scholar] [CrossRef]
- Kitazawa, M.; Hayashi, S.; Imamura, M.; Takeda, S.i.; Oishi, Y.; Kaneko-Ishino, T.; Ishino, F. Deficiency and overexpression of Rtl1 in the mouse cause distinct muscle abnormalities related to Temple and Kagami-Ogata syndromes. Development 2020, 147, dev185918. [Google Scholar] [CrossRef]
- Su, Z.-Z.; Goldstein, N.I.; Jiang, H.; Wang, M.-N.; Duigou, G.J.; Young, C.S.; Fisher, P.B. PEG-3, a nontransforming cancer progression gene, is a positive regulator of cancer aggressiveness and angiogenesis. Proc. Natl. Acad. Sci. USA 1999, 96, 15115–15120. [Google Scholar] [CrossRef]
- Nativio, R.; Sparago, A.; Ito, Y.; Weksberg, R.; Riccio, A.; Murrell, A. Disruption of genomic neighbourhood at the imprinted IGF2-H19 locus in Beckwith–Wiedemann syndrome and Silver–Russell syndrome. Hum. Mol. Genet. 2011, 20, 1363–1374. [Google Scholar] [CrossRef]
- Jurkiewicz, D.; Kugaudo, M.; Skórka, A.; Śmigiel, R.; Smyk, M.; Ciara, E.; Chrzanowska, K.; Krajewska-Walasek, M. A novel IGF2/H19 domain triplication in the 11p15. 5 imprinting region causing either Beckwith–Wiedemann or Silver–Russell syndrome in a single family. Am. J. Med. Genet. Part A 2017, 173, 72–78. [Google Scholar] [CrossRef]
- Hubertus, J.; Lacher, M.; Rottenkolber, M.; Müller-Höcker, J.; Berger, M.; Stehr, M.; Von Schweinitz, D.; Kappler, R. Altered expression of imprinted genes in Wilms tumors. Oncol. Rep. 2011, 25, 817–823. [Google Scholar] [CrossRef] [PubMed]
- Glaser, J.; Iranzo, J.; Borensztein, M.; Marinucci, M.; Gualtieri, A.; Jouhanneau, C.; Teissandier, A.; Gaston-Massuet, C.; Bourc’His, D. The imprinted Zdbf2 gene finely tunes control of feeding and growth in neonates. eLife 2022, 11, e65641. [Google Scholar] [CrossRef] [PubMed]
- Ferrón, S.R.; Charalambous, M.; Radford, E.; McEwen, K.; Wildner, H.; Hind, E.; Morante-Redolat, J.M.; Laborda, J.; Guillemot, F.; Bauer, S.R. Postnatal loss of Dlk1 imprinting in stem cells and niche astrocytes regulates neurogenesis. Nature 2011, 475, 381–385. [Google Scholar] [CrossRef]
- Weinberg-Shukron, A.; Ben-Yair, R.; Takahashi, N.; Dunjić, M.; Shtrikman, A.; Edwards, C.A.; Ferguson-Smith, A.C.; Stelzer, Y. Balanced gene dosage control rather than parental origin underpins genomic imprinting. Nat. Commun. 2022, 13, 4391. [Google Scholar] [CrossRef] [PubMed]
- Teixeira da Rocha, S.; Charalambous, M.; Lin, S.-P.; Gutteridge, I.; Ito, Y.; Gray, D.; Dean, W.; Ferguson-Smith, A.C. Gene dosage effects of the imprinted delta-like homologue 1 (dlk1/pref1) in development: Implications for the evolution of imprinting. PLoS Genet. 2009, 5, e1000392. [Google Scholar] [CrossRef]
- Montalbán-Loro, R.; Lassi, G.; Lozano-Ureña, A.; Perez-Villalba, A.; Jiménez-Villalba, E.; Charalambous, M.; Vallortigara, G.; Horner, A.E.; Saksida, L.M.; Bussey, T.J. Dlk1 dosage regulates hippocampal neurogenesis and cognition. Proc. Natl. Acad. Sci. USA 2021, 118, e2015505118. [Google Scholar] [CrossRef]
- Scagliotti, V.; Vignola, M.L.; Willis, T.; Howard, M.; Marinelli, E.; Gaston-Massuet, C.; Andoniadou, C.; Charalambous, M. Imprinted Dlk1 dosage as a size determinant of the mammalian pituitary gland. eLife 2023, 12, e84092. [Google Scholar] [CrossRef]
- Hillman, P.R.; Christian, S.G.; Doan, R.; Cohen, N.D.; Konganti, K.; Douglas, K.; Wang, X.; Samollow, P.B.; Dindot, S.V. Genomic imprinting does not reduce the dosage of UBE3A in neurons. Epigenet. Chromatin 2017, 10, 1–14. [Google Scholar] [CrossRef]
- Peters, S.; Beaudet, A.; Madduri, N.; Bacino, C. Autism in Angelman syndrome: Implications for autism research. Clin. Genet. 2004, 66, 530–536. [Google Scholar] [CrossRef]
- Smith, S.E.; Zhou, Y.-D.; Zhang, G.; Jin, Z.; Stoppel, D.C.; Anderson, M.P. Increased gene dosage of Ube3a results in autism traits and decreased glutamate synaptic transmission in mice. Sci. Transl. Med. 2011, 3, 103ra97. [Google Scholar] [CrossRef]
- Xu, X.; Li, C.; Gao, X.; Xia, K.; Guo, H.; Li, Y.; Hao, Z.; Zhang, L.; Gao, D.; Xu, C. Excessive UBE3A dosage impairs retinoic acid signaling and synaptic plasticity in autism spectrum disorders. Cell Res. 2018, 28, 48–68. [Google Scholar] [CrossRef] [PubMed]
- Copping, N.A.; Christian, S.G.; Ritter, D.J.; Islam, M.S.; Buscher, N.; Zolkowska, D.; Pride, M.C.; Berg, E.L.; LaSalle, J.M.; Ellegood, J. Neuronal overexpression of Ube3a isoform 2 causes behavioral impairments and neuroanatomical pathology relevant to 15q11. 2-q13. 3 duplication syndrome. Hum. Mol. Genet. 2017, 26, 3995–4010. [Google Scholar] [CrossRef] [PubMed]
- Zhao, D.; Lin, M.; Pedrosa, E.; Lachman, H.M.; Zheng, D. Characteristics of allelic gene expression in human brain cells from single-cell RNA-seq data analysis. BMC Genom. 2017, 18, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, S.; Totoki, Y.; Soma, M.; Matsumoto, K.; Fujihara, Y.; Toyoda, A.; Sakaki, Y.; Okabe, M.; Ishino, F. Identification of an imprinted gene cluster in the X-inactivation center. PLoS ONE 2013, 8, e71222. [Google Scholar] [CrossRef]
- Kay, G.F.; Penny, G.D.; Patel, D.; Ashworth, A.; Brockdorff, N.; Rastan, S. Expression of Xist during mouse development suggests a role in the initiation of X chromosome inactivation. Cell 1993, 72, 171–182. [Google Scholar] [CrossRef]
- Mahadevaiah, S.K.; Royo, H.; VandeBerg, J.L.; McCarrey, J.R.; Mackay, S.; Turner, J.M. Key features of the X inactivation process are conserved between marsupials and eutherians. Curr. Biol. 2009, 19, 1478–1484. [Google Scholar] [CrossRef]
- Ray, P.F.; Winston, R.M.; Handyside, A.H. XIST expression from the maternal X chromosome in human male preimplantation embryos at the blastocyst stage. Hum. Mol. Genet. 1997, 6, 1323–1327. [Google Scholar] [CrossRef]
- ten Boekel, E.; Melchers, F.; Rolink, A.G. Precursor B cells showing H chain allelic inclusion display allelic exclusion at the level of pre-B cell receptor surface expression. Immunity 1998, 8, 199–207. [Google Scholar] [CrossRef]
- Agata, Y.; Tamaki, N.; Sakamoto, S.; Ikawa, T.; Masuda, K.; Kawamoto, H.; Murre, C. Regulation of T cell receptor β gene rearrangements and allelic exclusion by the helix-loop-helix protein, E47. Immunity 2007, 27, 871–884. [Google Scholar] [CrossRef]
- Thamban, T.; Agarwaal, V.; Khosla, S. Role of genomic imprinting in mammalian development. J. Biosci. 2020, 45, 20. [Google Scholar] [CrossRef]
- Tomizawa, S.; Sasaki, H. Genomic imprinting and its relevance to congenital disease, infertility, molar pregnancy and induced pluripotent stem cell. J. Hum. Genet. 2012, 57, 84–91. [Google Scholar] [CrossRef]
- Dammann, R.H.; Kirsch, S.; Schagdarsurengin, U.; Dansranjavin, T.; Gradhand, E.; Schmitt, W.D.; Hauptmann, S. Frequent aberrant methylation of the imprinted IGF2/H19 locus and LINE1 hypomethylation in ovarian carcinoma. Int. J. Oncol. 2010, 36, 171–179. [Google Scholar] [CrossRef] [PubMed]
- Holm, T.M.; Jackson-Grusby, L.; Brambrink, T.; Yamada, Y.; Rideout, W.M.; Jaenisch, R. Global loss of imprinting leads to widespread tumorigenesis in adult mice. Cancer Cell 2005, 8, 275–285. [Google Scholar] [CrossRef]
- Goovaerts, T.; Steyaert, S.; Vandenbussche, C.A.; Galle, J.; Thas, O.; Van Criekinge, W.; De Meyer, T. A comprehensive overview of genomic imprinting in breast and its deregulation in cancer. Nat. Commun. 2018, 9, 4120. [Google Scholar] [CrossRef] [PubMed]
- DeBaun, M.R.; Niemitz, E.L.; McNeil, D.E.; Brandenburg, S.A.; Lee, M.P.; Feinberg, A.P. Epigenetic alterations of H19 and LIT1 distinguish patients with Beckwith-Wiedemann syndrome with cancer and birth defects. Am. J. Hum. Genet. 2002, 70, 604–611. [Google Scholar] [CrossRef] [PubMed]
- Kamrani, S.; Amirchaghmaghi, E.; Ghaffari, F.; Shahhoseini, M.; Ghaedi, K. Altered gene expression of VEGF, IGFs and H19 lncRNA and epigenetic profile of H19-DMR region in endometrial tissues of women with endometriosis. Reprod. Health 2022, 19, 100. [Google Scholar] [CrossRef] [PubMed]
- Lew, A.R.; Kellermayer, T.R.; Sule, B.P.; Szigeti, K. Copy number variations in adult-onset neuropsychiatric diseases. Curr. Genom. 2018, 19, 420–430. [Google Scholar] [CrossRef]
- Isles, A.R. Genomic Imprinting and Brain Function. In Neuroscience in the 21st Century: From Basic to Clinical; Springer: Cham, Switzerland, 2022; pp. 2655–2675. [Google Scholar]
- Tsang, S.Y.; Ullah, A.; Xue, H. GABRB2 in neuropsychiatric disorders: Genetic associations and functional evidences. Curr. Psychopharmacol. 2019, 8, 166–176. [Google Scholar] [CrossRef]
- Roldán, E.; Fuxa, M.; Chong, W.; Martinez, D.; Novatchkova, M.; Busslinger, M.; Skok, J.A. Locus’ decontraction’and centromeric recruitment contribute to allelic exclusion of the immunoglobulin heavy-chain gene. Nat. Immunol. 2005, 6, 31–41. [Google Scholar] [CrossRef]
- Magklara, A.; Yen, A.; Colquitt, B.M.; Clowney, E.J.; Allen, W.; Markenscoff-Papadimitriou, E.; Evans, Z.A.; Kheradpour, P.; Mountoufaris, G.; Carey, C. An epigenetic signature for monoallelic olfactory receptor expression. Cell 2011, 145, 555–570. [Google Scholar] [CrossRef]
- Tan, Y.-P.; Li, S.; Jiang, X.-J.; Loh, W.; Foo, Y.K.; Loh, C.-B.; Xu, Q.; Yuen, W.-H.; Jones, M.; Fu, J. Regulation of protocadherin gene expression by multiple neuron-restrictive silencer elements scattered in the gene cluster. Nucleic Acids Res. 2010, 38, 4985–4997. [Google Scholar] [CrossRef] [PubMed]
- Akbari, V.; Garant, J.-M.; O’Neill, K.; Pandoh, P.; Moore, R.; Marra, M.A.; Hirst, M.; Jones, S.J. Genome-wide detection of imprinted differentially methylated regions using nanopore sequencing. eLife 2022, 11, e77898. [Google Scholar] [CrossRef] [PubMed]
- Iwasaki, M.; Hyvärinen, L.; Piskurewicz, U.; Lopez-Molina, L. Non-canonical RNA-directed DNA methylation participates in maternal and environmental control of seed dormancy. eLife 2019, 8, e37434. [Google Scholar] [CrossRef] [PubMed]
- Montgomery, S.A.; Hisanaga, T.; Wang, N.; Axelsson, E.; Akimcheva, S.; Sramek, M.; Liu, C.; Berger, F. Polycomb-mediated repression of paternal chromosomes maintains haploid dosage in diploid embryos of Marchantia. eLife 2022, 11, e79258. [Google Scholar] [CrossRef] [PubMed]
- Santini, L.; Halbritter, F.; Titz-Teixeira, F.; Suzuki, T.; Asami, M.; Ma, X.; Ramesmayer, J.; Lackner, A.; Warr, N.; Pauler, F.; et al. Genomic imprinting in mouse blastocysts is predominantly associated with H3K27me3. Nat. Commun. 2021, 12, 3804. [Google Scholar] [CrossRef]
- Weisenberger, D.J.; Velicescu, M.; Preciado-Lopez, M.A.; Gonzales, F.A.; Tsai, Y.C.; Liang, G.; Jones, P.A. Identification and characterization of alternatively spliced variants of DNA methyltransferase 3a in mammalian cells. Gene 2002, 298, 91–99. [Google Scholar] [CrossRef]
- Hata, K.; Okano, M.; Lei, H.; Li, E. Dnmt3L cooperates with the Dnmt3 family of de novo DNA methyltransferases to establish maternal imprints in mice. Development 2002, 129, 1983–1993. [Google Scholar] [CrossRef]
- Davis, T.L.; Yang, G.J.; McCarrey, J.R.; Bartolomei, M.S. The H19 methylation imprint is erased and re-established differentially on the parental alleles during male germ cell development. Hum. Mol. Genet. 2000, 9, 2885–2894. [Google Scholar] [CrossRef]
- Gao, L.; Emperle, M.; Guo, Y.; Grimm, S.A.; Ren, W.; Adam, S.; Uryu, H.; Zhang, Z.-M.; Chen, D.; Yin, J. Comprehensive structure-function characterization of DNMT3B and DNMT3A reveals distinctive de novo DNA methylation mechanisms. Nat. Commun. 2020, 11, 3355. [Google Scholar] [CrossRef]
- Barau, J.; Teissandier, A.; Zamudio, N.; Roy, S.; Nalesso, V.; Hérault, Y.; Guillou, F.; Bourc’his, D. The DNA methyltransferase DNMT3C protects male germ cells from transposon activity. Science 2016, 354, 909–912. [Google Scholar] [CrossRef]
- Chedin, F.; Lieber, M.R.; Hsieh, C.-L. The DNA methyltransferase-like protein DNMT3L stimulates de novo methylation by Dnmt3a. Proc. Natl. Acad. Sci. USA 2002, 99, 16916–16921. [Google Scholar] [CrossRef] [PubMed]
- Hara, S.; Takano, T.; Fujikawa, T.; Yamada, M.; Wakai, T.; Kono, T.; Obata, Y. Forced expression of DNA methyltransferases during oocyte growth accelerates the establishment of methylation imprints but not functional genomic imprinting. Hum. Mol. Genet. 2014, 23, 3853–3864. [Google Scholar] [CrossRef]
- Bonthuis, P.J.; Huang, W.-C.; Hörndli, C.N.S.; Ferris, E.; Cheng, T.; Gregg, C. Noncanonical genomic imprinting effects in offspring. Cell Rep. 2015, 12, 979–991. [Google Scholar] [CrossRef] [PubMed]
- Bonthuis, P.J.; Steinwand, S.; Hörndli, C.N.S.; Emery, J.; Huang, W.-C.; Kravitz, S.; Ferris, E.; Gregg, C. Noncanonical genomic imprinting in the monoamine system determines naturalistic foraging and brain-adrenal axis functions. Cell Rep. 2022, 38, 110500. [Google Scholar] [CrossRef] [PubMed]
- Hanna, C.W.; Kelsey, G. Features and mechanisms of canonical and noncanonical genomic imprinting. Genes Dev. 2021, 35, 821–834. [Google Scholar] [CrossRef]
- Okae, H.; Hiura, H.; Nishida, Y.; Funayama, R.; Tanaka, S.; Chiba, H.; Yaegashi, N.; Nakayama, K.; Sasaki, H.; Arima, T. Re-investigation and RNA sequencing-based identification of genes with placenta-specific imprinted expression. Hum. Mol. Genet. 2012, 21, 548–558. [Google Scholar] [CrossRef]
- Tucci, V.; Isles, A.R.; Kelsey, G.; Ferguson-Smith, A.C.; Erice Imprinting, G. Genomic Imprinting and Physiological Processes in Mammals. Cell 2019, 176, 952–965. [Google Scholar] [CrossRef]
- Hanna, C.W. Placental imprinting: Emerging mechanisms and functions. PLoS Genet. 2020, 16, e1008709. [Google Scholar] [CrossRef]
- Kobayashi, H. Canonical and Non-canonical Genomic Imprinting in Rodents. Front. Cell. Dev. Biol. 2021, 9, 713878. [Google Scholar] [CrossRef]
- Chen, Z.; Zhang, Y. Maternal H3K27me3-dependent autosomal and X chromosome imprinting. Nat. Rev. Genet. 2020, 21, 555–571. [Google Scholar] [CrossRef]
- Albert, J.R.; Greenberg, M.V. Non-canonical imprinting in the spotlight. Development 2023, 150, dev201087. [Google Scholar] [CrossRef] [PubMed]
- Edwards, C.A.; Watkinson, W.M.; Telerman, S.B.; Hulsmann, L.C.; Hamilton, R.S.; Ferguson-Smith, A.C. Reassessment of weak parent-of-origin expression bias shows it rarely exists outside of known imprinted regions. eLife 2023, 12, e83364. [Google Scholar] [CrossRef]
- Li, B.; Carey, M.; Workman, J.L. The role of chromatin during transcription. Cell 2007, 128, 707–719. [Google Scholar] [CrossRef]
- Navarrete-Modesto, V.; Orozco-Suarez, S.; Feria-Romero, I.A.; Rocha, L. The molecular hallmarks of epigenetic effects mediated by antiepileptic drugs. Epilepsy Res. 2019, 149, 53–65. [Google Scholar] [CrossRef] [PubMed]
- Czarny, P.; Bialek, K.; Ziolkowska, S.; Talarowska, M.; Śliwiński, T. Epigenetics in depression. In The Neuroscience of Depression; Elsevier: Amsterdam, The Netherlands, 2021; pp. 3–13. [Google Scholar]
- McEwen, K.R.; Ferguson-Smith, A.C. Genomic imprinting—A model for roles of histone modifications in epigenetic control. In Epigenomics; Springer: Dordrecht, The Netherlands, 2009; pp. 235–258. [Google Scholar]
- Brekke, T.D.; Henry, L.A.; Good, J.M. Genomic imprinting, disrupted placental expression, and speciation. Evolution 2016, 70, 2690–2703. [Google Scholar] [CrossRef]
- Jirtle, R.L.; Sander, M.; Barrett, J.C. Genomic imprinting and environmental disease susceptibility. Environ. Health Perspect. 2000, 108, 271–278. [Google Scholar] [CrossRef] [PubMed]
- Vrana, P.B.; Guan, X.-J.; Ingram, R.S. Genomic imprinting is disrupted in interspecific Peromyscus hybrids. Nat. Genet. 1998, 20, 362–365. [Google Scholar] [CrossRef]
- Burton, G.J.; Jauniaux, E. What is the placenta? Am. J. Obstet. Gynecol. 2015, 213, S6.e1–S6.e4. [Google Scholar] [CrossRef]
- Roberts, R.M.; Green, J.A.; Schulz, L.C. The evolution of the placenta. Reproduction 2016, 152, R179. [Google Scholar] [CrossRef]
- Selwood, L.; Johnson, M. Trophoblast and hypoblast in the monotreme, marsupial and eutherian mammal: Evolution and origins. BioEssays News Rev. Mol. Cell. Dev. Biol. 2006, 28, 128–145. [Google Scholar] [CrossRef]
- Lambertini, L.; Marsit, C.J.; Sharma, P.; Maccani, M.; Ma, Y.; Hu, J.; Chen, J. Imprinted gene expression in fetal growth and development. Placenta 2012, 33, 480–486. [Google Scholar] [CrossRef]
- Ursini, G.; Di Carlo, P.; Mukherjee, S.; Chen, Q.; Han, S.; Kim, J.; Deyssenroth, M.; Marsit, C.J.; Chen, J.; Hao, K. Prioritization of potential causative genes for schizophrenia in placenta. Nat. Commun. 2023, 14, 2613. [Google Scholar] [CrossRef] [PubMed]
- Leiser, R.; Kaufmann, P. Placental structure: In a comparative aspect. Exp. Clin. Endocrinol. Diabetes 1994, 102, 122–134. [Google Scholar] [CrossRef]
- Monk, D.; Arnaud, P.; Apostolidou, S.; Hills, F.; Kelsey, G.; Stanier, P.; Feil, R.; Moore, G. Limited evolutionary conservation of imprinting in the human placenta. Proc. Natl. Acad. Sci. USA 2006, 103, 6623–6628. [Google Scholar] [CrossRef] [PubMed]
- Morison, I.M.; Paton, C.J.; Cleverley, S.D. The imprinted gene and parent-of-origin effect database. Nucleic Acids Res. 2001, 29, 275–276. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Miller, D.C.; Harman, R.; Antczak, D.F.; Clark, A.G. Paternally expressed genes predominate in the placenta. Proc. Natl. Acad. Sci. USA 2013, 110, 10705–10710. [Google Scholar] [CrossRef]
- Allach El Khattabi, L.; Backer, S.; Pinard, A.; Dieudonné, M.-N.; Tsatsaris, V.; Vaiman, D.; Dandolo, L.; Bloch-Gallego, E.; Jammes, H.; Barbaux, S. A genome-wide search for new imprinted genes in the human placenta identifies DSCAM as the first imprinted gene on chromosome 21. Eur. J. Hum. Genet. 2019, 27, 49–60. [Google Scholar] [CrossRef]
- Yuen, R.K.; Jiang, R.; Penaherrera, M.S.; McFadden, D.E.; Robinson, W.P. Genome-wide mapping of imprinted differentially methylated regions by DNA methylation profiling of human placentas from triploidies. Epigenet. Chromatin 2011, 4, 1–16. [Google Scholar] [CrossRef]
- Tayama, C.; Romanelli, V.; Martin-Trujillo, A.; Iglesias-Platas, I.; Okamura, K.; Sugahara, N.; Simón, C.; Moore, H.; Harness, J.V.; Keirstead, H. Genome-wide parent-of-origin DNA methylation analysis reveals the intricacies of human imprinting and suggests a germline methylation-independent mechanism of establishment. Genome Res. 2014, 24, 554–569. [Google Scholar]
- Hanna, C.W.; Peñaherrera, M.S.; Saadeh, H.; Andrews, S.; McFadden, D.E.; Kelsey, G.; Robinson, W.P. Pervasive polymorphic imprinted methylation in the human placenta. Genome Res. 2016, 26, 756–767. [Google Scholar] [CrossRef]
- Sanchez-Delgado, M.; Court, F.; Vidal, E.; Medrano, J.; Monteagudo-Sánchez, A.; Martin-Trujillo, A.; Tayama, C.; Iglesias-Platas, I.; Kondova, I.; Bontrop, R. Human oocyte-derived methylation differences persist in the placenta revealing widespread transient imprinting. PLoS Genet. 2016, 12, e1006427. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, S.; Renfree, M.B.; Pask, A.J.; Shaw, G.; Kobayashi, S.; Kohda, T.; Kaneko-Ishino, T.; Ishino, F. Genomic imprinting of IGF2, p57KIP2 and PEG1/MEST in a marsupial, the tammar wallaby. Mech. Dev. 2005, 122, 213–222. [Google Scholar] [CrossRef] [PubMed]
- Ager, E.; Suzuki, S.; Pask, A.; Shaw, G.; Ishino, F.; Renfree, M.B. Insulin is imprinted in the placenta of the marsupial, Macropus eugenii. Dev. Biol. 2007, 309, 317–328. [Google Scholar] [CrossRef]
- Das, R.; Anderson, N.; Koran, M.I.; Weidman, J.R.; Mikkelsen, T.S.; Kamal, M.; Murphy, S.K.; Linblad-Toh, K.; Greally, J.M.; Jirtle, R.L. Convergent and divergent evolution of genomic imprinting in the marsupial Monodelphis domestica. BMC Genom. 2012, 13, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Renfree, M.B.; Suzuki, S.; Kaneko-Ishino, T. The origin and evolution of genomic imprinting and viviparity in mammals. Philos. Trans. R Soc. Lond. B Biol. Sci. 2013, 368, 20120151. [Google Scholar] [CrossRef]
- Frost, J.M.; Moore, G.E. The importance of imprinting in the human placenta. PLoS Genet. 2010, 6, e1001015. [Google Scholar] [CrossRef]
- Varmuza, S.; Miri, K. What does genetics tell us about imprinting and the placenta connection? Cell. Mol. Life Sci. 2015, 72, 51–72. [Google Scholar] [CrossRef]
- Reik, W.; Lewis, A. Co-evolution of X-chromosome inactivation and imprinting in mammals. Nat. Rev. Genet. 2005, 6, 403–410. [Google Scholar] [CrossRef]
- Huynh, K.D.; Lee, J.T. Inheritance of a pre-inactivated paternal X chromosome in early mouse embryos. Nature 2003, 426, 857–862. [Google Scholar] [CrossRef]
- Mak, W.; Nesterova, T.B.; De Napoles, M.; Appanah, R.; Yamanaka, S.; Otte, A.P.; Brockdorff, N. Reactivation of the paternal X chromosome in early mouse embryos. Science 2004, 303, 666–669. [Google Scholar] [CrossRef]
- Braidotti, G.; Baubec, T.; Pauler, F.; Seidl, C.; Smrzka, O.; Stricker, S.; Yotova, I.; Barlow, D. The Air noncoding RNA: An imprinted cis-silencing transcript. Cold Spring Harb. Symp. Quant. Biol. 2004, 69, 55–66. [Google Scholar] [CrossRef] [PubMed]
- Donker, R.B.; Mouillet, J.-F.; Chu, T.; Hubel, C.A.; Stolz, D.B.; Morelli, A.E.; Sadovsky, Y. The expression profile of C19MC microRNAs in primary human trophoblast cells and exosomes. Mol. Hum. Reprod. 2012, 18, 417–424. [Google Scholar] [CrossRef] [PubMed]
- Reish, O.; Lerer, I.; Amiel, A.; Heyman, E.; Herman, A.; Dolfin, T.; Abeliovich, D. Wiedemann-Beckwith syndrome: Further prenatal characterization of the condition. Am. J. Med. Genet. 2002, 107, 209–213. [Google Scholar] [CrossRef] [PubMed]
- Kagami, M.; Kurosawa, K.; Miyazaki, O.; Ishino, F.; Matsuoka, K.; Ogata, T. Comprehensive clinical studies in 34 patients with molecularly defined UPD (14) pat and related conditions (Kagami–Ogata syndrome). Eur. J. Hum. Genet. 2015, 23, 1488–1498. [Google Scholar] [CrossRef]
- Kobayashi, E.H.; Shibata, S.; Oike, A.; Kobayashi, N.; Hamada, H.; Okae, H.; Arima, T. Genomic imprinting in human placentation. Reprod. Med. Biol. 2022, 21, e12490. [Google Scholar] [CrossRef]
- Sekita, Y.; Wagatsuma, H.; Nakamura, K.; Ono, R.; Kagami, M.; Wakisaka, N.; Hino, T.; Suzuki-Migishima, R.; Kohda, T.; Ogura, A. Role of retrotransposon-derived imprinted gene, Rtl1, in the feto-maternal interface of mouse placenta. Nat. Genet. 2008, 40, 243–248. [Google Scholar] [CrossRef]
- McMinn, J.; Wei, M.; Schupf, N.; Cusmai, J.; Johnson, E.; Smith, A.; Weksberg, R.; Thaker, H.; Tycko, B. Unbalanced placental expression of imprinted genes in human intrauterine growth restriction. Placenta 2006, 27, 540–549. [Google Scholar] [CrossRef]
- Diplas, A.I.; Lambertini, L.; Lee, M.-J.; Sperling, R.; Lee, Y.L.; Wetmur, J.G.; Chen, J. Differential expression of imprinted genes in normal and IUGR human placentas. Epigenetics 2009, 4, 235–240. [Google Scholar] [CrossRef]
- Monk, D. Genomic imprinting in the human placenta. Am. J. Obstet. Gynecol. 2015, 213, S152–S162. [Google Scholar] [CrossRef]
- Sachs, M.; Brohmann, H.; Zechner, D.; Müller, T.; Hülsken, J.; Walther, I.; Schaeper, U.; Birchmeier, C.; Birchmeier, W. Essential role of Gab1 for signaling by the c-Met receptor in vivo. J. Cell Biol. 2000, 150, 1375–1384. [Google Scholar] [CrossRef]
- Miri, K.; Latham, K.; Panning, B.; Zhong, Z.; Andersen, A.; Varmuza, S. The imprinted polycomb group gene Sfmbt2 is required for trophoblast maintenance and placenta development. Development 2013, 140, 4480–4489. [Google Scholar] [CrossRef]
- Zadora, J.; Singh, M.; Herse, F.; Przybyl, L.; Haase, N.; Golic, M.; Yung, H.W.; Huppertz, B.; Cartwright, J.E.; Whitley, G.; et al. Disturbed Placental Imprinting in Preeclampsia Leads to Altered Expression of DLX5, a Human-Specific Early Trophoblast Marker. Circulation 2017, 136, 1824–1839. [Google Scholar] [CrossRef] [PubMed]
- Deyssenroth, M.A.; Li, Q.; Escudero, C.; Myatt, L.; Chen, J.; Roberts, J.M. Differences in placental imprinted gene expression across preeclamptic and non-preeclamptic pregnancies. Genes 2020, 11, 1146. [Google Scholar] [CrossRef] [PubMed]
- Wilkinson, L.S.; Davies, W.; Isles, A.R. Genomic imprinting effects on brain development and function. Nat. Rev. Neurosci. 2007, 8, 832–843. [Google Scholar] [CrossRef]
- Gregg, C.; Zhang, J.; Butler, J.E.; Haig, D.; Dulac, C. Sex-specific parent-of-origin allelic expression in the mouse brain. Science 2010, 329, 682–685. [Google Scholar] [CrossRef]
- Perez, J.D.; Rubinstein, N.D.; Fernandez, D.E.; Santoro, S.W.; Needleman, L.A.; Ho-Shing, O.; Choi, J.J.; Zirlinger, M.; Chen, S.-K.; Liu, J.S. Quantitative and functional interrogation of parent-of-origin allelic expression biases in the brain. eLife 2015, 4, e07860. [Google Scholar] [CrossRef]
- Keverne, E.B.; Fundele, R.; Narasimha, M.; Barton, S.C.; Surani, M.A. Genomic imprinting and the differential roles of parental genomes in brain development. Dev. Brain Res. 1996, 92, 91–100. [Google Scholar] [CrossRef]
- Champagne, F.A.; Curley, J.P.; Swaney, W.T.; Hasen, N.; Keverne, E.B. Paternal influence on female behavior: The role of Peg3 in exploration, olfaction, and neuroendocrine regulation of maternal behavior of female mice. Behav. Neurosci. 2009, 123, 469. [Google Scholar] [CrossRef] [PubMed]
- Green, B.B.; Kappil, M.; Lambertini, L.; Armstrong, D.A.; Guerin, D.J.; Sharp, A.J.; Lester, B.M.; Chen, J.; Marsit, C.J. Expression of imprinted genes in placenta is associated with infant neurobehavioral development. Epigenetics 2015, 10, 834–841. [Google Scholar] [CrossRef]
- Frim, D.; Emanuel, R.; Robinson, B.; Smas, C.; Adler, G.; Majzoub, J. Characterization and gestational regulation of corticotropin-releasing hormone messenger RNA in human placenta. J. Clin. Investig. 1988, 82, 287–292. [Google Scholar] [CrossRef]
- Petraglia, F.; Potter, E.; Cameron, V.; Sutton, S.; Behan, D.; Woods, R.; Sawchenko, P.; Lowry, P.; Vale, W. Corticotropin-releasing factor-binding protein is produced by human placenta and intrauterine tissues. J. Clin. Endocrinol. Metab. 1993, 77, 919–924. [Google Scholar] [PubMed]
- Lambertini, L. Genomic imprinting: Sensing the environment and driving the fetal growth. Curr. Opin. Pediatr. 2014, 26, 237–242. [Google Scholar] [CrossRef] [PubMed]
- Woods, R.M.; Lorusso, J.M.; Fletcher, J.; ElTaher, H.; McEwan, F.; Harris, I.; Kowash, H.M.; D’Souza, S.W.; Harte, M.; Hager, R. Maternal immune activation and role of placenta in the prenatal programming of neurodevelopmental disorders. Neuronal Signal. 2023, 7, NS20220064. [Google Scholar] [CrossRef] [PubMed]
- Puelles, L.; Sandoval, J.; Ayad, A.; Del Corral, R.; Alonso, A.; Ferran, J.; Martínez-De-La-Torre, M. The pallium in reptiles and birds in the light of the updated tetrapartite pallium model. Evol. Nerv. Syst. 2017, 1, 519–555. [Google Scholar]
- Reiner, A.; Perkel, D.J.; Bruce, L.L.; Butler, A.B.; Csillag, A.; Kuenzel, W.; Medina, L.; Paxinos, G.; Shimizu, T.; Striedter, G. Revised nomenclature for avian telencephalon and some related brainstem nuclei. J. Comp. Neurol. 2004, 473, 377–414. [Google Scholar] [CrossRef]
- Ji, L.; Bishayee, K.; Sadra, A.; Choi, S.; Choi, W.; Moon, S.; Jho, E.-H.; Huh, S.-O. Defective neuronal migration and inhibition of bipolar to multipolar transition of migrating neural cells by Mesoderm-Specific Transcript, Mest, in the developing mouse neocortex. Neuroscience 2017, 355, 126–140. [Google Scholar] [CrossRef]
- Krubitzer, L.; Manger, P.; Pettigrew, J.; Calford, M. Organization of somatosensory cortex in monotremes: In search of the prototypical plan. J. Comp. Neurol. 1995, 351, 261–306. [Google Scholar] [CrossRef]
- Cheung, A.F.; Kondo, S.; Abdel-Mannan, O.; Chodroff, R.A.; Sirey, T.M.; Bluy, L.E.; Webber, N.; DeProto, J.; Karlen, S.J.; Krubitzer, L. The subventricular zone is the developmental milestone of a 6-layered neocortex: Comparisons in metatherian and eutherian mammals. Cereb. Cortex 2010, 20, 1071–1081. [Google Scholar] [CrossRef]
- Kaas, J.H. Neocortex in early mammals and its subsequent variations. Ann. N. Y. Acad. Sci. 2011, 1225, 28–36. [Google Scholar] [CrossRef]
- Franchini, L.F. Genetic mechanisms underlying cortical evolution in mammals. Front. Cell Dev. Biol. 2021, 9, 591017. [Google Scholar] [CrossRef]
- Azevedo, F.A.; Carvalho, L.R.; Grinberg, L.T.; Farfel, J.M.; Ferretti, R.E.; Leite, R.E.; Filho, W.J.; Lent, R.; Herculano-Houzel, S. Equal numbers of neuronal and nonneuronal cells make the human brain an isometrically scaled-up primate brain. J. Comp. Neurol. 2009, 513, 532–541. [Google Scholar] [CrossRef] [PubMed]
- Changizi, M.A.; Shimojo, S. Parcellation and area-area connectivity as a function of neocortex size. Brain Behav. Evol. 2005, 66, 88–98. [Google Scholar] [CrossRef] [PubMed]
- Suárez, R.; Paolino, A.; Fenlon, L.R.; Morcom, L.R.; Kozulin, P.; Kurniawan, N.D.; Richards, L.J. A pan-mammalian map of interhemispheric brain connections predates the evolution of the corpus callosum. Proc. Natl. Acad. Sci. USA 2018, 115, 9622–9627. [Google Scholar] [CrossRef] [PubMed]
- Paul, L.K.; Van Lancker-Sidtis, D.; Schieffer, B.; Dietrich, R.; Brown, W.S. Communicative deficits in agenesis of the corpus callosum: Nonliteral language and affective prosody. Brain Lang. 2003, 85, 313–324. [Google Scholar] [CrossRef]
- Badaruddin, D.H.; Andrews, G.L.; Bölte, S.; Schilmoeller, K.J.; Schilmoeller, G.; Paul, L.K.; Brown, W.S. Social and behavioral problems of children with agenesis of the corpus callosum. Child Psychiatry Hum. Dev. 2007, 38, 287–302. [Google Scholar] [CrossRef]
- Isles, A.R. The contribution of imprinted genes to neurodevelopmental and neuropsychiatric disorders. Transl. Psychiatry 2022, 12, 210. [Google Scholar] [CrossRef]
- Imaizumi, Y.; Furutachi, S.; Watanabe, T.; Miya, H.; Kawaguchi, D.; Gotoh, Y. Role of the imprinted allele of the Cdkn1c gene in mouse neocortical development. Sci. Rep. 2020, 10, 1884. [Google Scholar] [CrossRef]
- Adnani, L.; Langevin, L.M.; Gautier, E.; Dixit, R.; Parsons, K.; Li, S.; Kaushik, G.; Wilkinson, G.; Wilson, R.; Childs, S. Zac1 regulates the differentiation and migration of neocortical neurons via Pac1. J. Neurosci. 2015, 35, 13430–13447. [Google Scholar] [CrossRef]
- Rraklli, V.; Södersten, E.; Nyman, U.; Hagey, D.W.; Holmberg, J. Elevated levels of ZAC1 disrupt neurogenesis and promote rapid in vivo reprogramming. Stem Cell Res. 2016, 16, 1–9. [Google Scholar] [CrossRef]
- Martinez, M.E.; Charalambous, M.; Saferali, A.; Fiering, S.; Naumova, A.K.; St Germain, D.; Ferguson-Smith, A.C.; Hernandez, A. Genomic imprinting variations in the mouse type 3 deiodinase gene between tissues and brain regions. Mol. Endocrinol. 2014, 28, 1875–1886. [Google Scholar] [CrossRef]
- Ziegler, M.; Russell, B.E.; Eberhardt, K.; Geisel, G.; D’Amore, A.; Sahin, M.; Kornblum, H.I.; Ebrahimi-Fakhari, D. Blended phenotype of Silver-Russell syndrome and SPG50 caused by maternal isodisomy of chromosome 7. Neurol. Genet. 2020, 7, e544. [Google Scholar] [CrossRef]
- Tunster, S.J.; Creeth, H.; John, R.M. The imprinted Phlda2 gene modulates a major endocrine compartment of the placenta to regulate placental demands for maternal resources. Dev. Biol. 2016, 409, 251–260. [Google Scholar] [CrossRef] [PubMed]
- Apostolidou, S.; Abu-Amero, S.; O’donoghue, K.; Frost, J.; Olafsdottir, O.; Chavele, K.; Whittaker, J.; Loughna, P.; Stanier, P.; Moore, G. Elevated placental expression of the imprinted PHLDA2 gene is associated with low birth weight. J. Mol. Med. 2007, 85, 379–387. [Google Scholar] [CrossRef] [PubMed]
- Creeth, H.D.J.; McNamara, G.I.; Tunster, S.J.; Boque-Sastre, R.; Allen, B.; Sumption, L.; Eddy, J.B.; Isles, A.R.; John, R.M. Maternal care boosted by paternal imprinting in mammals. PLoS Biol. 2018, 16, e2006599. [Google Scholar] [CrossRef] [PubMed]
- Stringer, J.M.; Suzuki, S.; Pask, A.J.; Shaw, G.; Renfree, M.B. Selected imprinting of INS in the marsupial. Epigenet. Chromatin 2012, 5, 1–12. [Google Scholar] [CrossRef]
- Cowley, M.; Garfield, A.S.; Madon-Simon, M.; Charalambous, M.; Clarkson, R.W.; Smalley, M.J.; Kendrick, H.; Isles, A.R.; Parry, A.J.; Carney, S. Developmental programming mediated by complementary roles of imprinted Grb10 in mother and pup. PLoS Biol. 2014, 12, e1001799. [Google Scholar] [CrossRef]
- Garfield, A.S.; Cowley, M.; Smith, F.M.; Moorwood, K.; Stewart-Cox, J.E.; Gilroy, K.; Baker, S.; Xia, J.; Dalley, J.W.; Hurst, L.D. Distinct physiological and behavioural functions for parental alleles of imprinted Grb10. Nature 2011, 469, 534–538. [Google Scholar] [CrossRef]
- McNamara, G.I.; Davis, B.A.; Browne, M.; Humby, T.; Dalley, J.W.; Xia, J.; John, R.M.; Isles, A.R. Dopaminergic and behavioural changes in a loss-of-imprinting model of Cdkn1c. Genes Brain Behav. 2018, 17, 149–157. [Google Scholar] [CrossRef]
| DNMTs | Function | References |
|---|---|---|
| DNMT1 | mainly maintains methylation and therefore imprints | [32,118] |
| DNMT2 | transfer RNA methylation | |
| DNMT3 | de novo methylation of unmethylated DNA | |
| DNMT3A | imprinting and methylation at major satellite repeats; maternal-specific germline DMR methylation occurs postnatally in growing oocytes but prenatally in prospermatogonia before onset of meiosis in male germline | [119,120] |
| DNMT3B | minor satellite repeat methylation | [121] |
| Dnmt3c (rodents) | methylates young retrotransposons in male germ line | [122] |
| DNMT3L | contains no catalytic domain for methylation, instead, it co-orchestrates de novo methylation; establishes genomic imprints in oocyte | [123,124] |
| Canonical Imprinting | Non-Canonical Imprinting | ASE | |
|---|---|---|---|
| Establishment of imprinting | DNA methylation in oocyte or sperm at imprinting control region | H3K27me3 H2AK119ub | Allele-specific histone modifications (H3K9ac; H3K9me3) |
| Established by | Oocyte: Dnmt3a, Dnmt3L, Kdm1b, Zfp57; Sperm: Dnmt3a, Dnmt3b, Dnmt3L, piRNA, CTCFL/Prm7 | PRC2, Pcg1/6-PRC1 | Unclear |
| Maintenance of imprinting | ZFP57 and/or ZNF445 required to maintain DNA-methylation of imprinting control regions | H3K27me3 replaced by monoallelic DNA methylation | Unclear |
| Regulated by | Dnmt1, Dppa3, Zfp57 | PRC2, Smchd1 | Unclear |
| Set of imprinting | During oogenesis and spermatogenesis | During oogenesis | Unclear |
| Location | Widely distributed over the body, predominantly in neural and placental tissues | Extraembryonic and embryonic cell lineages | Widely distributed over the body, highly tissue-specific |
| Conservation between species | High | Low | Unclear |
| Clinical picture of imprinting abnormalities | Infertility, moles, miscarriage, classical clinical pictures of imprinting disorders such as Angelman, Beckwith–Wiedemann or Prader–Willi syndrome | Placental defects in murine loss of function/loss of prominent imprinting mutants with (sub)-lethality | Unclear |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Schuff, M.; Strong, A.D.; Welborn, L.K.; Ziermann-Canabarro, J.M. Imprinting as Basis for Complex Evolutionary Novelties in Eutherians. Biology 2024, 13, 682. https://doi.org/10.3390/biology13090682
Schuff M, Strong AD, Welborn LK, Ziermann-Canabarro JM. Imprinting as Basis for Complex Evolutionary Novelties in Eutherians. Biology. 2024; 13(9):682. https://doi.org/10.3390/biology13090682
Chicago/Turabian StyleSchuff, Maximillian, Amanda D. Strong, Lyvia K. Welborn, and Janine M. Ziermann-Canabarro. 2024. "Imprinting as Basis for Complex Evolutionary Novelties in Eutherians" Biology 13, no. 9: 682. https://doi.org/10.3390/biology13090682
APA StyleSchuff, M., Strong, A. D., Welborn, L. K., & Ziermann-Canabarro, J. M. (2024). Imprinting as Basis for Complex Evolutionary Novelties in Eutherians. Biology, 13(9), 682. https://doi.org/10.3390/biology13090682








