Purifying Selection Influences the Comparison of Heterozygosities between Populations
Abstract
Simple Summary
Abstract
1. Introduction
2. Methods
2.1. Quantifying the Excess Fraction of Deleterious Variants Segregating in Small Populations
2.2. Genome and Gene Expression Data
2.3. Data Analysis
2.4. Statistical Analysis
3. Results
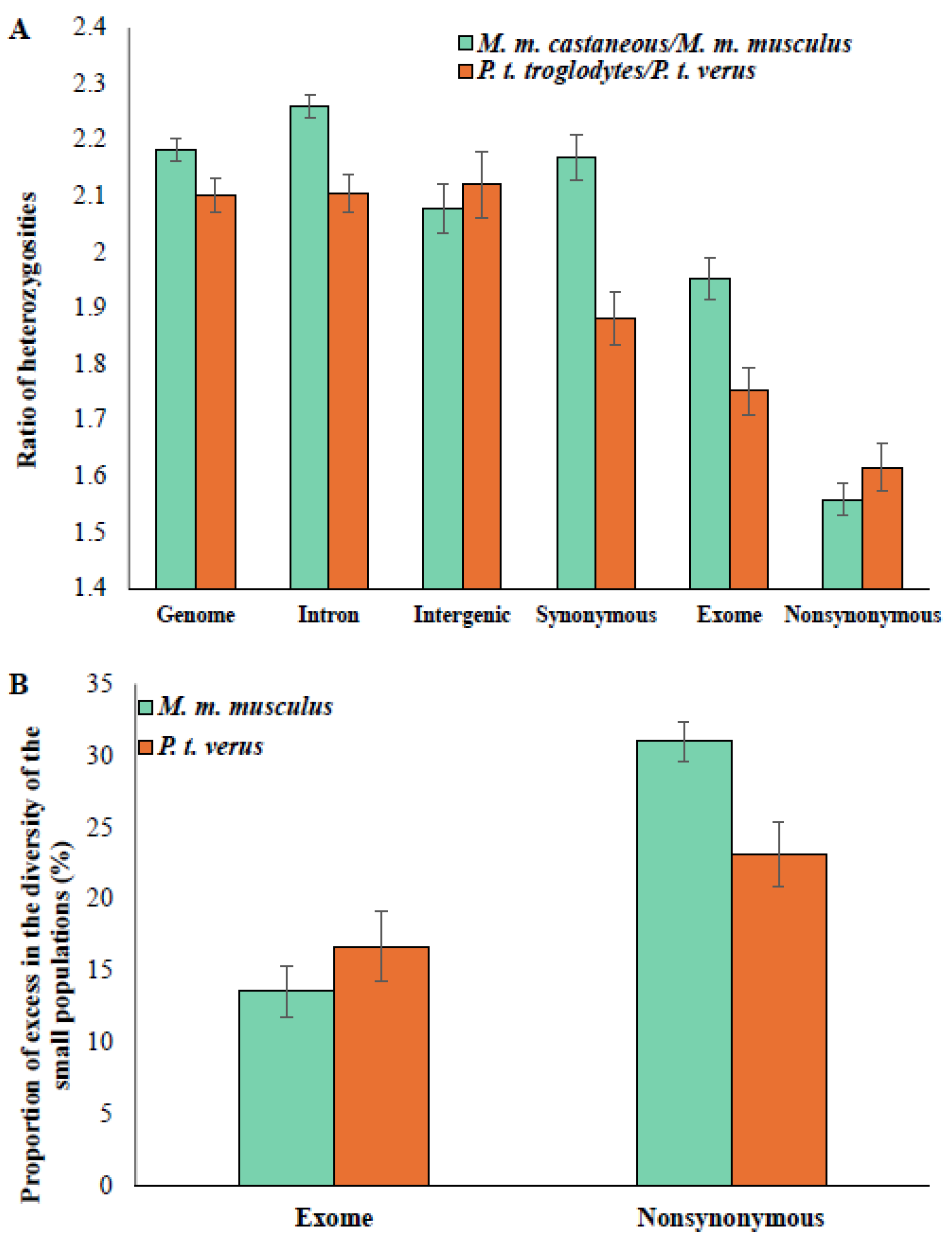
3.1. The Excess in the Constrained-Site Diversity of Small Populations
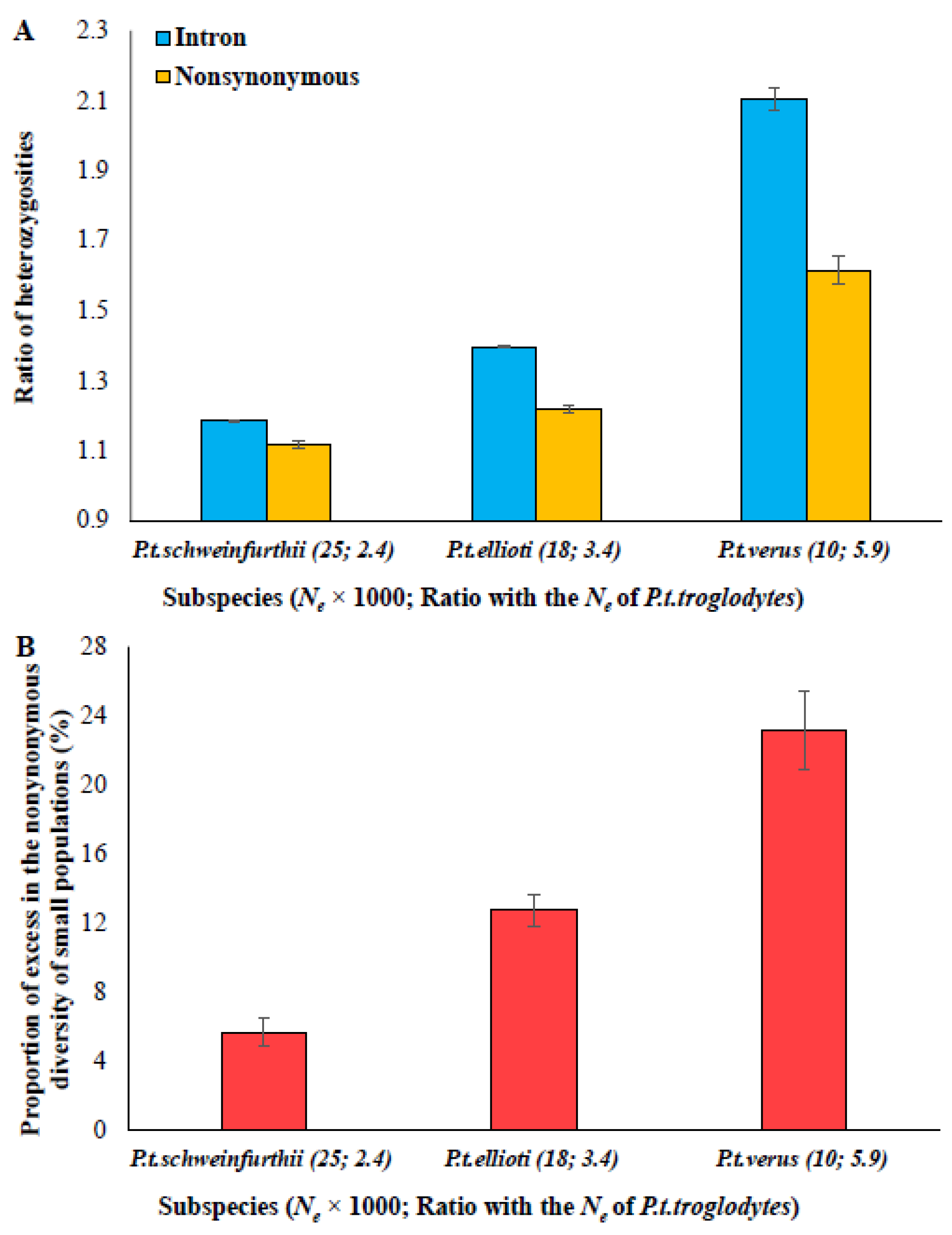
3.2. The Magnitude of the Excess Fraction Correlates with Effective Population Size (Ne)
3.3. Positive Relationship between the Magnitude of the Excess and Selection Intensity (s)
4. Discussion
5. Conclusions
Supplementary Materials
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hartl, D.L.; Clark, G.C. Principles of Population Genetics, 4th ed.; Sinauer and Associates: Sunderland, MA, USA, 2007. [Google Scholar]
- Nei, M.; Kumar, S. Molecular Evolution and Phylogenetics; Oxford University Press: New York, NY, USA, 2000. [Google Scholar]
- Kimura, M. The Neutral Theory of Molecular Evolution; Oxford University Press: Oxford, UK, 1983. [Google Scholar]
- Briggs, A.W.; Good, J.M.; Green, R.E.; Krause, J.; Maricic, T.; Stenzel, U.; Lalueza-Fox, C.; Rudan, P.; Brajkovic, D.; Kucan, Z.; et al. Targeted retrieval and analysis of five Neandertal mtDNA genomes. Science 2009, 325, 318–321. [Google Scholar] [CrossRef] [PubMed]
- Green, R.E.; Malaspinas, A.S.; Krause, J.; Briggs, A.W.; Johnson, P.L.; Uhler, C.; Meyer, M.; Good, J.M.; Maricic, T.; Stenzel, U.; et al. A complete Neandertal mitochondrial genome sequence determined by high-throughput sequencing. Cell 2008, 134, 416–426. [Google Scholar] [CrossRef] [PubMed]
- Prado-Martinez, J.; Sudmant, P.H.; Kidd, J.M.; Li, H.; Kelley, J.L.; Lorente-Galdos, B.; Veeramah, K.R.; Woerner, A.E.; O’Connor, T.D.; Santpere, G.; et al. Great ape genetic diversity and population history. Nature 2013, 499, 471–475. [Google Scholar] [CrossRef]
- Robinson, J.A.; Ortega-Del Vecchyo, D.; Fan, Z.; Kim, B.Y.; vonHoldt, B.M.; Marsden, C.D.; Lohmueller, K.E.; Wayne, R.K. Genomic Flatlining in the Endangered Island Fox. Curr. Biol. 2016, 26, 1183–1189. [Google Scholar] [CrossRef]
- Rogers, R.L.; Slatkin, M. Excess of genomic defects in a woolly mammoth on Wrangel island. PLoS Genet. 2017, 13, e1006601. [Google Scholar] [CrossRef]
- Subramanian, S.; Kumar, M. Genomic footprints of bottleneck in landlocked salmon population. Sci. Rep. 2023, 13, 6706. [Google Scholar] [CrossRef] [PubMed]
- Kumar, M.; Conroy, G.; Ogbourne, S.; Cairns, K.; Borburgh, L.; Subramanian, S. Genomic signatures of bottleneck and founder effects in dingoes. Ecol. Evol. 2023, 13, e10525. [Google Scholar] [CrossRef]
- Leroy, T.; Rousselle, M.; Tilak, M.K.; Caizergues, A.E.; Scornavacca, C.; Recuerda, M.; Fuchs, J.; Illera, J.C.; De Swardt, D.H.; Blanco, G.; et al. Island songbirds as windows into evolution in small populations. Curr. Biol. 2021, 31, 1303–1310.e4. [Google Scholar] [CrossRef]
- Bosse, M.; Megens, H.J.; Derks, M.F.L.; de Cara, A.M.R.; Groenen, M.A.M. Deleterious alleles in the context of domestication, inbreeding, and selection. Evol. Appl. 2019, 12, 6–17. [Google Scholar] [CrossRef]
- Marsden, C.D.; Ortega-Del Vecchyo, D.; O’Brien, D.P.; Taylor, J.F.; Ramirez, O.; Vila, C.; Marques-Bonet, T.; Schnabel, R.D.; Wayne, R.K.; Lohmueller, K.E. Bottlenecks and selective sweeps during domestication have increased deleterious genetic variation in dogs. Proc. Natl. Acad. Sci. USA 2016, 113, 152–157. [Google Scholar] [CrossRef]
- Makino, T.; Rubin, C.J.; Carneiro, M.; Axelsson, E.; Andersson, L.; Webster, M.T. Elevated Proportions of Deleterious Genetic Variation in Domestic Animals and Plants. Genome Biol. Evol. 2018, 10, 276–290. [Google Scholar] [CrossRef] [PubMed]
- Xie, X.; Yang, Y.; Ren, Q.; Ding, X.; Bao, P.; Yan, B.; Yan, X.; Han, J.; Yan, P.; Qiu, Q. Accumulation of deleterious mutations in the domestic yak genome. Anim. Genet. 2018, 49, 384–392. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Tang, T.; Tang, H.; Huang, J.; Shi, S.; Wu, C.I. The accumulation of deleterious mutations in rice genomes: A hypothesis on the cost of domestication. Trends Genet. 2006, 22, 126–131. [Google Scholar] [CrossRef]
- Renaut, S.; Rieseberg, L.H. The Accumulation of Deleterious Mutations as a Consequence of Domestication and Improvement in Sunflowers and Other Compositae Crops. Mol. Biol. Evol. 2015, 32, 2273–2283. [Google Scholar] [CrossRef]
- Subramanian, S. Deleterious protein-coding variants in diverse cattle breeds of the world. Genet. Sel. Evol. 2021, 53, 80. [Google Scholar] [CrossRef]
- Fujiwara, K.; Kawai, Y.; Takada, T.; Shiroishi, T.; Saitou, N.; Suzuki, H.; Osada, N. Insights into Mus musculus Population Structure across Eurasia Revealed by Whole-Genome Analysis. Genome Biol. Evol. 2022, 14, evac068. [Google Scholar] [CrossRef] [PubMed]
- Pal, C.; Papp, B.; Hurst, L.D. Highly expressed genes in yeast evolve slowly. Genetics 2001, 158, 927–931. [Google Scholar] [CrossRef]
- Subramanian, S.; Kumar, S. Gene expression intensity shapes evolutionary rates of the proteins encoded by the vertebrate genome. Genetics 2004, 168, 373–381. [Google Scholar] [CrossRef]
- McDonald, J.H.; Kreitman, M. Adaptive protein evolution at the Adh locus in Drosophila. Nature 1991, 351, 652–654. [Google Scholar] [CrossRef]
- Hasegawa, M.; Cao, Y.; Yang, Z. Preponderance of slightly deleterious polymorphism in mitochondrial DNA: Nonsynonymous/synonymous rate ratio is much higher within species than between species. Mol. Biol. Evol. 1998, 15, 1499–1505. [Google Scholar] [CrossRef][Green Version]
- Nachman, M.W.; Brown, W.M.; Stoneking, M.; Aquadro, C.F. Nonneutral mitochondrial DNA variation in humans and chimpanzees. Genetics 1996, 142, 953–963. [Google Scholar] [CrossRef]
- Rand, D.M.; Kann, L.M. Excess amino acid polymorphism in mitochondrial DNA: Contrasts among genes from Drosophila, mice, and humans. Mol. Biol. Evol. 1996, 13, 735–748. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, S. High proportions of deleterious polymorphisms in constrained human genes. Mol. Biol. Evol. 2011, 28, 49–52. [Google Scholar] [CrossRef]
- De Baets, G.; Van Durme, J.; Reumers, J.; Maurer-Stroh, S.; Vanhee, P.; Dopazo, J.; Schymkowitz, J.; Rousseau, F. SNPeffect 4.0: On-line prediction of molecular and structural effects of protein-coding variants. Nucleic Acids Res. 2012, 40, D935–D939. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z. PAML 4: Phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 2007, 24, 1586–1591. [Google Scholar] [CrossRef]
- Pollard, K.S.; Hubisz, M.J.; Rosenbloom, K.R.; Siepel, A. Detection of nonneutral substitution rates on mammalian phylogenies. Genome Res. 2010, 20, 110–121. [Google Scholar] [CrossRef]
- Su, A.I.; Wiltshire, T.; Batalov, S.; Lapp, H.; Ching, K.A.; Block, D.; Zhang, J.; Soden, R.; Hayakawa, M.; Kreiman, G.; et al. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc. Natl. Acad. Sci. USA 2004, 101, 6062–6067. [Google Scholar] [CrossRef]
- Tajima, F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 1989, 123, 585–595. [Google Scholar] [CrossRef]
- Geraldes, A.; Basset, P.; Gibson, B.; Smith, K.L.; Harr, B.; Yu, H.T.; Bulatova, N.; Ziv, Y.; Nachman, M.W. Inferring the history of speciation in house mice from autosomal, X-linked, Y-linked and mitochondrial genes. Mol. Ecol. 2008, 17, 5349–5363. [Google Scholar] [CrossRef]
- Halligan, D.L.; Oliver, F.; Eyre-Walker, A.; Harr, B.; Keightley, P.D. Evidence for pervasive adaptive protein evolution in wild mice. PLoS Genet. 2010, 6, e1000825. [Google Scholar] [CrossRef]
- Phifer-Rixey, M.; Bonhomme, F.; Boursot, P.; Churchill, G.A.; Pialek, J.; Tucker, P.K.; Nachman, M.W. Adaptive evolution and effective population size in wild house mice. Mol. Biol. Evol. 2012, 29, 2949–2955. [Google Scholar] [CrossRef] [PubMed]
- Salcedo, T.; Geraldes, A.; Nachman, M.W. Nucleotide variation in wild and inbred mice. Genetics 2007, 177, 2277–2291. [Google Scholar] [CrossRef] [PubMed]
- Rogers, A.R. Genetic Evidence for Geographic Structure within the Neanderthal Population. Peer Community J. 2024, 4, e68. [Google Scholar] [CrossRef]
- Parmley, J.L.; Chamary, J.V.; Hurst, L.D. Evidence for purifying selection against synonymous mutations in mammalian exonic splicing enhancers. Mol. Biol. Evol. 2006, 23, 301–309. [Google Scholar] [CrossRef]
- Shields, D.C.; Sharp, P.M.; Higgins, D.G.; Wright, F. “Silent” sites in Drosophila genes are not neutral: Evidence of selection among synonymous codons. Mol. Biol. Evol. 1988, 5, 704–716. [Google Scholar] [CrossRef]
- Charlesworth, B. Background selection 20 years on: The Wilhelmine E. Key 2012 invitational lecture. J. Hered. 2013, 104, 161–171. [Google Scholar] [CrossRef] [PubMed]
- Charlesworth, D.; Charlesworth, B.; Morgan, M.T. The pattern of neutral molecular variation under the background selection model. Genetics 1995, 141, 1619–1632. [Google Scholar] [CrossRef]
- McVicker, G.; Gordon, D.; Davis, C.; Green, P. Widespread genomic signatures of natural selection in hominid evolution. PLoS Genet. 2009, 5, e1000471. [Google Scholar] [CrossRef]
- Nam, K.; Munch, K.; Mailund, T.; Nater, A.; Greminger, M.P.; Krutzen, M.; Marques-Bonet, T.; Schierup, M.H. Evidence that the rate of strong selective sweeps increases with population size in the great apes. Proc. Natl. Acad. Sci. USA 2017, 114, 1613–1618. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Subramanian, S. Purifying Selection Influences the Comparison of Heterozygosities between Populations. Biology 2024, 13, 810. https://doi.org/10.3390/biology13100810
Subramanian S. Purifying Selection Influences the Comparison of Heterozygosities between Populations. Biology. 2024; 13(10):810. https://doi.org/10.3390/biology13100810
Chicago/Turabian StyleSubramanian, Sankar. 2024. "Purifying Selection Influences the Comparison of Heterozygosities between Populations" Biology 13, no. 10: 810. https://doi.org/10.3390/biology13100810
APA StyleSubramanian, S. (2024). Purifying Selection Influences the Comparison of Heterozygosities between Populations. Biology, 13(10), 810. https://doi.org/10.3390/biology13100810





