Identification of Cuproptosis-Associated Prognostic Gene Expression Signatures from 20 Tumor Types
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Compilation of Cuproptosis-Associated Genes
2.2. Kaplan–Meier Survival Statistics
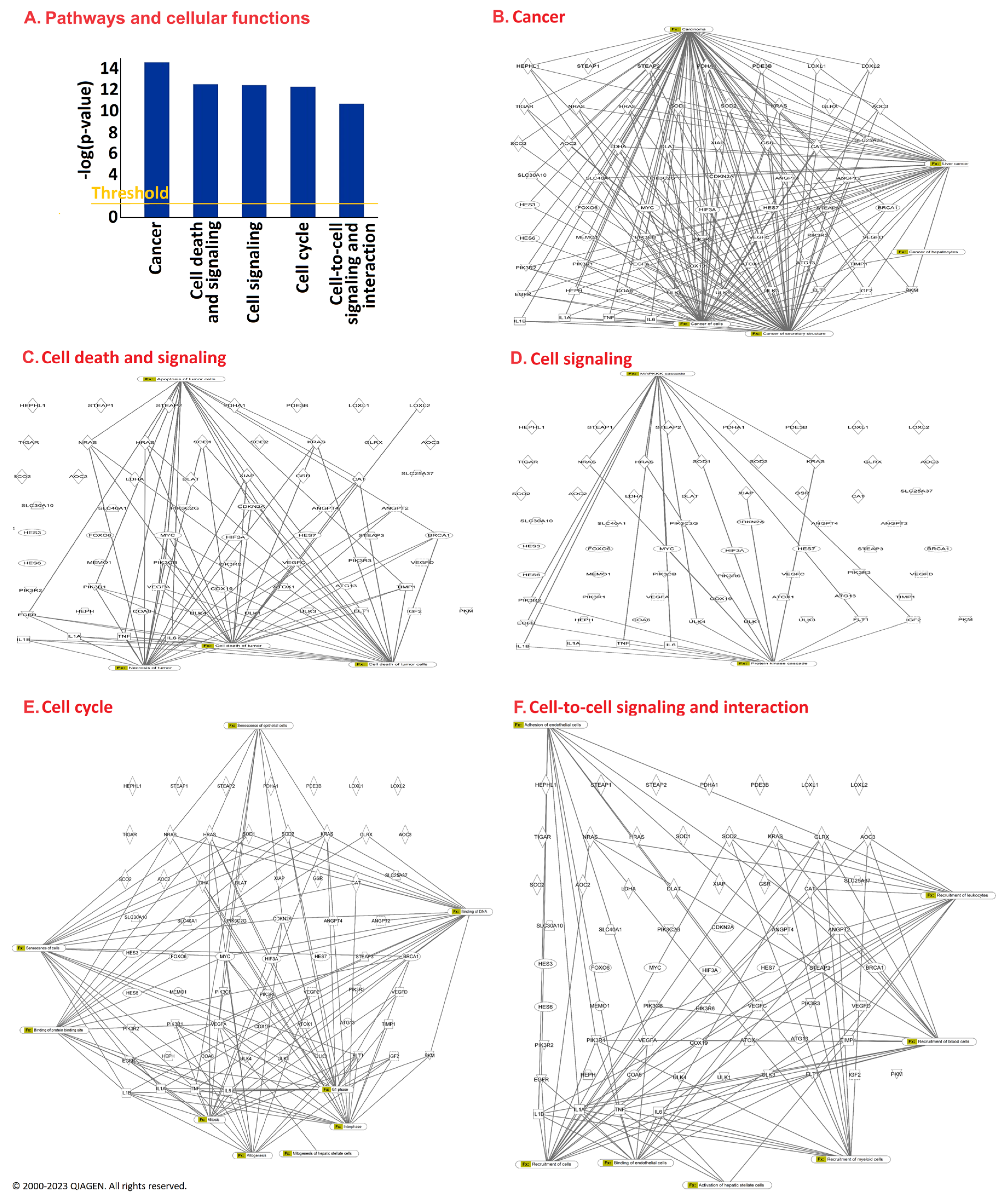
2.3. Ingenuity Pathway Analysis
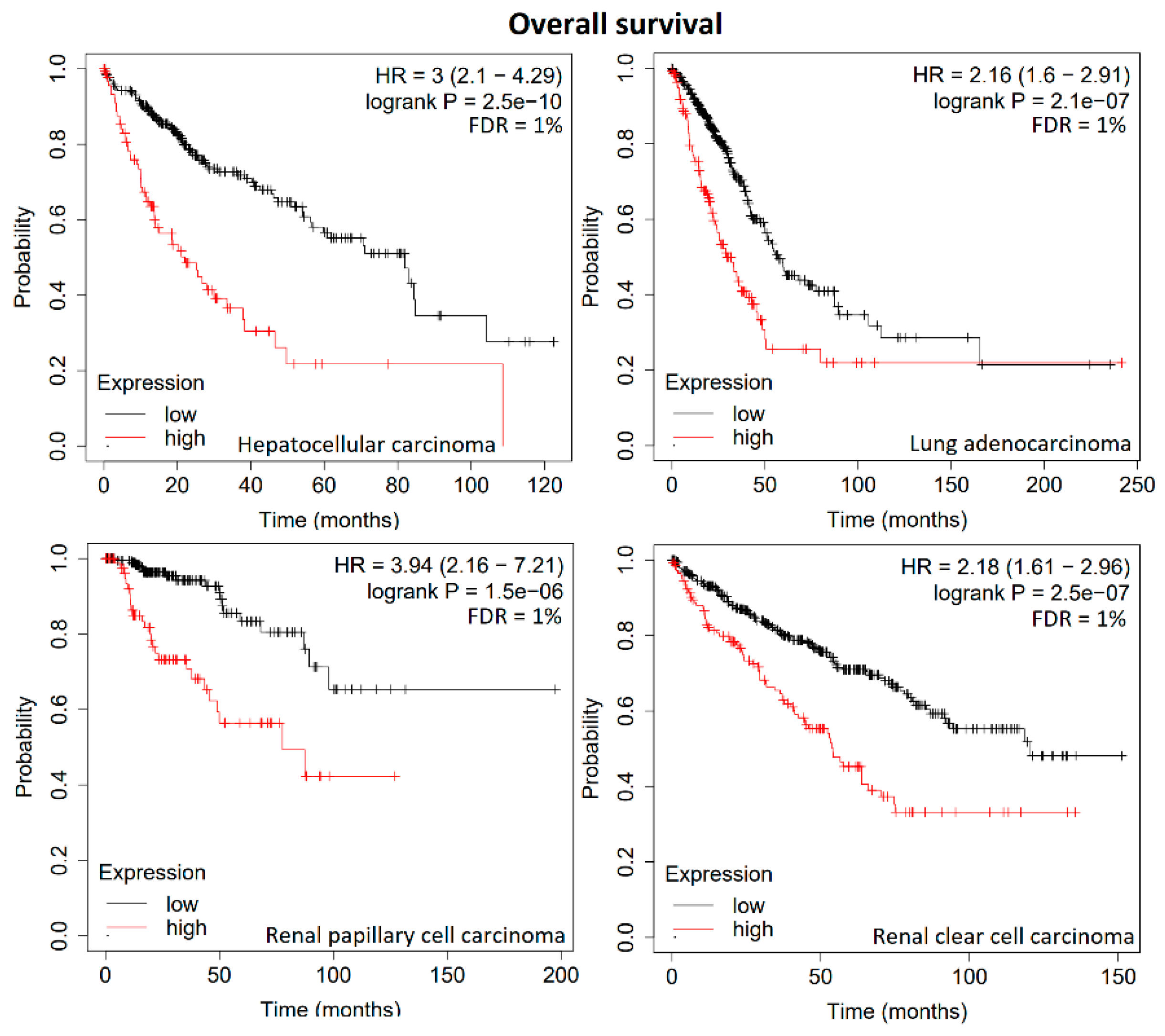
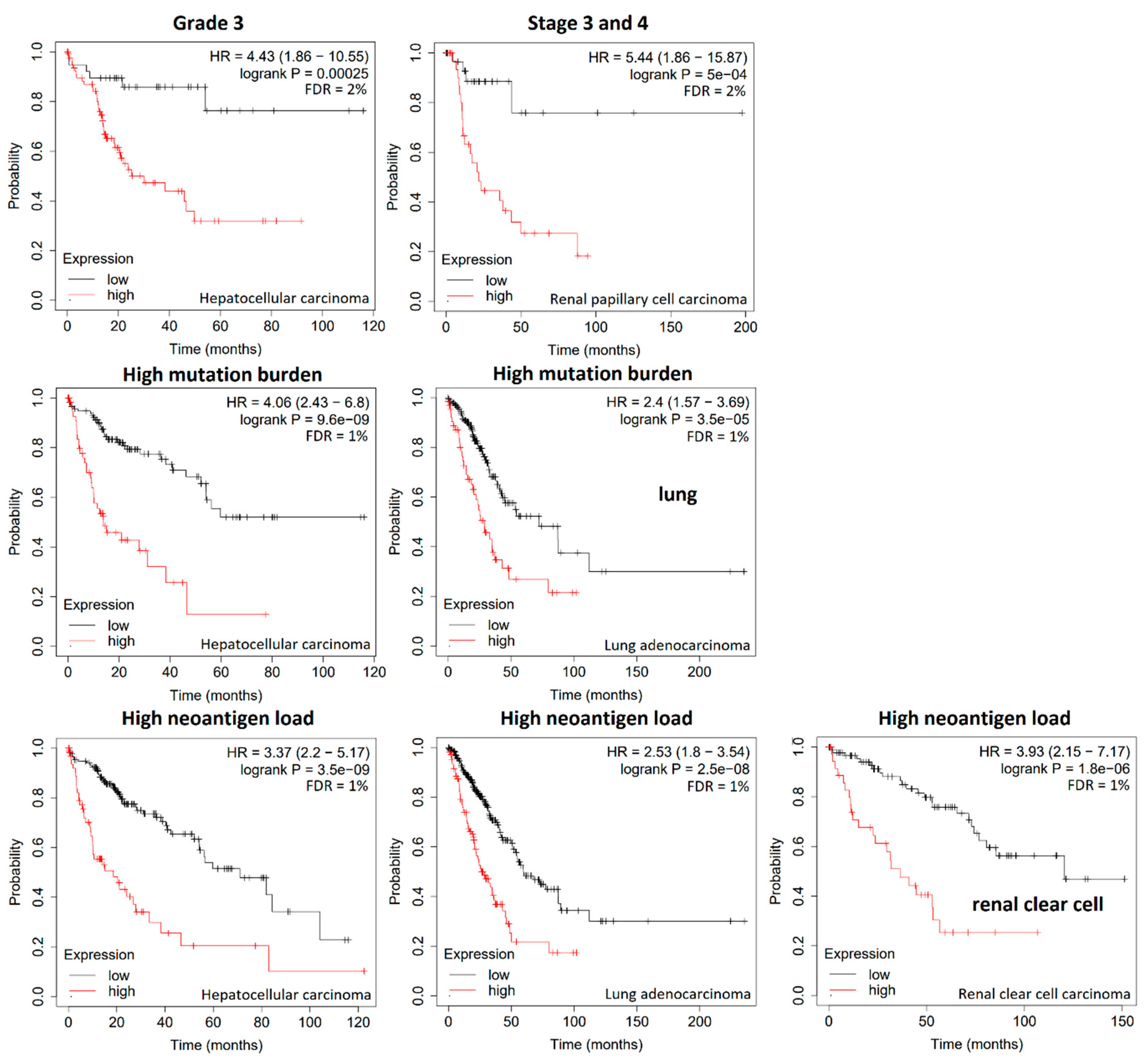
3. Results
4. Discussion
- Cell growth and gene expression (CDKN2A, FOXO6, HES6, HES7, IGF2, LOXL1, MEMO1, TIMP1);
- Oncogenes and tumor suppressors (BRCA1, HRAS, NRAS);
- Signal transduction (MAP2K2, PIK3R1, PIK3R2, PIK3R3, PIK3R6, ULK1, ULK6);
- Angiogenesis (ANGPT4, FLT1, PDE3B, VEGFC);
- Metabolism (AOC2, AOC3, COA6, DLAT, PKM, SCO2, TIGAR);
- Transporters and channels (HEPH, RYR1, SCL2A1, SLC40A1, STEAP2).
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Jomova, K.; Makova, M.; Alomar, S.Y.; Alwasel, S.H.; Nepovimova, E.; Kuca, K.; Rhodes, C.J.; Valko, M. Essential metals in health and disease. Chem. Biol. Interact. 2022, 367, 110173. [Google Scholar] [CrossRef] [PubMed]
- Galaris, D.; Barbouti, A.; Pantopoulos, K. Iron homeostasis and oxidative stress: An intimate relationship. Biochim. Biophys. Acta Mol. Cell Res. 2019, 1866, 118535. [Google Scholar] [CrossRef]
- Hu, H.; Xu, Q.; Mo, Z.; Hu, X.; He, Q.; Zhang, Z.; Xu, Z. New anti-cancer explorations based on metal ions. J. Nanobiotechnol. 2022, 20, 457. [Google Scholar] [CrossRef]
- Jomova, K.; Valko, M. Advances in metal-induced oxidative stress and human disease. Toxicology 2011, 283, 65–87. [Google Scholar] [CrossRef]
- Dev, S.; Babitt, J.L. Overview of iron metabolism in health and disease. Hemodial. Int. 2017, 21, S6–S20. [Google Scholar] [CrossRef]
- Leung, A.K.C.; Lam, J.M.; Wong, A.H.C.; Hon, K.L.; Li, X. Iron Deficiency Anemia: An Updated Review. Curr. Pediatr. Rev. 2024, 20, 339–356. [Google Scholar] [CrossRef]
- Rubino, F.M. Toxicity of Glutathione-Binding Metals: A Review of Targets and Mechanisms. Toxics 2015, 3, 20–62. [Google Scholar] [CrossRef]
- Fu, Z.; Xi, S. The effects of heavy metals on human metabolism. Toxicol. Mech. Methods 2020, 30, 167–176. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Y.; Niu, F.; Liu, Y.; Lu, N. Zinc and its effects on oxidative stress in Alzheimer’s disease. Neurol. Sci. 2014, 35, 923–938. [Google Scholar] [CrossRef] [PubMed]
- Choi, S.; Liu, X.; Pan, Z. Zinc deficiency and cellular oxidative stress: Prognostic implications in cardiovascular diseases. Acta Pharmacol. Sin. 2018, 39, 1120–1132. [Google Scholar] [CrossRef]
- do Perpétuo Socorro Carvalho Martins, M.; da Silva Santos Oliveira, A.S.; do Carmo de Carvalho e Martins, M.; de Carvalho, V.B.L.; Rodrigues, L.A.R.L.; Arcanjo, D.D.R.; dos Santos, M.A.P.; Machado, J.S.R.; de Moura Rocha, M. Effects of zinc supplementation on glycemic control and oxidative stress in experimental diabetes: A systematic review. Clin. Nutr. ESPEN 2022, 51, 28–36. [Google Scholar] [CrossRef]
- Chen, Q.Y.; DesMarais, T.; Costa, M. Metals and Mechanisms of Carcinogenesis. Annu. Rev. Pharmacol. Toxicol. 2019, 59, 537–554. [Google Scholar] [CrossRef]
- Zhu, Y.; Costa, M. Metals and molecular carcinogenesis. Carcinogenesis 2020, 41, 1161–1172. [Google Scholar] [CrossRef]
- Torti, S.V.; Torti, F.M. Ironing out cancer. Cancer Res. 2011, 71, 1511–1514. [Google Scholar] [CrossRef]
- Denoyer, D.; Masaldan, S.; La Fontaine, S.; Cater, M.A. Targeting copper in cancer therapy: ‘Copper That Cancer’. Metallomics 2015, 7, 1459–1476. [Google Scholar] [CrossRef]
- Capriotti, G.; Piccardo, A.; Giovannelli, E.; Signore, A. Targeting Copper in Cancer Imaging and Therapy: A New Theragnostic Agent. J. Clin. Med. 2022, 12, 223. [Google Scholar] [CrossRef] [PubMed]
- da Silva, D.A.; De Luca, A.; Squitti, R.; Rongioletti, M.; Rossi, L.; Machado, C.M.L.; Cerchiaro, G. Copper in tumors and the use of copper-based compounds in cancer treatment. J. Inorg. Biochem. 2022, 226, 111634. [Google Scholar] [CrossRef] [PubMed]
- Stockwell, B.R.; Friedmann Angeli, J.P.; Bayir, H.; Bush, A.I.; Conrad, M.; Dixon, S.J.; Fulda, S.; Gascón, S.; Hatzios, S.K.; Kagan, V.E.; et al. Ferroptosis: A Regulated Cell Death Nexus Linking Metabolism, Redox Biology, and Disease. Cell 2017, 171, 273–285. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Min, J.; Wang, F. Copper homeostasis and cuproptosis in health and disease. Signal Transduct. Target. Ther. 2022, 7, 378. [Google Scholar] [CrossRef]
- Hadian, K.; Stockwell, B.R. The therapeutic potential of targeting regulated non-apoptotic cell death. Nat. Rev. Drug Discov. 2023, 22, 723–742. [Google Scholar] [CrossRef]
- Tang, D.; Kroemer, G.; Kang, R. Targeting cuproplasia and cuproptosis in cancer. Nat. Rev. Clin. Oncol. 2024, 21, 370–388. [Google Scholar] [CrossRef]
- Chen, X.; Kang, R.; Kroemer, G.; Tang, D. Broadening horizons: The role of ferroptosis in cancer. Nat. Rev. Clin. Oncol. 2021, 18, 280–296. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.; Stockwell, B.R.; Conrad, M. Ferroptosis: Mechanisms, biology and role in disease. Nat. Rev. Mol. Cell Biol. 2021, 22, 266–282. [Google Scholar] [CrossRef] [PubMed]
- Cao, S.; Wang, Q.; Sun, Z.; Zhang, Y.; Liu, Q.; Huang, Q.; Ding, G.; Jia, Z. Role of cuproptosis in understanding diseases. Hum. Cell 2023, 36, 1244–1252. [Google Scholar] [CrossRef]
- Xie, J.; Yang, Y.; Gao, Y.; He, J. Cuproptosis: Mechanisms and links with cancers. Mol. Cancer 2023, 22, 46. [Google Scholar] [CrossRef] [PubMed]
- Mookerjee, A.; Mookerjee Basu, J.; Dutta, P.; Majumder, S.; Bhattacharyya, S.; Biswas, J.; Pal, S.; Mukherjee, P.; Raha, S.; Baral, R.N.; et al. Overcoming drug-resistant cancer by a newly developed copper chelate through host-protective cytokine-mediated apoptosis. Clin. Cancer Res. 2006, 12, 4339–4349. [Google Scholar] [CrossRef]
- Majumder, S.; Dutta, P.; Mukherjee, P.; Datta, E.R.; Efferth, T.; Bhattacharya, S.; Choudhuri, S.K. Reversal of drug resistance in P-glycoprotein-expressing T-cell acute lymphoblastic CEM leukemia cells by copper N-(2-hydroxy acetophenone) glycinate and oxalyl bis (N-phenyl) hydroxamic acid. Cancer Lett. 2006, 244, 16–23. [Google Scholar] [CrossRef]
- Denoyer, D.; Clatworthy, S.A.S.; Cater, M.A. Copper Complexes in Cancer Therapy. Met. Ions Life Sci. 2018, 18, 469–506. [Google Scholar] [CrossRef]
- Hartinger, E.M.; Mahringer, A.; Choudhuri, S.K.; Fricker, G.; Efferth, T. Modulatory Activity of the Copper Chelate, Copper N-(2-Hydroxy Acetophenone) Glycinate, in ABC-transporter-expressing Cell Lines. Anticancer. Res. 2023, 43, 1031–1041. [Google Scholar] [CrossRef]
- Mascia, M.; Villano, C.; De Francesco, V.; Schips, L.; Marchioni, M.; Cindolo, L. Efficacy and Safety of the 64Cu(II)Cl2 PET/CT for Urological Malignancies: Phase IIa Clinical Study. Clin. Nucl. Med. 2021, 46, 443–448. [Google Scholar] [CrossRef]
- Werlenius, K.; Kinhult, S.; Solheim, T.S.; Magelssen, H.; Löfgren, D.; Mudaisi, M.; Hylin, S.; Bartek, J., Jr.; Strandéus, M.; Lindskog, M.; et al. Effect of Disulfiram and Copper Plus Chemotherapy vs Chemotherapy Alone on Survival in Patients With Recurrent Glioblastoma: A Randomized Clinical Trial. JAMA Netw. Open 2023, 6, e234149. [Google Scholar] [CrossRef] [PubMed]
- Zeng, H.; You, C.; Zhao, L.; Wang, J.; Ye, X.; Yang, T.; Wan, C.; Deng, L. Ferroptosis-Associated Classifier and Indicator for Prognostic Prediction in Cutaneous Melanoma. J. Oncol. 2021, 2021, 3658196. [Google Scholar] [CrossRef] [PubMed]
- Lin, L.; Li, X.; Zhu, S.; Long, Q.; Hu, Y.; Zhang, L.; Liu, Z.; Li, B.; Li, X. Ferroptosis-related NFE2L2 and NOX4 Genes are Potential Risk Prognostic Biomarkers and Correlated with Immunogenic Features in Glioma. Cell Biochem. Biophys. 2023, 81, 7–17. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Wu, Y.; Song, M.; Yang, Y.; Yu, Y.; Xu, S. Establishment and validation of a ferroptosis-related prognostic signature for hepatocellular carcinoma. Front. Oncol. 2023, 13, 1149370. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Zhao, T.; Wu, X.; Tian, H.; Gao, P.; Chen, Q.; Chen, C.; Zhang, Y.; Wang, S.; Qi, X.; et al. Construction of a ferroptosis-based prognostic model for breast cancer helps to discriminate high/low risk groups and treatment priority. Front. Immunol. 2023, 14, 1264206. [Google Scholar] [CrossRef]
- Bao, J.H.; Lu, W.C.; Duan, H.; Ye, Y.Q.; Li, J.B.; Liao, W.T.; Li, Y.C.; Sun, Y.P. Identification of a novel Cuproptosis-related gene signature and integrative analyses in patients with lower-grade gliomas. Front. Immunol. 2022, 13, 933973. [Google Scholar] [CrossRef]
- Jawed, R.; Bhatti, H. Cuproptosis in lung cancer: Therapeutic options and prognostic models. Apoptosis 2024, 29, 1393–1398. [Google Scholar] [CrossRef]
- Bian, Z.; Fan, R.; Xie, L. A Novel Cuproptosis-Related Prognostic Gene Signature and Validation of Differential Expression in Clear Cell Renal Cell Carcinoma. Genes 2022, 13, 851. [Google Scholar] [CrossRef]
- Chen, Y.; Tang, L.; Huang, W.; Abisola, F.H.; Zhang, Y.; Zhang, G.; Yao, L. Identification of a prognostic cuproptosis-related signature in hepatocellular carcinoma. Biol. Direct. 2023, 18, 4. [Google Scholar] [CrossRef]
- Liu, G.M.; Zeng, H.D.; Zhang, C.Y.; Xu, J.W. Identification of a six-gene signature predicting overall survival for hepatocellular carcinoma. Cancer Cell Int. 2019, 19, 1–13. [Google Scholar] [CrossRef]
- He, L.; Chen, J.; Xu, F.; Li, J.; Li, J. Prognostic Implication of a Metabolism-Associated Gene Signature in Lung Adenocarcinoma. Mol. Ther. Oncolytics 2020, 19, 265–277. [Google Scholar] [CrossRef] [PubMed]
- Sun, S.; Guo, W.; Wang, Z.; Wang, X.; Zhang, G.; Zhang, H.; Li, R.; Gao, Y.; Qiu, B.; Tan, F.; et al. Development and validation of an immune-related prognostic signature in lung adenocarcinoma. Cancer Med. 2020, 9, 5960–5975. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Jiang, L.; Gao, F.; Zhang, E.; Wang, T.; Zhang, N.; Wang, X.; Zheng, J. Machine learning-based pathomics signature could act as a novel prognostic marker for patients with clear cell renal cell carcinoma. Br. J. Cancer 2022, 126, 771–777. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Tang, L.; Huang, W.; Zhang, Y.; Abisola, F.H.; Li, L. Identification and validation of a novel cuproptosis-related signature as a prognostic model for lung adenocarcinoma. Front. Endocrinol. 2022, 13, 963220. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.; Luo, Y.; Chen, Y.; Qu, H.; Zheng, L.; Yao, J. Development of a prognostic gene signature for hepatocellular carcinoma. Cancer Treat. Res. Commun. 2022, 31, 100511. [Google Scholar] [CrossRef]
- Pang, Y.; Wang, Y.; Zhou, X.; Ni, Z.; Chen, W.; Liu, Y.; Du, W. Cuproptosis-Related LncRNA-Based Prediction of the Prognosis and Immunotherapy Response in Papillary Renal Cell Carcinoma. Int. J. Mol. Sci. 2023, 24, 1464. [Google Scholar] [CrossRef]
- Zhang, W.; Qu, H.; Ma, X.; Li, L.; Wei, Y.; Wang, Y.; Zeng, R.; Nie, Y.; Zhang, C.; Yin, K.; et al. Identification of cuproptosis and immune-related gene prognostic signature in lung adenocarcinoma. Front. Immunol. 2023, 14, 1179742. [Google Scholar] [CrossRef]
- Cai, X.; Lin, J.; Liu, L.; Zheng, J.; Liu, Q.; Ji, L.; Sun, Y. A novel TCGA-validated programmed cell-death-related signature of ovarian cancer. BMC Cancer 2024, 24, 515. [Google Scholar] [CrossRef]
- Clayton, E.A.; Pujol, T.A.; McDonald, J.F.; Qiu, P. Leveraging TCGA gene expression data to build predictive models for cancer drug response. BMC Bioinformatics 2020, 21 (Suppl. S14), 364. [Google Scholar] [CrossRef]
- Danaher, P.; Warren, S.; Lu, R.; Samayoa, J.; Sullivan, A.; Pekker, I.; Wallden, B.; Marincola, F.M.; Cesano, A. Pan-cancer adaptive immune resistance as defined by the Tumor Inflammation Signature (TIS): Results from The Cancer Genome Atlas (TCGA). J. Immunother. Cancer 2018, 6, 63. [Google Scholar] [CrossRef]
- Gao, W.; Yuan, Z.; Zhao, X.; Wang, S.; Lai, S.; Ni, K.; Zhan, Y.; Liu, Z.; Liu, L.; Xin, R.; et al. The prognostic and clinical value of p53 upregulated modulator of apoptosis expression in solid tumors: A meta-analysis and TCGA data review. Expert Rev. Mol. Diagn. 2022, 22, 811–819. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Dai, Z.; Wu, X.; Zhang, N.; Zhang, H.; Wang, Z.; Zhang, X.; Liang, X.; Luo, P.; Zhang, J.; et al. The Comprehensive Analysis Identified an Autophagy Signature for the Prognosis and the Immunotherapy Efficiency Prediction in Lung Adenocarcinoma. Front. Immunol. 2022, 13, 749241. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Ge, M.; Mo, S.; Shi, M.; Zhang, J.; Liu, J. Construction of a New Ferroptosis-related Prognosis Model for Survival Prediction in Colorectal Cancer. Curr. Med. Chem. 2024. [Google Scholar] [CrossRef] [PubMed]
- Győrffy, B. Integrated analysis of public datasets for the discovery and validation of survival-associated genes in solid tumors. Innovation 2024, 5, 100625. [Google Scholar] [CrossRef]
- Győrffy, B. Transcriptome-level discovery of survival-associated biomarkers and therapy targets in non-small-cell lung cancer. Br. J. Pharmacol. 2024, 181, 362–374. [Google Scholar] [CrossRef]
- Lánczky, A.; Györffy, B. Web-based survival analysis tool tailored for medical research (KMplot): Development and implementation. J. Med. Internet Res. 2021, 23, e27633. [Google Scholar] [CrossRef]
- Nagy, Á.; Munkácsy, G.; Györffy, B. Pancancer survival analysis of cancer hallmark genes. Sci. Rep. 2021, 11, 6047. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Statist. Soc. B 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Ge, E.J.; Bush, A.I.; Casini, A.; Cobine, P.A.; Cross, J.R.; DeNicola, G.M.; Dou, Q.P.; Franz, K.J.; Gohil, V.M.; Gupta, S.; et al. Connecting copper and cancer: From transition metal signalling to metalloplasia. Nat. Rev. Cancer 2022, 22, 102–113. [Google Scholar] [CrossRef]
- Zhang, L.; Shao, J.; Tan, S.W.; Ye, H.P.; Shan, X.Y. Association between serum copper/zinc ratio and lung cancer: A systematic review with meta-analysis. J. Trace Elem. Med. Biol. 2022, 74, 127061. [Google Scholar] [CrossRef]
- Zhou, C.; Yang, J.; Liu, T.; Jia, R.; Yang, L.; Sun, P.; Zhao, W. Copper metabolism and hepatocellular carcinoma: Current insights. Front. Oncol. 2023, 13, 1186659. [Google Scholar] [CrossRef] [PubMed]
- Berglund, A.; Amankwah, E.K.; Kim, Y.C.; Spiess, P.E.; Sexton, W.J.; Manley, B.; Park, H.Y.; Wang, L.; Chahoud, J.; Chakrabarti, R.; et al. Influence of gene expression on survival of clear cell renal cell carcinoma. Cancer Med. 2020, 9, 8662–8675. [Google Scholar] [CrossRef] [PubMed]
- Song, P.; Li, W.; Wu, X.; Qian, Z.; Ying, J.; Gao, S.; He, J. Integrated analysis of single-cell and bulk RNA-sequencing identifies a signature based on B cell marker genes to predict prognosis and immunotherapy response in lung adenocarcinoma. Cancer Immunol. Immunother. 2022, 71, 2341–2354. [Google Scholar] [CrossRef]
- Lu, J.; Chen, Y.; Zhang, X.; Guo, J.; Xu, K.; Li, L. A novel prognostic model based on single-cell RNA sequencing data for hepatocellular carcinoma. Cancer Cell Int. 2022, 22, 38. [Google Scholar] [CrossRef]
- Li, K.; Tan, L.; Li, Y.; Lyu, Y.; Zheng, X.; Jiang, H.; Zhang, X.; Wen, H.; Feng, C. Cuproptosis identifies respiratory subtype of renal cancer that confers favorable prognosis. Apoptosis 2022, 27, 1004–1014. [Google Scholar] [CrossRef]
- Mei, W.; Liu, X.; Jia, X.; Jin, L.; Xin, S.; Sun, X.; Zhang, J.; Zhang, B.; Chen, Y.; Che, J.; et al. A Cuproptosis-Related Gene Model For Predicting the Prognosis of Clear Cell Renal Cell Carcinoma. Front. Genet. 2022, 13, 905518. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; Jiang, S.; Liu, Y.; Zhang, T.; Zhu, C.; Zhang, L.; Sang, X.; Lu, X.; Wei, J.; Deng, K.; et al. A novel cuproptosis-related gene signature for overall survival prediction in patients with hepatocellular carcinoma. Heliyon 2022, 8, e11768. [Google Scholar] [CrossRef]
- Li, Y.; Zeng, X. A novel cuproptosis-related prognostic gene signature and validation of differential expression in hepatocellular carcinoma. Front. Pharmacol. 2023, 13, 1081952. [Google Scholar] [CrossRef] [PubMed]
- Lv, Y.; Xiao, Y.; Cui, X.; Luo, H.; Xu, L. Identification of cuproptosis-related gene signature to predict prognosis in lung adenocarcinoma. Front. Genet. 2022, 13, 1016871. [Google Scholar] [CrossRef]
- Zhang, H.; Shi, Y.; Yi, Q.; Wang, C.; Xia, Q.; Zhang, Y.; Jiang, W.; Qi, J. A novel defined cuproptosis-related gene signature for predicting the prognosis of lung adenocarcinoma. Front. Genet. 2022, 13, 975185. [Google Scholar] [CrossRef]
- Zhou, J.; Chen, D.; Zhang, S.; Wang, C.; Zhang, L. Identification of two molecular subtypes and a novel prognostic model of lung adenocarcinoma based on a cuproptosis-associated gene signature. Front. Genet. 2023, 13, 1039983. [Google Scholar] [CrossRef] [PubMed]
- Xie, N.; Shen, G.; Gao, W.; Huang, Z.; Huang, C.; Fu, L. Neoantigens: Promising targets for cancer therapy. Signal Transduct. Target. Ther. 2023, 8, 9. [Google Scholar] [CrossRef] [PubMed]
- Hopkins, A.L. Network pharmacology. Nat. Biotechnol. 2007, 25, 1110–1111. [Google Scholar] [CrossRef]
- Hopkins, A.L. Network pharmacology: The next paradigm in drug discovery. Nat. Chem. Biol. 2008, 4, 682–690. [Google Scholar] [CrossRef]
- Li, S.; Zhang, Z.Q.; Wu, L.J.; Zhang, X.G.; Li, Y.D.; Wang, Y.Y. Understanding ZHENG in traditional Chinese medicine in the context of neuro-endocrine-immune network. IET Syst. Biol. 2007, 1, 51–60. [Google Scholar] [CrossRef]
- Zhang, P.; Zhang, D.; Zhou, W.; Wang, L.; Wang, B.; Zhang, T.; Li, S. Network pharmacology: Towards the artificial intelligence-based precision traditional Chinese medicine. Brief. Bioinform. 2023, 25, bbad518. [Google Scholar] [CrossRef]
- van de Vijver, M.J.; He, Y.D.; van’t Veer, L.J.; Dai, H.; Hart, A.A.; Voskuil, D.W.; Schreiber, G.J.; Peterse, J.L.; Roberts, C.; Marton, M.J.; et al. A gene-expression signature as a predictor of survival in breast cancer. N. Engl. J. Med. 2002, 347, 1999–2009. [Google Scholar] [CrossRef] [PubMed]
- Zuo, S.; Wei, M.; Zhang, H.; Chen, A.; Wu, J.; Wei, J.; Dong, J. A robust six-gene prognostic signature for prediction of both disease-free and overall survival in non-small cell lung cancer. J. Transl. Med. 2019, 17, 152. [Google Scholar] [CrossRef]
- Feng, Z.; Qian, H.; Li, K.; Lou, J.; Wu, Y.; Peng, C. Development and Validation of a 7-Gene Prognostic Signature to Improve Survival Prediction in Pancreatic Ductal Adenocarcinoma. Front. Mol. Biosci. 2021, 8, 676291. [Google Scholar] [CrossRef]
- Yue, T.; Chen, S.; Zhu, J.; Guo, S.; Huang, Z.; Wang, P.; Zuo, S.; Liu, Y. The aging-related risk signature in colorectal cancer. Aging 2021, 13, 7330–7349. [Google Scholar] [CrossRef]
- Gao, Z.; Zhang, D.; Duan, Y.; Yan, L.; Fan, Y.; Fang, Z.; Liu, Z. A five-gene signature predicts overall survival of patients with papillary renal cell carcinoma. PLoS ONE 2019, 14, e0211491. [Google Scholar] [CrossRef] [PubMed]
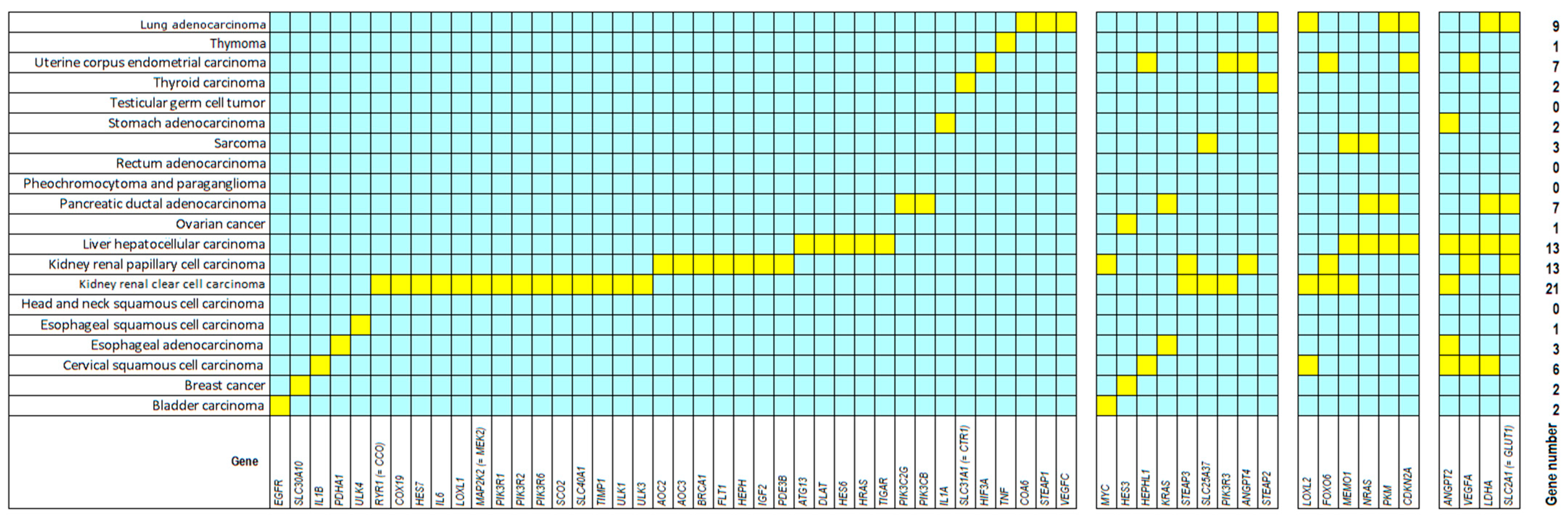
| Symbol | Gene Name | Function | Tumor Type | Sample No. | p-Value | FDR |
|---|---|---|---|---|---|---|
| RYR1 (=CCO) | Ryanodine receptor 1 (skeletal) | Calcium release channel | KIRC | 530 | 2.1 × 10−4 | 5% |
| HES7 | Hairy and enhancer of split (Drosophila) family BHLH transcription factor 7 | Transcriptional repressor | KIRC | 530 | 6.7 × 10−7 | 1% |
| IL6 | Interleukin 6 | Proinflammatory cytokine | KIRC | 530 | 5.3 × 10−8 | 1% |
| LOXL1 | Lysine oxidase-like | Biogenesis of connective tissue | KIRC | 530 | 3.0 × 10−7 | 1% |
| MAP2K2(=MEK2) | Mitogen-activated protein kinase kinase 2 | Mitogenic growth factor, signal transduction | KIRC | 530 | 3.7 × 10−4 | 1% |
| PIK3R1 | Phosphoinositide-3-kinase regulatory subunit 1 | Role in the metabolic insulin action | KIRC | 530 | 4.9 × 10−5 | 1% |
| PIK3R2 | Phosphoinositide-3-kinase regulatory subunit 2 | Regulatory component of PI3K, growth signaling pathways | KIRC | 530 | 1.0 × 10−4 | 3% |
| PIK3R6 | Phosphoinositide-3-kinase regulatory subunit 6 | Regulatory component of PI3K, growth signaling pathways | KIRC | 530 | 5.3 × 10−5 | 1% |
| SCO2 | Synthesis of cytochrome C oxidase 1 | Role in aerobic ATP production | KIRC | 530 | 1.6 × 10−4 | 1% |
| SLC40A1 | Solute carrier family 40 member 1 | Iron export | KIRC | 530 | 8.5 × 10−13 | 1% |
| TIMP1 | Tissue inhibitor of metalloproteinases 1 | Degradation of extracellular matrix, cell proliferation | KIRC | 530 | 2.1 × 10−4 | 1% |
| ULK1 | Unc-51-like autophagy- activating kinase 1 | Serine/threonine kinase, autophagosome assembly | KIRC | 530 | 5.6 × 10−6 | 1% |
| ULK3 | Unc-51-like autophagy- activating kinase 3 | Serine/threonine kinase, fibroblast activation | KIRC | 530 | 1.0 × 10−4 | 3% |
| AOC2 | Amine oxidase copper- containing 2 | Oxidative conversion of amines to aldehydes and ammonia | KIRP | 287 | 3.0 × 10−4 | 5% |
| AOC3 | Amine oxidase copper- containing 3 | Adhesive properties, leukocyte trafficking | KIRP | 287 | 1.6 × 10−4 | 2% |
| BRCA1 | Breast and ovarian cancer susceptibility protein 1 | Tumor suppressor | KIRP | 287 | 4.1 × 10−5 | 1% |
| FLT1 | Fms-related receptor tyrosine kinase 1 | Role in angiogenesis | KIRP | 287 | 4.1 × 10−6 | 1% |
| HEPH | Hephaestin | Copper and iron transport and homeostasis | KIRP | 287 | 5.0 × 10−4 | 5% |
| IGF2 | Insulin-like growth factor 2 | Cell development and growth | KIRP | 287 | 7.2 × 10−5 | 1% |
| PDE3B | Phosphodiesterase 3B | Negative regulation of angiogenesis and cell adhesion | KIRP | 287 | 2.4 × 10−4 | 3% |
| ATG13 | Autophagy-related 13 | Autophagosome formation and mitophagy | LIHC | 370 | 6.0 × 10−5 | 1% |
| DLAT | Dihydrolipoamide S-acetyltransferase | Component of the pyruvate dehydrogenase complex | LIHC | 370 | 5.0 × 10−5 | 1% |
| HES6 | Hairy and enhancer of split (Drosophila) family BHLH transcription factor 6 | Regulation of cell differentiation | LIHC | 370 | 2.1 × 10−4 | 3% |
| HRAS | Harvey rat sarcoma viral oncogene homolog | Oncogenic GTPase | LIHC | 370 | 2.3 × 10−4 | 5% |
| TIGAR | TP53-induced glycolysis regulatory phosphatase | Blockage of glycolysis | LIHC | 370 | 3.2 × 10−4 | 5% |
| COA6 | Cytochrome c oxidase assembly factor 6 | Mitochondrial respiration | LUAD | 504 | 1.1 × 10−4 | 3% |
| STEAP1 | Six-transmembrane epithelial antigen of prostate metalloreductase 1 | Cell surface antigen at cell–cell junctions | LUAD | 504 | 1.4 × 10−6 | 1% |
| VEGFC | Vascular endothelial growth | Angiogenesis and endothelial | LUAD | 504 | 3.0 × 10−4 | 1% |
| factor C | cell growth | KIRP | 287 | 4.1 × 10−4 | 5% | |
| STEAP3 | STEAP3 metalloreductase | Iron and copper transporter | KIRC | 530 | 3.7 × 10−2 | 1% |
| in p53-mediated apoptosis | KIRP | 287 | 1.3 × 10−5 | 1% | ||
| SLC25A37 | Solute carrier family 25 member 37 | Imports iron for the synthesis of mitochondrial heme | KIRP | 530 | 3.3 × 10−7 | 1% |
| PIK3R3 | Phosphoinositide-3-kinase regulatory subunit 3 | Second messenger in growth signaling pathways | KIRC | 530 | 4.6 × 10−8 | 1% |
| ANGPT4 | Angiopoietin 4 | Involved in angiogenesis | KIRP | 287 | 3.1 × 10−4 | 5% |
| STEAP2 | STEAP2 metalloreductase | Iron and copper uptake | LUAD | 504 | 1.2 × 10−4 | 3% |
| KIRC | 530 | 3.5 × 10−5 | 1% | |||
| LUAD | 504 | 3.7 × 10−5 | 1% | |||
| FOXO6 | Forkhead box O6 | Regulation of transcription | KIRC | 530 | 5.4 × 10−5 | 1% |
| by RNA polymerase II | KIRP | 287 | 3.6 × 10−4 | 5% | ||
| MEMO1 | Mediator of cell motility 1 | Microtubule-based processes | KIRC | 530 | 3.3 × 10−7 | 1% |
| LIHC | 370 | 1.7 × 10−4 | 3% | |||
| NRAS | Neuroblastoma RAS viral oncogene homolog | Oncogenic GTPase | LIHC | 370 | 3.4 × 10−5 | 1% |
| PKM | Pyruvate kinase M | Glycolysis | LIHC | 370 | 2.7 × 10−6 | 1% |
| LUAD | 504 | 1.5 × 10−4 | 3% | |||
| CDKN2A | Cyclin-dependent kinase | Regulation of the G1 phase of | LIHC | 370 | 2.2 × 10−4 | 5% |
| inhibitor 2A | the cell cycle | LUAD | 504 | 2.4 × 10−4 | 5% | |
| KIRP | 287 | 7.1 × 10−8 | 1% | |||
| LIHC | 370 | 6.3 × 10−5 | 1% | |||
| KIRP | 287 | 1.6 × 10−7 | 1% | |||
| LIHC | 370 | 2.3 × 10−5 | 1% | |||
| LIHC | 370 | 3.9 × 10−7 | 1% | |||
| LUAD | 504 | 1.8 × 10−6 | 1% | |||
| SLC2A1 | Glucose transporter type 1 | Glucose transport in the blood | KIRP | 287 | 4.5 × 10−4 | 5% |
| (=GLUT1) | brain barrier | LIHC | 370 | 2.7 × 10−8 | 1% | |
| LUAD | 504 | 3.6 × 10−7 | 1% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ooko, E.; Ali, N.T.; Efferth, T. Identification of Cuproptosis-Associated Prognostic Gene Expression Signatures from 20 Tumor Types. Biology 2024, 13, 793. https://doi.org/10.3390/biology13100793
Ooko E, Ali NT, Efferth T. Identification of Cuproptosis-Associated Prognostic Gene Expression Signatures from 20 Tumor Types. Biology. 2024; 13(10):793. https://doi.org/10.3390/biology13100793
Chicago/Turabian StyleOoko, Ednah, Nadeen T. Ali, and Thomas Efferth. 2024. "Identification of Cuproptosis-Associated Prognostic Gene Expression Signatures from 20 Tumor Types" Biology 13, no. 10: 793. https://doi.org/10.3390/biology13100793
APA StyleOoko, E., Ali, N. T., & Efferth, T. (2024). Identification of Cuproptosis-Associated Prognostic Gene Expression Signatures from 20 Tumor Types. Biology, 13(10), 793. https://doi.org/10.3390/biology13100793








