Modulation of the Cellular microRNA Landscape: Contribution to the Protective Effects of High-Density Lipoproteins (HDL)
Abstract
Simple Summary
Abstract
1. Introduction
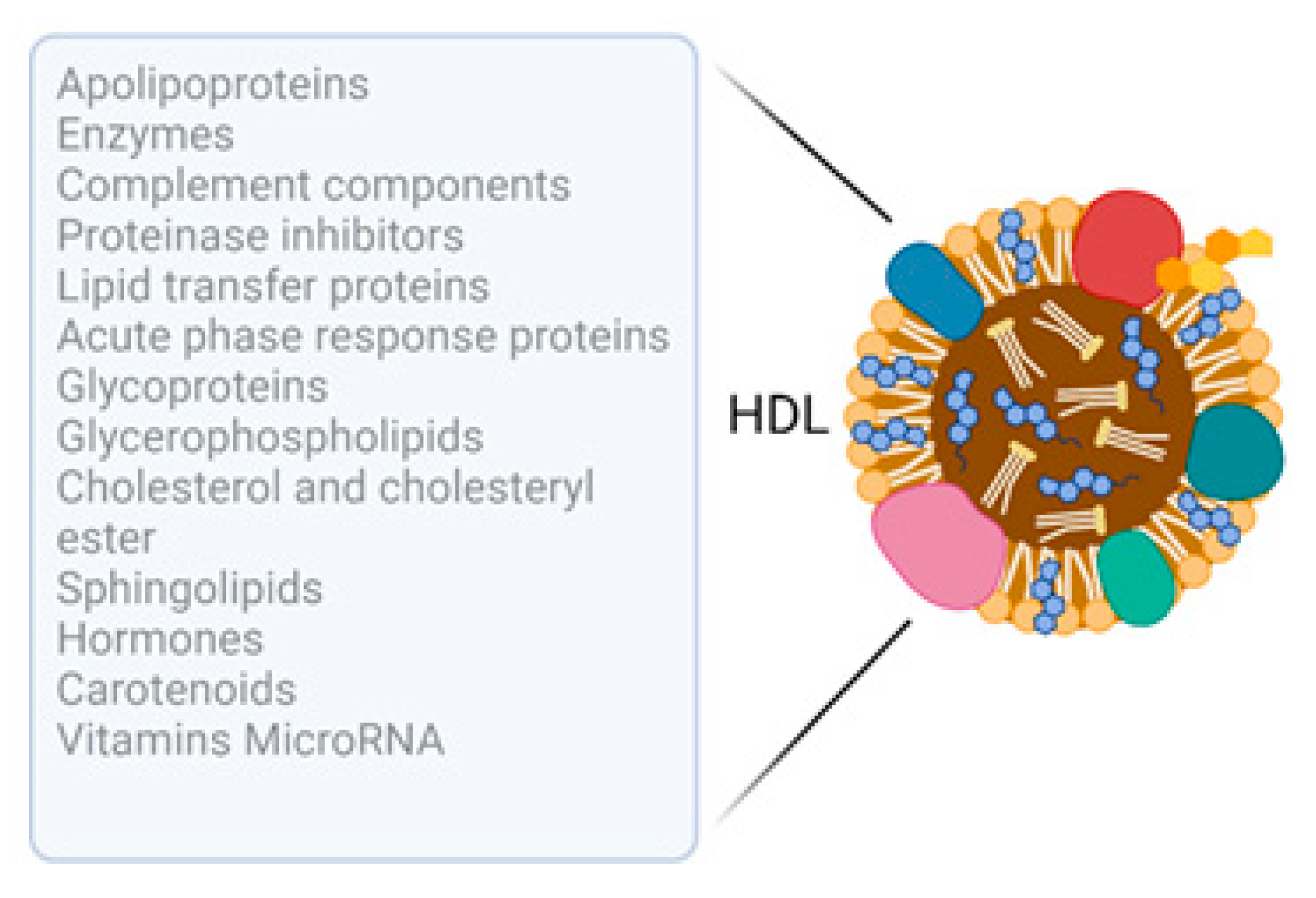
1.1. HDL: Composition and Cargo
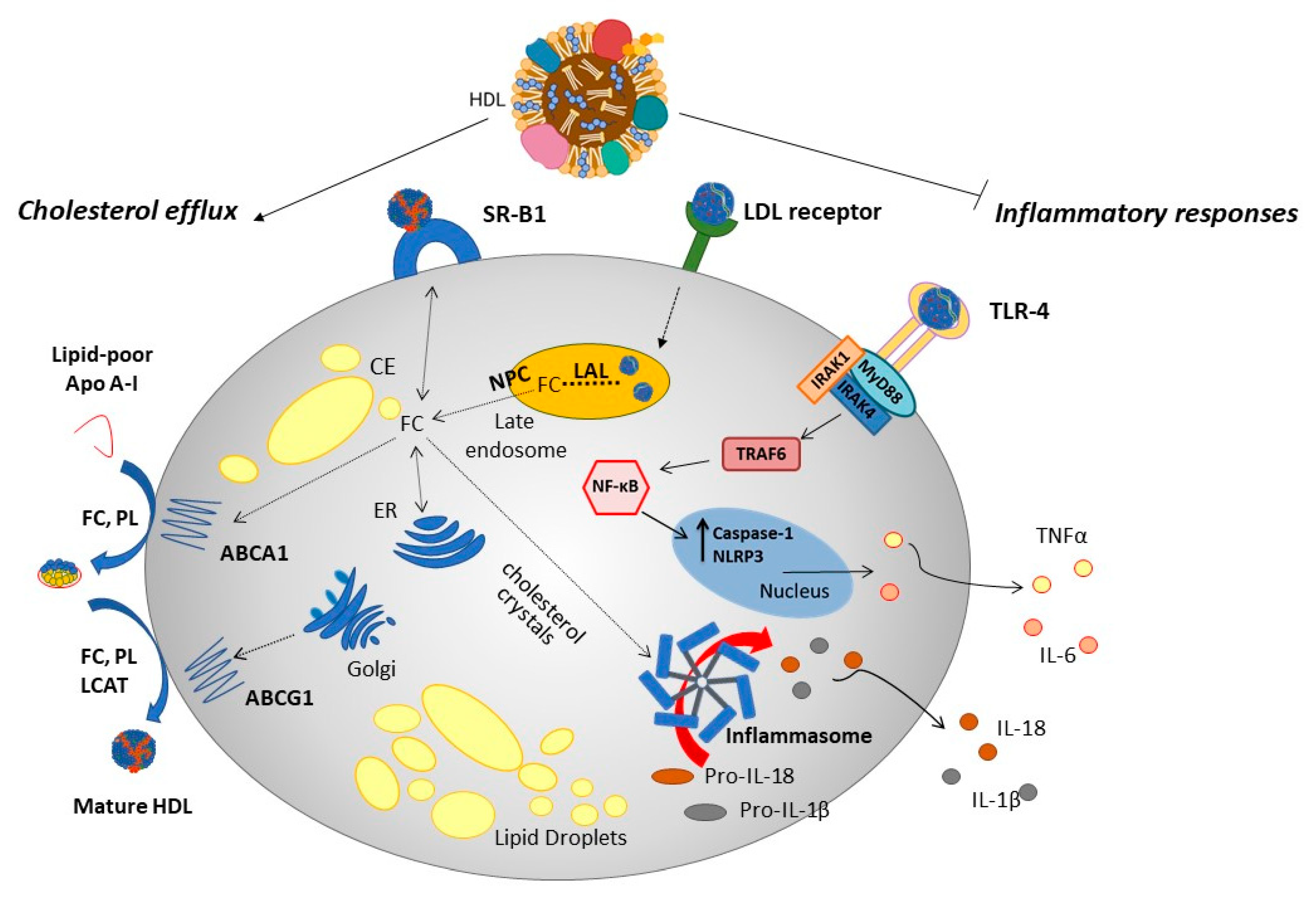
1.2. Assembly, Maturation and Uptake of HDL Particles
1.3. HDL: Protective Biological Functions
2. MicroRNA
2.1. MicroRNA Biogenesis and Function
2.2. HDL microRNA Cargo: Cellular Export and Import of microRNA to HDL
2.3. Regulation of Endogenous microRNA Expression by HDL
3. Modification of the Functional Cellular microRNA Landscape by HDL
4. Conclusions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sirtori, C.R.; Corsini, A.; Ruscica, M. The role of high-density lipoprotein cholesterol in 2022. Curr. Atheroscler. Rep. 2022, 24, 365–377. [Google Scholar] [CrossRef] [PubMed]
- Pownall, H.J.; Rosales, C.; Gillard, B.K.; Gotto, A.M., Jr. High-density lipoproteins, reverse cholesterol transport and atherogenesis. Nat. Rev. Cardiol. 2021, 18, 712–723. [Google Scholar] [CrossRef] [PubMed]
- Ouimet, M.; Barrett, T.J.; Fisher, E.A. HDL and reverse cholesterol transport. Circ. Res. 2019, 124, 1505–1518. [Google Scholar] [CrossRef]
- Rodriguez, A. High HDL-cholesterol paradox: SCARB1-LAG3-HDL axis. Curr. Atheroscler. Rep. 2021, 23, 5. [Google Scholar] [CrossRef]
- Zhong, G.-C.; Huang, S.-Q.; Peng, Y.; Wan, L.; Wu, Y.-Q.-L.; Hu, T.-Y.; Hu, J.-J.; Hao, F.-B. HDL-C is associated with mortality from all causes, cardiovascular disease and cancer in a J-shaped dose-response fashion: A pooled analysis of 37 prospective cohort studies. Eur. J. Prev. Cardiol. 2020, 27, 1187–1203. [Google Scholar] [CrossRef] [PubMed]
- Madsen, C.M.; Varbo, A.; NOrdestgaard, B.G. Extreme high high-density lipoprotein cholesterol is paradoxically associated with high mortality in men and women: Two prospective cohort studies. Eur. Heart J. 2017, 38, 2478–2486. [Google Scholar] [CrossRef]
- Voight, B.F.; Peloso, G.M.; Orho-Melander, M.; Frikke-Schmidt, R.; Barbalic, M.; Jensen, M.K.; Hindy, G.; Hólm, H.; Ding, E.L.; Johnson, T.; et al. Plasma HDL cholesterol and risk of myocardial infarction: A Mendelian Randomisation Study. Lancet 2012, 380, 572–580. [Google Scholar] [CrossRef]
- Geller, A.S.; Polisecki, E.Y.; Diffenderfer, M.R.; Asztalos, B.F.; Karathanasis, S.K.; Hegele, R.A.; Schaefer, E.J. Genetic and secondary causes of severe HDL deficiency and cardiovascular disease. J. Lipid Res. 2018, 59, 2421–2435. [Google Scholar] [CrossRef]
- Thomas, D.G.; Wei, Y.; Tall, A.R. Lipid and metabolic syndrome traits in coronary artery disease: A Mendelian randomization study. J. Lipid Res. 2021, 62, 100044. [Google Scholar] [CrossRef]
- White, J.; Swerdlow, D.I.; Preiss, D.; Fairhust-Hunter, Z.; Keating, B.J.; Asselbergs, F.W.; Sattar, N.; Humphries, S.E.; Hingorani, A.D.; Holmes, M.V. Association of lipid fractions with risks for coronary artery disease and diabetes. JAMA Cardiol. 2016, 1, 692–699. [Google Scholar] [CrossRef]
- Vickers, K.C.; Remaley, A.T. HDL and cholesterol: Life after the divorce? J. Lipid Res. 2014, 55, 4–12. [Google Scholar] [CrossRef] [PubMed]
- Cho, K.-H. The current status of research on high-density lipoproteins (HDL): A paradigm shift from HDL quantity to HDL quality and HDL functionality. Int. J. Mol. Sci. 2022, 23, 3967. [Google Scholar] [CrossRef] [PubMed]
- Kontush, A.; Lindahl, M.; Lhomme, M.; Calabresi, L.; Chapman, M.J.; Davidson, W.S. Structure of HDL: Particle subclasses and molecular components. Handb. Exp. Pharmacol. 2015, 224, 3–51. [Google Scholar] [CrossRef] [PubMed]
- Collins, L.A.; Mirza, S.P.; Kissebah, A.H.; Olivier, M. Integrated approach for the comprehensive characterization of lipoproteins from human plasma using FPLC and nano-HPLC-tandem mass spectrometry. Physiol. Genom. 2010, 40, 208–2015. [Google Scholar] [CrossRef] [PubMed]
- von Eckardstein, A.; Sibler, R.A. Possible contributions of lipoproteins and cholesterol to the pathogenesis of diabetes mellitus type 2. Curr. Opin. Lipidol. 2011, 22, 26–32. [Google Scholar] [CrossRef]
- Santana, M.F.M.; Lira, A.L.A.; Pinto, R.S.; Minanni, C.A.; Silva, A.R.M.; Sawada, M.I.B.A.C.; Nakandakare, E.R.; Correa-Giannella, M.L.C.; Queiroz, M.S.; Ronsein, G.E.; et al. Enrichment of apolipoprotein A-IV and apolipoprotein D in the HDL proteome is associated with HDL functions in diabetic kidney disease without dialysis. Lipids Health Dis. 2020, 19, 205. [Google Scholar] [CrossRef]
- Stasi, A.; Franzin, R.; Fiorentino, M.; Eqioccomarro, E.; Castellano, G.; Gesualdo, L. Multifaceted roles of HDL in sepsis and SARS-CoV-2 infection: Renal implications. Int. J. Mol. Sci. 2021, 22, 5980. [Google Scholar] [CrossRef]
- Zimetti, F.; Adorni, M.P.; Marsillach, J.; Marchi, C.; Trentini, A.; Valacchi, G.; Cervellati, C. Connection between the altered HDL antioxidant and anti-inflammatory properties and risk to develop Alzheimer’s disease: A narrative review. Oxid. Med. Cell Longev. 2021, 2021, 6695796. [Google Scholar] [CrossRef]
- Wiesner, P.; Lield, K.; Boettcher, A.; Schmitz, G.; Liebisch, G. Lipid profiling of FPLC-separated lipoprotein fractions by electrospray ionization tandem mass spectrometer. J. Lipid Res. 2009, 50, 574–585. [Google Scholar] [CrossRef]
- Camont, L.; Lhomme, M.; Rached, F.; Le Goff, W.; Negre-Salvaryre, A.; Salvarye, R.; Calazada, C.; Largarde, M.; Chapman, M.J.; Kontush, A. Small, dense high-density lipoprotein-3 particles are enriched in negatively charged phospholipids; relevance to cellular cholesterol efflux, antioxidative, antithrombotic, anti-inflammatory, and antiapoptotic functions. Arterioscler. Thromb. Vasc. Biol. 2013, 33, 2714–2723. [Google Scholar] [CrossRef]
- Vickers, K.C.; Palmisano, B.T.; Shoucri, B.M.; Shamburek, R.D.; Remaley, A.T. MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat. Cell Biol. 2011, 13, 423–433. [Google Scholar] [CrossRef] [PubMed]
- Vickers, K.C.; Landstreet, S.R.; Levin, M.G.; Shoucr, B.M.; Toth, C.L.; Taylor, R.C.; Palmisano, B.T.; Tabet, F.; Cui, H.; Rue, K.-A.; et al. MicroRNA-223 coordinates cholesterol homeostasis. Proc. Natl. Acad. Sci. USA 2014, 111, 14518–14523. [Google Scholar] [CrossRef] [PubMed]
- Vickers, K.C.; Michell, D.L. HDL-small RNA export, transport, and functional delivery in atherosclerosis. Curr. Atheroscler Rep. 2021, 23, 38. [Google Scholar] [CrossRef] [PubMed]
- Zannis, V.I.; Chroni, A.; Krieger, M.J. Role of apoA-I, ABCA1, LCAT and SR-B1 in the biogenesis of HDL. Mol. Med. 2006, 84, 276–294. [Google Scholar] [CrossRef] [PubMed]
- Shen, W.J.; Azhar, S.; Kraemer, F.B. SR-B1, A unique multifunctional receptor for cholesterol influx and efflux. Annu. Rev. Physiol. 2018, 10, 80–95. [Google Scholar] [CrossRef]
- Groenen, A.G.; Halmos, B.; Tall, A.R.; Westerterp, M. Cholesterol efflux pathways, inflammation, and atherosclerosis. Crit. Rev. Biochem. Mol. Biol. 2021, 56, 426–439. [Google Scholar] [CrossRef]
- Zhu, X.; Lee, J.Y.; Timmins, J.M.; Brown, J.M.; Boudyguina, E.; Mulya, A.; Gebre, A.K.; Willingham, M.C.; Hiltbold, E.M.; Mishra, N.; et al. Increased cellular free cholesterol in macrophage-specific ABCA1 knock-out mice enhances pro-inflammatory response of macrophages. J. Biol. Chem. 2008, 283, 22930–22941. [Google Scholar] [CrossRef]
- Yvan-Charvet, L.; Welch, C.; Pagler, T.A.; Ranalletta, M.; Lamkanfi, M.; Han, S.; Ishibashi, M.; Li, R.; Wang, N.; Tall, A.R. Increased inflammatory gene expression in ABC transporter-deficient macrophages: Free cholesterol accumulation, increased signaling via toll-like receptors, and neutrophil infiltration of atherosclerotic lesions. Circulation 2008, 118, 1837–1847. [Google Scholar] [CrossRef]
- Fernandes das Neves, M.; Batuca, J.R.; Delgado Alves, J. The role of high-density lipoprotein in the regulation of the immune response: Implications for atherosclerosis and autoimmunity. Immunology 2021, 164, 231–241. [Google Scholar] [CrossRef]
- Westerterp, M.; Fotakis, P.; Ouimet, M.; Bochem, A.E.; Zhang, H.; Molusky, M.M.; Wang, W.; Abramowicz, S.; la Bastide-van Gemert, S.; Wang, N.; et al. Cholesterol efflux pathways suppress inflammasome activation, NETosis and atherogenesis. Circulation 2018, 138, 898–912. [Google Scholar] [CrossRef]
- Thurm, C.; Schraven, B.; Kahlfuss, S. ABC transporters in T cell-mediated physiological and pathological immune responses. Int. J. Mol. Sci. 2021, 22, 9186. [Google Scholar] [CrossRef]
- Armstrong, A.J.; Gebre, A.K.; Parks, J.S.; Hedrick, C.C. ATP binding cassette transporter G1 negatively regulates thymocyte and peripheral lymphocyte proliferation. J. Immunol. 2010, 184, 173–183. [Google Scholar] [CrossRef] [PubMed]
- Bensinger, S.J.; Bradley, M.N.; Joseph, S.B.; Zelcern NJanssen, E.M.; Hausner, M.A.; Shish, R.; Parks, J.S.; Edwards, P.A.; Jamieson, B.D.; Tontonez, P. LXR signalling couples sterol metabolism to proliferation in the acquired immune response. Cell 2008, 134, 97–111. [Google Scholar] [CrossRef] [PubMed]
- Sag, D.; Wingender, G.; Nowyhed, H.; Wu, R.; Gebre, A.K.; Parks, J.S.; Kronenberg, M.; Hedrick, C.C. ATP-binding cassette transporter G1 intrinsically regulates invariant NKT development. J. Immunol. 2012, 189, 5129–5138. [Google Scholar] [CrossRef]
- Cheng, H.Y.; Gaddis, D.E.; Wu, R.; McSkimming, C.; Haynes, L.D.; Taylor, A.M.; McNamara, C.A.; Sorci-Thomas, M.; Hedrick, C.C. Loss of ABCG1 influences T cell differentiation and atherosclerosis. J. Clin. Invest. 2016, 126, 3236–3246. [Google Scholar] [CrossRef]
- Pierce, S.K. Lipid rafts and B-cell activation. Nat. Rev. Immunol. 2002, 2, 96–105. [Google Scholar] [CrossRef]
- Blery, M.; Tze, L.; Miosge, L.A.; Jun, J.E.; Goodnow, C.C. Essential role of membrane cholesterol in accelerated BCR internalisation and uncoupling from NF-κB in B cell clonal anergy. J. Exp. Med. 2006, 203, 1773–1783. [Google Scholar] [CrossRef] [PubMed]
- Viaud, M.; Abdel-Wahab, O.; Gall, J.; Ivanov, S.; Guinamard, R.; Sore, S.; Merlin, J.; Ayrault, M.; Guilbaud, E.; Jacquel, A.; et al. ABCA1 exerts tumor suppressor function in myeloproliferative neoplasma. Cell Rep. 2020, 30, 3397–3410.e5. [Google Scholar] [CrossRef]
- von Eckardstein, A.; Widmann, C. High-density lipoprotein, beta cells, and diabetes. Cardiovasc. Res. 2014, 103, 384–394. [Google Scholar] [CrossRef]
- Rutti, S.; Ehses, J.A.; Sibler, R.A.; Prazak, R.; Rohrer, L.; Georgopoulos, S.; Meier, D.R.; Niclauss, N.; Berney, T.; Donath, M.Y.; et al. Low- and high-density lipoproteins modulate function, apoptosis, and proliferation of primary human and murine pancreatic beta cells. Endocrinology 2009, 150, 4521–4530. [Google Scholar] [CrossRef]
- Fryirs, M.A.; Barter, P.J.; Appavoo, M.; Tuch, B.E.; Tabet, F.; Heather, A.K.; Rye, K.-A. Effects of high-density lipoproteins on pancreatic beta-cell insulin secretion. Arterioscler. Thromb. Vasc. Biol. 2010, 30, 1642–1648. [Google Scholar] [CrossRef] [PubMed]
- Yalcinkaya, M.; Kerksiek, A.; Gebert, K.; Annema, W.; Sibler, R.; Radosavljevic, S.; Lutjohann, D.; Rohrer, L.; von Eckardstein, A. HDL inhibits endoplasmic reticulum stress-induced apoptosis of pancreatic β-cells in vitro by activation of Smoothened. J. Lipid Res. 2020, 61, 492–504. [Google Scholar] [CrossRef] [PubMed]
- Cochran, B.J.; Bisoendial, R.J.; Hou, L.; Glaros, E.N.; Rossy, J.; Thomas, S.R.; Barter, P.J.; Rye, K.-A. Apolipoprotein A-I increases insulin secretion and production from pancreatic β-cells via a G-protein-cAMP-PKA-FoxO1-dependent mechanism. Arterioscler. Thromb. Vasc. Biol. 2014, 34, 2261–2267. [Google Scholar] [CrossRef] [PubMed]
- Hou, L.; Tang, S.; Wu, B.J.; Ong, K.-L.; Westerterp, M.; Barter, P.J.; Cochran, B.J.; Tabet, F.; Rye, K.-A. Apolipoprotein A-I improves pancreatic β-cell function independent of the ATP-binding cassette transporters ABCA1 and ABCG1. FASEB J. 2019, 33, 8479–8489. [Google Scholar] [CrossRef]
- Nilsson, O.; Del Giudice, R.; Nago, M.; Gronberg, C.; Eliasson, L.; Lagerstedt, J.O. Apolipoprotein A-I primes beta cells to increase glucose-stimulated insulin secretion. Biochim. Biophys. Acta Mol. Basis Dis. 2020, 1866, 165613. [Google Scholar] [CrossRef] [PubMed]
- Brunham, L.R.; Kruit, J.R.; Pape, T.D.; Timmins, J.M.; Reuwer, A.Q.; Vasanji, Z.; Marsh, B.J.; Rodrigues, B.; Johnson, J.D.; Parks, J.S.; et al. Beta-cell ABCA1 influences insulin secretion, glucose homeostasis and response to thiazolidine treatment. Nat. Med. 2007, 13, 340–347. [Google Scholar] [CrossRef]
- Kruit, J.K.; Kremer, P.H.C.; Dai, L.; Tang, R.; Ruddle, P.; de Haan, W.; Brunham, L.R.; Verchere, C.B.; Hayden, M.R. Cholesterol efflux via ATP-binding cassette transporter A1 (ABCA1) and cholesterol uptake via the LDL receptor influences cholesterol-induced impairment of beta cell function in mice. Diabetologia 2010, 53, 1110–1119. [Google Scholar] [CrossRef]
- Kruit, J.K.; Wijesekara, N.; Fox, J.E.M.; Dai, X.-Q.; Brunham, L.R.; Searle, G.J.; Morgan, G.P.; Costin, A.J.; Tang, R.; Bhattacharjee, A.; et al. Islet cholesterol accumulation due to loss of ABCA1 leads to impaired exocytosis of insulin granules. Diabetes 2011, 60, 3186–3196. [Google Scholar] [CrossRef]
- Wijesekara, N.; Zhang, L.-H.; Kang, M.H.; Abraham, T.; Bhattacharjee, A.; Warnock, G.L.; Verchere, C.B.; Hayden, M.R. miR-33a modulates ABCA1 expression, cholesterol accumulation, and insulin secretion in pancreatic islets. Diabetes 2012, 61, 653–658. [Google Scholar] [CrossRef]
- Kruit, J.K.; Wijesekara, N.; Westwell-Roper, C.; Vanmierlo, T.; de Haan, W.; Bhattacharjee, A.; Tang, R.; Wellington, C.L.; LutJohann, D.; Johnson, J.D.; et al. Loss of ABCA1 and ABCG1 results in increased disturbances in islet sterol homeostasis, inflammation and impaired β-cell function. Diabetes 2012, 61, 659–664. [Google Scholar] [CrossRef]
- Dullaart, R.P.F.; Annema, W.; de Boer, J.F.; Tietge, U.J.F. Pancreatic β-cell function relates positively to HDL functionality in well-controlled type 2 diabetes mellitus. Atherosclerosis 2012, 222, 567–573. [Google Scholar] [CrossRef]
- Bardini, G.; Dicembrini, I.; Rotella, C.M.; Giannini, S. Correlation between HDL cholesterol levels and beta-cell function in subjects with various degree of glucose tolerance. Acta Diabetol. 2013, 50, 277–281. [Google Scholar] [CrossRef] [PubMed]
- Zheng, S.; Xu, H.; Zhou, H.; Ren, X.; Han, T.; Chen, Y.; Qui, H.; Wu, P.; Zheng, J.; Wang, L.; et al. Associations of lipid profiles with insulin resistance and beta cell function in adults with normal glucose tolerance and different categories of impaired glucose regulation. PLoS ONE 2017, 12, e0172221. [Google Scholar] [CrossRef]
- Kumar, H.; Mishra, M.; Bajpai, S.; Pokhria, D.; Arya, A.K.; Singh, R.K.; Tripathi, K. Correlation of insulin resistance, beta cell function and insulin sensitivity with serum sFas and sFasL in newly diagnosed type 2 diabetes. Acta Diabetol. 2013, 50, 511–518. [Google Scholar] [CrossRef] [PubMed]
- Fiorentino, T.V.; Succurro, E.; Marini, M.A.; Pedace, E.; Andreozzi, F.; Perticone, M.; Sciacqua, A.; Perticone, F.; Sesti, G. HDL cholesterol is an independent predictor of β-cell function decline and incident type 2 diabetes: A longitudinal study. Diabetes Metab. Res. Rev. 2020, 36, e3289. [Google Scholar] [CrossRef]
- Tarlton, M.R.; Patterson, S.; Graham, A. MicroRNA sequences modulated by beta cell lipid metabolism: Implications for type 2 diabetes mellitus. Biology 2021, 10, 534. [Google Scholar] [CrossRef]
- Pan, B.; May, Y.; Ren, H.; He, Y.; Wang, Y.; Lv, X.; Lui, D.; Yu, B.; Wang, Y.; Chen, Y.E.; et al. Diabetic HDL is dysfunctional in stimulating endothelial cell migration and proliferation due to down regulation of SR-B1 expression. PLoS ONE 2012, 7, e48530. [Google Scholar] [CrossRef]
- Zanoni, P.; Khetarpal, S.A.; Larach, D.B.; Hancock-Cerutti, W.F.; Millar, J.S.; Cuchel, M.; Der Ohannessian, S.; Kontush, A.; Surendran, P.; Saleheen, D.; et al. Rare variant in scavenger receptor B1 raises HDL cholesterol and increases risk of coronary heart disease. Science 2016, 351, 1166–1171. [Google Scholar] [CrossRef]
- Tao, H.; Yancey, P.G.; Blakemore, J.L.; Zhang, Y.; Ding, L.; Jerome, W.G.; Brown, J.D.; Vickers, K.C.; Linton, M.F.J. Macrophage SR-B1 modulates autophagy via VPS34 complex and PPARα transcription of Tfeb in atherosclerosis. J. Clin. Investig. 2021, 131, e94229. [Google Scholar] [CrossRef]
- Plebanek, M.P.; Bhaumik, D.; Bryce, P.J.; Thaxton, C.S. Scavenger receptor type B1 and lipoprotein nanoparticle inhibit myeloid derived suppressor cells. Mol. Cancer Ther. 2018, 17, 686–697. [Google Scholar] [CrossRef]
- Jin, F.; Hagemann, N.; Sun, L.; Wu, J.; Doeppner, T.R.; Dai, Y.; Hermann, D.M. High-density lipoprotein (HDL) promotes angiogenesis via S1P3-dependent VEGFR2 activation. Angiogenesis 2018, 21, 381–394. [Google Scholar] [CrossRef]
- Primer, K.R.; Psaltis, P.J.; Tan, J.T.M.; Bursill, C.A. The role of high-density lipoproteins in endothelial cell metabolism and diabetes-impaired angiogenesis. Int. J. Mol. Sci. 2020, 21, 3633. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Lei, Z.; Han, W.-Q.; Wang, Q.-R.; Wu, H.-Y.; Liu, X.-H.; Xing, K.; Cheng, G.; Chang, F.-J. The alteration of HDL in patients with AMI inhibited angiogenesis by blocking ERK1/2 activation. Cardiovasc. Ther. 2022, 2022, 1057772. [Google Scholar] [CrossRef] [PubMed]
- Lotfollahi, Z.; Dawson, J.; Fitridge, R.; Bursill, C. The anti-inflammatory and proangiogenic properties of high-density lipoproteins: An emerging role in diabetic wound healing. Adv. Wound Care 2021, 10, 370–380. [Google Scholar] [CrossRef] [PubMed]
- Gordts, S.C.; Muthuramu, I.; Amin, R.; Jacobs, F.; De Geest, B. The impact of lipoproteins on would healing: Topical HDL therapy corrects delayed wound healing in apolipoprotein E deficient mice. Pharmaceuticals 2014, 7, 419–432. [Google Scholar] [CrossRef]
- Tsatralis, T.; Ridiandries, A.; Robertson, S.; Vanags, L.Z.; Lam, Y.T.; Tan, J.T.M.; Ng, M.K.C.; Bursill, C.A. Reconstituted high-density lipoproteins promote wound repair and blood flow recovery in response to ischaemia in aged mice. Lipids Health Dis. 2016, 15, 150. [Google Scholar] [CrossRef]
- Nofer, J.-R.; Brodde, M.R.; Kehrel, B.E. High-density lipoproteins, platelets and the pathogenesis of atherosclerosis. Clin. Exp. Pharmacol. Physiol. 2010, 37, 726–735. [Google Scholar] [CrossRef]
- Negre-Salvayre, A.; Dousset, N.; Ferretti, G.; Bacchetti, T.; Curatola, G.; Salvayre, R. Antioxidant and cytoprotective properties of high-density lipoproteins in vascular cells. Free Radic. Biol. Med. 2006, 41, 1031–1040. [Google Scholar] [CrossRef]
- Karlsson, H.; KoKarlntush, A.; James, R.W. Functionality of HDL: Antioxidation and detoxifying effects. In High Density Lipoproteins; Handbook of Experimental Pathology; von Eckardstein, A., Kardassis, D., Eds.; Springer: Cham, Switzerland, 2015; Volume 224, pp. 209–221. [Google Scholar] [CrossRef]
- Nofer, J.-R. Signal transduction by HDL: Agonists, receptors and signalling cascades. Handb. Exp. Pharmacol. 2015, 224, 229–256. [Google Scholar] [CrossRef]
- Rayner, K.J.; Moore, K.J. microRNA control of HDL metabolism and function. Circ. Res. 2014, 114, 183–192. [Google Scholar] [CrossRef]
- Canfran-Duque, A.; Lin, C.-S.; Goedeke, L.; Suarez, Y.; Fernandez-Hernando, C. MicroRNAs and HDL metabolism. Arterioscler. Thromb. Vasc. Biol. 2016, 36, 1076–1084. [Google Scholar] [CrossRef] [PubMed]
- Citrin, K.M.; Fernandez-Hernando, C.; Suarez, Y. MicroRNA regulation of cholesterol metabolism. Ann. N. Y. Acad. Sci. 2021, 1495, 55–77. [Google Scholar] [CrossRef]
- Rodriguez, A.; Griffiths-Jones, S.; Ashurst, J.; Bradley, A. Identification of mammalian microRNA host genes and transcription units. Genome Res. 2004, 14, 1902–1910. [Google Scholar] [CrossRef] [PubMed]
- Baskerville, S.; Bartel, D. Microarray profiling of microRNAs reveals frequent coexpression with neighboring miRNAs and host genes. RNA 2005, 11, 241–247. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.; Kim, M.; Han, J.; Yeom, K.; Lee, S.; Baek, S.; Kim, V. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004, 23, 4051–4060. [Google Scholar] [CrossRef]
- Borchert, G.; Lanier, W.; Davidson, B. RNA polymerase III transcribes human microRNAs. Nat. Struct. Mol. Biol. 2006, 13, 1097–1101. [Google Scholar] [CrossRef]
- Monteys, A.; Spengler, R.; Wan, J.; Tecedor, L.; Lennox, K.; Xing, Y.; Davidson, B. Structure and activity of putative intronic miRNA promoters. RNA 2010, 16, 495–505. [Google Scholar] [CrossRef]
- Ramalingam, P.; Palanichamy, J.; Singh, A.; Das, P.; Bhagat, M.; Kassab, M.; Sinha, S.; Chattopadhyay, P. Biogenesis of intronic miRNAs located in clusters by independent transcription and alternative splicing. RNA 2014, 20, 76–87. [Google Scholar] [CrossRef]
- Abdelfattah, A.; Park, C.; Choi, M. Update on non-canonical microRNAs. Biomol. Concepts 2014, 5, 275–287. [Google Scholar] [CrossRef]
- Cai, X.; Hagedorn, C.; Cullen, B. Human microRNAs are processed from capped, polyadenylated transcripts that can function as mRNAs. RNA 2004, 10, 1957–1966. [Google Scholar] [CrossRef]
- Han, J.; Lee, Y.; Yeom, K.; Kim, Y.; Jin, H.; Kim, V. The Drosha-DGCR8 complex in primary microRNA processing. Genes. Dev. 2004, 18, 3016–3027. [Google Scholar] [CrossRef]
- Alarcon, C.; Lee, H.; Goodarzi, H.; Halberg, N.; Tavazoie, S. N6-methyladenosine (m6A) marks primary microRNAs for processing. Nature 2015, 519, 482–485. [Google Scholar] [CrossRef]
- Yeom, K.; Lee, Y.; Han, J.; Suh, M.; Kim, V. Characterization of DGCR8/Pasha, the essential cofactor for Drosha in primary miRNA processing. Nucleic Acids Res. 2006, 34, 4622–4629. [Google Scholar] [CrossRef] [PubMed]
- Han, J.; Lee, Y.; Yeom, K.; Nam, J.; Heo, I.; Rhee, J.; Sohn, S.; Cho, Y.; Zhang, B.; Kim, V. Molecular basis for the recognition of primary microRNAs by the Drosha-DGCR8 complex. Cell 2006, 125, 887–901. [Google Scholar] [CrossRef]
- Lee, Y.; Ahn, C.; Han, J.; Choi, H.; Kim, J.; Yim, J.; Lee, J.; Provost, P.; Rådmark, O.; Kim, S.; et al. The nuclear RNase III Drosha initiates microRNA processing. Nature 2003, 425, 415–419. [Google Scholar] [CrossRef] [PubMed]
- Bohnsack, M.; Czaplinski, K.; Gorlich, D. Exportin 5 is a RanGTP-dependent dsRNA-binding protein that mediates nuclear export of pre-miRNAs. RNA 2004, 10, 185–191. [Google Scholar] [CrossRef]
- Hutvágner, G.; McLachlan, J.; Pasquinelli, A.; Bálint, E.; Tuschl, T.; Zamore, P. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science 2001, 293, 834–838. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Kolb, F.; Jaskiewicz, L.; Westhof, E.; Filipowicz, W. Single processing center models for human Dicer and bacterial RNase III. Cell 2004, 118, 57–68. [Google Scholar] [CrossRef]
- Gregory, R.; Chendrimada, T.; Cooch, N.; Shiekhattar, R. Human RISC couples microRNA biogenesis and posttranscriptional gene silencing. Cell 2005, 123, 631–640. [Google Scholar] [CrossRef]
- Lewis, B.; Shih, I.; Jones-Rhoades, M.; Bartel, D.; Burge, C. Prediction of mammalian microRNA targets. Cell 2003, 115, 787–798. [Google Scholar] [CrossRef]
- Lewis, B.; Burge, C.; Bartel, D. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 2005, 120, 15–20. [Google Scholar] [CrossRef] [PubMed]
- Grimson, A.; Farh, K.; Johnston, W.; Garrett-Engele, P.; Lim, L.; Bartel, D. MicroRNA targeting specificity in mammals: Determinants beyond seed pairing. Mol. Cell 2007, 27, 91–105. [Google Scholar] [CrossRef] [PubMed]
- Moore, M.; Scheel, T.; Luna, J.; Park, C.; Fak, J.; Nishiuchi, E.; Rice, C.; Darnell, R. miRNA–target chimeras reveal miRNA 3′-end pairing as a major determinant of Argonaute target specificity. Nat. Commun. 2015, 6, 8864. [Google Scholar] [CrossRef]
- Friedman, R.C.; Farh, K.K.; Burge, C.B.; Bartel, D.P. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009, 139, 466–472. [Google Scholar] [CrossRef]
- Bushati, N.; Cohen, S.M. MicroRNA functions. Annu. Rev. Cell Dev. Biol. 2007, 23, 175–205. [Google Scholar] [CrossRef]
- Deng, Q.; Hu, H.; Yu, X.; Liu, S.; Wang, L.; Chen, W.; Zhang, C.; Zeng, Z.; Cao, Y.; Xu-Monette, Z.Y.; et al. Tissue–specific microRNA expression alters cancer susceptibility conferred by a TP53 noncoding variant. Nat. Commun. 2019, 10, 5061. [Google Scholar] [CrossRef] [PubMed]
- Schulte, C.; Zeller, T. MicroRNA-based diagnostics and therapy in cardiovascular disease—Summing up the facts. Cardiovasc. Diagn. Ther. 2015, 5, 17–26. [Google Scholar] [CrossRef]
- Genemaras, A.A.; Ennis, H.; Kaplan, L.; Huang, C.Y. Inflammatory cytokines induced specific time- and concentration-dependent microRNA release by chondrocytes, synoviocytes and meniscus cells. J. Orthop. Res. 2016, 34, 779–790. [Google Scholar] [CrossRef]
- De Rosa, R.; De Rosa, S.; Leistner, D.; Boeckel, J.N.; Keller, T.; Fichtlscherer, S.; Dimmeler, S.; Zeiher, A.M. Transcoronary concentration gradient of microRNA-133a and outcome in patients with coronary artery disease. Am. J. Cardiol. 2017, 120, 15–24. [Google Scholar] [CrossRef]
- Jung, H.J.; Suh, Y. Circulating miRNAs in ageing and ageing-related diseases. J. Genet. Genom. 2014, 41, 465–472. [Google Scholar] [CrossRef]
- Solis-Toro, D.; Escudero, M.M.; Garcia-Perdomo, H.A. Association between circulating microRNAs and the metabolic syndrome in adult populations: A systematic review. Diabetes Metab. Syndr. Clin. Res. Rev. 2022, 16, 102376. [Google Scholar] [CrossRef]
- Cheleschi, S.; Tenti, C.; Bedogni, G.; Fioraventi, A. Circulating miR-140 and leptin improve the accuracy of the differential diagnosis between psoriatic arthritis and rheumatioid arthritis: A case-control study. Transl. Res. 2022, 239, 18–34. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Li, Z.; Zhao, M.; Ye, W.; Wu, H.; Liao, Q.; Bu, S.; Zhang, Y. Circulating miRNAs miR-574-5p and miR-3135b are potential metabolic regulators for serum lipids and blood glucose in gestational diabetes mellitus. Gynecol. Endocrinol. 2021, 37, 665–671. [Google Scholar] [CrossRef]
- Higuchi, C.; Nakatsuka, A.; Eguchi, J.; Tesigawa, S.; Kanzaki, M.; Katayama, A.; Yamaguchi, S.; Takahashi, N.; Murakami, K.; Ogawa, D.; et al. Identification of circulating miR-101, miR-375 and miR-802 as biomarkers for type 2 diabetes. Metabolism 2015, 64, 489–497. [Google Scholar] [CrossRef] [PubMed]
- Enwald, M.; Lehtimaki, T.; Mishra, P.P.; Mononen, N.; Murtola, T.J.; Raioharju, E. Human prostate tissue microRNAs and their predicted target pathways linked to prostate cancer risk factors. Cancers 2021, 13, 3537. [Google Scholar] [CrossRef] [PubMed]
- Michell, D.L.; Vickers, K.C. Lipoprotein carriers of microRNAs. Biochim. Biophys. Acta 2016, 1861, 2069–2074. [Google Scholar] [CrossRef]
- Mathivanan, S.; Ji, H.; Simpson, R.J. Exosomes: Extracellular organelles important in intercellular communication. J. Proteom. 2010, 73, 1907–1920. [Google Scholar] [CrossRef]
- Cantaluppi, V.; Gatti, S.; Medica, D.; Figiolini, F.; Bruno, S.; Deregibus, M.C.; Sordi, A.; Biancone, L.; Tetta, C.; Camussi, G. Microvesicles derived from endothelial progenitor cells protect the kidney from ischaemia-reperfusion injury by microRNA-dependent reprogramming of resident renal cells. Kidney Int. 2012, 82, 412–427. [Google Scholar] [CrossRef]
- Zernecke, A.; Bidzehkov, K.; Noels, H.; Shagdarsuren, E.; Gan, L.; Deneceke, B.; Hirstove, M.; Koppel, T.; Janantigh, M.N.; Lutgens, E.; et al. Delivery of microRNA-126 by apoptotic bodies induces CXCL12-dependent vascular protection. Sci. Signal. 2009, 2, rra81. [Google Scholar] [CrossRef]
- Arroyo, J.D.; Chevillet, J.R.; Kroh, E.M.; Ruf, I.K.; Pritchard, C.C.; Gibson, D.F.; Mitchell, P.S.; Bennett, C.F.; Pogosova-Agadjanyan, E.L.; Stirewalt, D.; et al. Argonaute 2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc. Natl. Acad. Sci. USA 2011, 108, 5003–5008. [Google Scholar] [CrossRef]
- Turchinov, A.; Burwinkel, B. Distinct AGO1 and AGO2 associated miRNA profiles in human cells and blood plasma. RNA Biol. 2012, 9, 1066–1075. [Google Scholar] [CrossRef] [PubMed]
- Colombo, M.; Raposo, G.; Thery, C. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu. Rev. Cell Dev. Biol. 2014, 30, 255–289. [Google Scholar] [CrossRef]
- Seneshaw, M.; Mirshahi, F.; Min, H.-K.; Asgharpour, A.; Mirshahi, S.; Daita, K.; Boyett, S.; Santekadur, P.K.; Fuchs, M.; Sanyal, A.J. Fast and simplified method for high throughput isolation of miRNA from highly purified High Density Lipoprotein. J. Vis. Exp. 2016, 113, 54257. [Google Scholar] [CrossRef]
- Ishikawa, H.; Yamada, H.; Taromaru, N.; Kondo, K.; Nagura, A.; Yamazaki, M.; Ano, Y.; Munetsuna, E.; Suzuki, K.; Ohashi, K.; et al. Stability of serum high-density lipoprotein-microRNAs for preanalytical conditions. Ann. Clin. Biochem. 2017, 54, 134–142. [Google Scholar] [CrossRef] [PubMed]
- Torres LFC, Zhu W, Ohrling G, Larsson R, Patel M, Wiese CB, Rye K-A, Vickers KC, Tabet F High density lipoproteins induce miR-223-3p biogenesis and export from myeloid cells: Role of scavenger receptor B1-mediated lipid transfer. Atherosclerosis 2019, 286, 20–39. [CrossRef]
- Desgagne, V.; Guerin, R.; Guay, S.P.; Boyer, M.; Hutchins, E.; Picard, S.; Marechal, A.; Corbin, F.; Keuren-Jensen, K.V.; Arsenault, B.J.; et al. Human high-density lipoprotein microtranscriptome is unique and suggests an extended role in lipid metabolism. Epigenomics 2019, 11, 917–934. [Google Scholar] [CrossRef]
- Michell, D.L.; Allen, R.M.; Cavnar, A.B.; Contreras, D.M.; Yu, M.; Semler, E.M.; Massick, C.; Raby, C.A.; Castleberry, M.; Ramirez, M.A.; et al. Elucidation of physio-chemical principles of high-density lipoprotein-small RNA binding interactions. J. Biol. Chem. 2022, 298, 101952. [Google Scholar] [CrossRef]
- Sedgeman, L.R.; Beysen, C.; Salonao, M.A.R.; Michell, D.L.; Sheng, Q.; Zhao, S.; Turner, S.; Linton, M.F.; Vickers, K.C. Beta cell secretion of miR-375-3p to HDL is inversely associated with insulin secretion. Sci. Rep. 2019, 9, 3803. [Google Scholar] [CrossRef]
- Veremeyko, T.; Kuznetsova, I.S.; Dukhiova, M.; Yung, A.W.Y.; Kopeikina, E.; Bartneva, N.S.; Ponomarev, E.D. Neuronal extracellular microRNAs miR-124 and miR-9 mediate cell-cell communication between neurons and microglia. J. Neurosci. Res. 2019, 97, 162–184. [Google Scholar] [CrossRef]
- Peng, Y.; Wang, X.; Guo, Y.; Peng, F.; Zheng, N.; He, B.; Ge, H.; Tao, L.; Wang, Q. Pattern of cell-to-cell transfer of microRNA by gap junction and its effect on the proliferation of glioma cells. Cancer Sci. 2019, 110, 1945–1958. [Google Scholar] [CrossRef]
- Morel, S.; Frias, M.A.; Rosker, C.; James, R.W.; Rohr, S.; Kwak, B.R. The natural cardioprotective particle HDL modulates connexin 43 gap junction channels. Cardiovasc. Res. 2012, 93, 41–49. [Google Scholar] [CrossRef] [PubMed]
- Wolfrum, C.; Shi, S.; Jayaprakash, K.N.; Jayaraman, M.; Wang, G.; Pandey, R.K.; Rajeev, K.G.; Nakayama, T.; Charrise, K.; Ndungo, E.M.; et al. Mechanisms and optimization of in vivo delivery of lipophilic siRNAs. Nat. Biotechnol. 2007, 25, 1149–1157. [Google Scholar] [CrossRef] [PubMed]
- Ben-Aicha, S.; Escate, R.; Casani, L.; Padro, T.; Pena, E.; Arderiu, G.; Mendieta, G.; Badimon, L.; Vilahur, G. High-density lipoprotein remodelled in hypercholesterolaemic blood induce epigenetically driven downregulation of endothelial HIF-1alpha expression in a preclinical animal model. Cardiovasc. Res. 2020, 116, 1288–1299. [Google Scholar] [CrossRef]
- Allen, R.M.; Zhao, S.; Ramirez Solano, M.A.; Zhu, W.; Michell, D.L.; Wang, Y.; Shyr, Y.; Sethupathy, P.; Linton, M.F.; Graf, G.A.; et al. Bioinformatic analysis of endogenous and exogenous small RNAs on lipoproteins. J. Extracell. Vesicles. 2018, 7, 1506198. [Google Scholar] [CrossRef]
- Finnegan, E.F.; Pasquinelli, A.E. MicroRNA biogenesis: Regulating the regulators. Crit. Rev. Biochem. Mol. Biol. 2013, 48, 51–68. [Google Scholar] [CrossRef] [PubMed]
- Krol, J.; Loedige, I.; Filipowicz, W. The widespread regulation of microRNA biogenesis, function and decay. Nat. Rev. Genet. 2010, 11, 597–610. [Google Scholar] [CrossRef]
- Rayner, K.J.; Suarez, Y.; Davalos, A.; Parathath, S.; Fitzgerald, M.L.; Tamehiro, N.; Fisher, E.A.; Moore, K.J.; Fernandez-Hernando, C. MiR-33 contributes to the regulation of cholesterol homeostasis. Science 2010, 328, 1570–1573. [Google Scholar] [CrossRef]
- Najafi-Shoushtari, S.H.; Kristo, F.; Li, Y.; Shioda, T.; Cohen, D.E.; Gerszten, R.E.; Naar, A.M. MicroRNA-33 and the SREBP host genes cooperate to control cholesterol homeostasis. Science 2010, 328, 1566–1569. [Google Scholar] [CrossRef]
- Martinez, N.J.; Ow, M.C.; Barrasa, M.I.; Hammell, M.; Sequerra, R.; Doucette-Stamm, L.; Roth, F.P.; Ambros, V.R.; Walhout, A.J. A C. elegans genome-scale microRNA network contains composite feedback motifs with high flux capacity. Genes. Dev. 2008, 22, 2535–2549. [Google Scholar] [CrossRef]
- Lu Lf Thai, T.H.; Calado, D.P.; Chaudry, A.; Kubo, M.; Tanaka, K.; Loeb, G.B.; Lee, H.; Yoshimura, A.; Rajewsky, K.; Rudensky, A.Y. Foxp3-dependent microRNA155 confers competitive fitness to regulatory T cells by targeting SOCS1 protein. Immunity 2009, 30, 80–91. [Google Scholar] [CrossRef]
- Gatto, G.; Rossi, A.; Rossi, D.; Kroening, S.; Bonatti, S.; Mallardo, M. Epstein-Barr virus latent membrane protein 1 trans-activates miR-155 transcription through the NF-kappaB pathway. Nucleic Acids Res. 2008, 36, 6608–6619. [Google Scholar] [CrossRef] [PubMed]
- Yin, Q.; Wang, X.; McBride, J.; Fewell, C.; Flemington, E. B-cell receptor activation induces BIC/miR-155 expression through a conserved AP-1 element. J. Biol. Chem. 2008, 283, 2654–2662. [Google Scholar] [CrossRef] [PubMed]
- Barbalata, T.; Moraru, O.E.; Stancu, C.S.; Sima, A.V.; Niculescu, L.S. MiR-223-3p levels in the plasma and atherosclerotic plaques are increased in aged patients with carotid artery stenosis; association with HDL-related proteins. Mol. Rep. 2022, 49, 6779–6788. [Google Scholar] [CrossRef]
- Simionescu, N.; Niculescu, L.S.; Sanda, G.M.; Margina, D.; Sima, A.V. Analysis of circulating microRNAs that are specifically increased in hyperlipidaemia and/or hyperglycemic sera. Mol. Biol. Rep. 2014, 41, 5765–5773. [Google Scholar] [CrossRef]
- Kawahara, Y.; Mieda-Sato, A. TDP-43 promotes microRNA biogenesis as a component of the Drosha and Dicer complexes. Proc. Natl. Acad. Sci. USA 2012, 109, 3347–3352. [Google Scholar] [CrossRef]
- Stribl, C.; Samara, A.; Trumbach, D.; Peis, R.; Neumann, M.; Guchs, H.; Gailus-Durner, V.; de Angelis, M.J.; Rathkolb, B.; Wolf, E.; et al. Mitochondrial dysfunction and decrease in body weight of a transgenic knock-in mouse model for TDP-43. J. Biol. Chem. 2014, 289, 10769–10784. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Liu, Y.; Zhang, T.; Yin, C.; Kang, S.Y.; Kim, S.J.; Park, Y.-K.; Jung, H.W. Effects of root extract of Morinda officinalis in mice with high-fat-diet/streptozotocin-induced diabetes and c2C12 myoblast differentiation. ACS Omega 2021, 6, 26959–26968. [Google Scholar] [CrossRef]
- Xia, P.; Vadas, M.A.; Rye, K.A.; Barter, P.G.; Gamble, J.R. High densitiy lipoproteins (HDL) interrupt the sphingosine kinase signaling pathway. A possible mechanism for protection against atherosclerosis by HDL. J. Biol. Chem. 1999, 274, 33134307. [Google Scholar] [CrossRef]
- McGrath, K.C.Y.; Li, X.H.; Puranik, R.; Liong, E.C.; Tan, J.T.M.; Dy, V.M.; DiBartolo, V.A.; Barter, P.J.; Rye, K.A.; Heather, A.K. Role of 3 beta-hydroxysteroid-delta 24 reductase in mediating antiinflammatory effects of high-density lipoproteins in endothelial cells. Arterioscler. Thromb. Vasc. Biol. 2009, 29, 877–882. [Google Scholar] [CrossRef]
- Park, S.-H.; Park, J.H.Y.; Kang, J.-S.; Kang, Y.-H. Involvement of transcription factors in plasma HDL protection against TNF alpha-induced vascular cell adhesion molecule-1 expression. Int. J. Biochem. Cell Biol. 2003, 35, 168–182. [Google Scholar] [CrossRef]
- Van der Vorst, E.P.C.; Vanags, L.Z.; Dunn, D.L.; Prosser, H.C.; Rye, K.-A.; Bursill, C.A. High-density lipoproteins suppress chemokine expression and proliferation in human vascular smooth muscle cells. FASEB J. 2013, 27, 1413025. [Google Scholar] [CrossRef]
- McGrath, K.C.; Li, X.H.; Whitworth, P.T.; Kasz, R.; Tan, J.T.; McLennan, S.V.; Celermajer, D.S.; Barter, P.J.; Rye, K.-A.; Heather, A.K. High density lipoproteins improve insulin sensitivity in high-fat diet-fed mice by suppressing hepatic inflammation. J. Lipid Res. 2014, 55, 421–430. [Google Scholar] [CrossRef] [PubMed]
- Spirig, R.; Shaub, A.; Kropf, A.; Miescher, S.; Spycher, M.O.; Rieben, R. Reconstituted high-density lipoprotein modulates activation of human leukocytes. PLoS ONE 2013, 8, e71235. [Google Scholar] [CrossRef]
- Hong, S.; Niu, M.; Meng, D.; Li, A.; Dong, Q.; Zhang, J.; Tian, X.; Lu, S.; Wang, Y. High-density lipoprotein reduces microglia activation and protects against experimental autoimmune encephalomyelitis in mice. Int. Immunopharmacol. 2022, 105, 108566. [Google Scholar] [CrossRef] [PubMed]
- Spillmann, F.; Miteva, K.; Pieske, B.; Tschope, C.; Van Linthout, S. High-density lipoproteins reduce endothelial-to-mesenchymal transition. Arterioscler. Thromb. Vasc. Biol. 2015, 35, 1771–1777. [Google Scholar] [CrossRef]
- Lin, F.Y.; Lin, Y.W.; Shih, C.M.; Lin, S.J.; Tung, Y.T.; Li, C.Y.; Chen, Y.H.; Lin, C.Y.; Tsai, Y.T.; Huang, C.Y. A novel relative high-density lipoprotein index to predict the structural changes in high-density lipoprotein and its ability to inhibit endothelial-mesenchymal transition. Int. J. Mol. Sci. 2021, 22, 5210. [Google Scholar] [CrossRef]
- Feng, J.; Zhang, J.; Jackson, A.O.; Zhu, X.; Chen, H.; Chen, W.; Gui, Q.; Yin, K. Apolipoprotein A-I inhibits the TGF-β1-induced endothelial-to-mesenchymal transition of human coronary artery endothelial cells. Cardiology 2017, 37, 179–187. [Google Scholar] [CrossRef] [PubMed]
- Ng, K.M.; Lee, Y.K.; Lai, W.H.; Chan, Y.C.; Fung, M.L.; Tse, H.F.; Siu, C.W. Exogenous expression of human apoA-I enhances cardiac differentiation of pluripotent stem cells. PLoS ONE 2011, 6, e19787. [Google Scholar] [CrossRef]
- Muller, M.; Schoeberlein, A.; Zhou, J.; Joerger-Messerli, M.; Oppiger, B.; Reinhart, U.; Bordey, A.; Surbek, D.; Barnea, E.R.; Huang, Y.; et al. Preimplantation factor bolsters neuroprotection via modulating protein kinase A and protein kinase C signalling. Cell Death Differ. 2015, 22, 2078–2086. [Google Scholar] [CrossRef]
- Shi, Z.; To, S.K.Y.; Zhang, S.; Deng, S.; Artemenko, M.; Zhang, M.; Tang, J.; Zeng, J.Z.; Wong, A.S.T. Hypoxia-induced Nur77 activates PI3K/Akt signalling via suppression of Dicer/let-7i-5p to induce epithelial to mesenchymal transition. Theranostics 2021, 11, 3376–3391. [Google Scholar] [CrossRef]
- Brandao, B.B.; Madsen, S.; Rabjee, A.; Oliverio, M.; Ruiz, G.P.; Ferrucci, D.L.; Branquinho, J.L.; Tazolli, D.; Pinto, S.; Nielsen, T.S.; et al. Dynamic changes in DICER levels in adipose tissue control metabolic adaptations to exercise. Proc. Natl. Acad. Sci. USA 2020, 117, 23932–23941. [Google Scholar] [CrossRef] [PubMed]
- Frias, M.A.; James, R.W.; Gerber-Wicht, C.; Lang, U. Native and reconstituted HDL activate Stat3 in ventricular cardiomyocytes via ERK1/2, role of sphingosine-1-phosphate. Cardiovasc. Res. 2009, 82, 313–323. [Google Scholar] [CrossRef]
- Pedretti, S.; Brulhart-Meynet, M.C.; Montecucco, F.; Lecour, S.; James, R.W.; Frias, M.A. HDL protects against myocardial ischemia reperfusion injury via miR-34b and miR-337 expression which requires STAT3. PLoS ONE 2019, 14, e0218432. [Google Scholar] [CrossRef] [PubMed]
- Frias, M.A.; Lecour, S.; James, R.W.; Pedretti, S. High density lipoprotein/sphingosine-1-phosphate induced cardioprotection: Role of STAT3 as part of the SAFE pathway. JAKSTAT 2012, 1, 92–100. [Google Scholar] [CrossRef]
- Jozefczuk, E.; Guzik, T.J.; Sidelinski, M. Significance of sphingosine-1-phosphate in cardiovascular physiology and pathology. Pharmacol. Res. 2020, 156, 104793. [Google Scholar] [CrossRef]
- Haghikia, A.; Hoch, M.; Stapel, B.; Hilfiker-Kleiner, D. STAT3 regulation of and by microRNAs in development and disease. JAKSTAT 2012, 1, 143–150. [Google Scholar] [CrossRef] [PubMed]
- Sadrkhanloo, M.; Entezari, M.; Orouei, S.; Ghollasi, M.; Fathi, N.; Rezaei, S.; Hejazi, E.S.; Kakavand, A.; Saebfar, H.; Hashemi, M.; et al. STAT3-EMT axis in tumors: Modulation of cancer metastasis, stemness and therapy response. Pharmacol. Res. 2022, 182, 106311. [Google Scholar] [CrossRef]
- Loffler, D.; Brocke-Heidrich, K.; Pfeifer, G.; Stocsits, C.; Hackermuller, J.; Kretzschmar, A.K.; Burger, R.; Gramatzki, M.; Blumert, C.; Bauer, K.; et al. Interleukin-6 dependent survival of multiple myeloma cells involves the Stat3-mediated induction of miR-21 through a highly conserved enhancer. Blood 2007, 114, 1330–1333. [Google Scholar] [CrossRef] [PubMed]
- Haghikia, A.; Missol-Kolka, E.; Tsikas, D.; Venturini, L.; Brundiers, S.; Castoldi, M.; Muckenthaler, M.U.; Eder, M.; Stapel, B.; Thum, T.; et al. Signal transducer and activator of transcription 3-mediated regulation of miR-99a-5p links cardiomyocyte and endothelial cell function in the heart: A key role for ubiquitin-conjugating enzymes. Eur. Heart J. 2011, 32, 1287–1297. [Google Scholar] [CrossRef]
- Moradi, H.; Vaziri, N.D.; Kashyap, M.L.; Said, H.M.; Kalantar-Zalh, K. Role of HDL dysfunction in end-stage renal disease: A double-edged sword. J. Ren. Nutr. 2013, 23, 203–206. [Google Scholar] [CrossRef]
- Pirillo, A.; Catapano, A.L.; Norata, G.D. Biological consequences of dysfunctional HDL. Curr. Med. Chem. 2019, 26, 1644–1664. [Google Scholar] [CrossRef]
- Rosenson, R.S.; Brewer, H.B., Jr.; Ansell, B.J.; Barter, P.; Chapman, M.J.; Heinecke, J.W.; Kontush, A.; Tall, A.R.; Webb, N.R. Dysfunctional HDL and atherosclerotic cardiovascular disease. Nat. Rev. Cardiol. 2016, 13, 48–60. [Google Scholar] [CrossRef]
- Ortiz-Munoz, G.; Couret, D.; Lapergue, B.; Bruckert, E.; Meseguer, E.; Amarenco, P.; Meilhad, O. Dysfunctional HDL in acute stroke. Atherosclerosis 2016, 253, 75–80. [Google Scholar] [CrossRef] [PubMed]
- Tan, K.C.B.; Chow, W.-S.; Lam, J.C.M.; Lam, B.; Wong, W.-K.; Tam, S.; Ip, M.S.M. HDL dysfunction in obstructive sleep apnea. Atherosclerosis 2006, 184, 377–382. [Google Scholar] [CrossRef]
- Srivastava, R.A.K. Dysfunctional HDL in diabetes mellitus and its role in the pathogenesis of cardiovascular disease. Mol. Cell Biochem. 2017, 440, 167–187. [Google Scholar] [CrossRef]
- Lu, X.; Yang, B.; Yang, H.; Wang, L.; Li, H.; Chen, S.; Lu, X.; Gu, D. MicroRNA-320b modulates cholesterol efflux and atherosclerosis. J. Atheroscler. Thromb. 2022, 29, 200–220. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.W.; Yun, K.S.; Ban, H.S.; Choe, W.; Lee, S.K.; Lee, K.Y. Preparation and characterization of chitosan/polyguluronate nanoparticles for siRNA delivery. J. Control Release 2009, 139, 146–152. [Google Scholar] [CrossRef]
- Lee, H.; Kim, S.I.; Shin, D.; Yoon, Y.; Choi, T.H.; Cheon, G.J.; Kim, M. Hepatic siRNA delivery using recombinant human apolipoprotein A-I in mice. Biochem. Biophys. Res. Commun. 2009, 378, 192–196. [Google Scholar] [CrossRef]
- Kuwahara, H.; Nishina, K.; Yoshida, K.; Nishina, T.; Yamamoto, M.; Saito, Y.; Piao, W.; Masayuki, Y.; Mizusawa, H.; Yokoto, T. Efficient in vivo delivery of SiRNA into brain capillary endothelial cells along with endogenous lipoprotein. Mol. Ther. 2011, 19, 2213–2221. [Google Scholar] [CrossRef] [PubMed]
- Tabet, F.; Vickers, K.C.; Cuesta Torres, L.F.; Wises, C.V.; Shoucri, B.M.; Lambert, G.; Catherinet, C.; Prado-Lourenco, L.; Levin, M.G.; Thacker, S.; et al. HDL-transferred microRNA-223 regulates ICAM-1 expression in endothelial cells. Nat. Commun. 2014, 5, 3292. [Google Scholar] [CrossRef]
- Jiao, P.; Wang, X.-P.; Luoreng, Z.-M.; Yang, J.; Jia, L.; Ma, Y.; Wei, D.-W. MiR-223, an effective regulator of immune cell differentiation and function. Int. J. Biol. Sci. 2021, 17, 2308–2322. [Google Scholar] [CrossRef] [PubMed]
- Wagner, J.; Riwanto, M.; Besler, C.; Knau, A.; Fischtscherer, S.; Roxe, T.; Zeiher, A.M.; Landmesser, U.; Dimmeler, S. Characterisation of levels and cellular transfer of circulating lipoprotein-bound microRNAs. Arterioscler. Thromb. Vasc. Biol. 2013, 33, 1292–1400. [Google Scholar] [CrossRef] [PubMed]
- Axmann, M.; Meier, S.M.; Karner, A.; Strobl, W.; Stangl, H.; Plochberger, B. Serum and lipoprotein particle miRNA profile in uremia patients. Genes 2018, 9, 533. [Google Scholar] [CrossRef]
- Riedel, S.; Radzanowski, S.; Bowen, T.S.; Werner, S.; Erbs, S.; Schuler, G.; Adams, V. Exercise training improves high-density lipoprotein-mediated transcription of proangiogenic microRNA in endothelial cells. Eur. J. Prev. Cardiol. 2015, 22, 899–903. [Google Scholar] [CrossRef]
- Morrison, K.R.; Solly, E.L.; Shemesh, T.; Psaltis, P.J.; Nicholls, S.J.; Brown, A.; Bursill, C.A.; Tan, J.T.M. Elevated HDL-bound miR-181c-5p level is associated with diabetic vascular complications in Australian Aboriginal people. Diabetologia 2021, 64, 1402–1411. [Google Scholar] [CrossRef]
- Choteau, S.A.; Cuesta Torres, L.F.; Barraclough, J.Y.; Elder, A.M.M.; Martinez, G.J.; Chen Fan, W.Y.; Shretsha, S.; Ong, K.L.; Barter, P.J.; Celermajer, D.S.; et al. Transcoronary gradients of HDL-associated microRNAs in unstable coronary artery disease. Int. J. Cardiol. 2018, 253, 138–144. [Google Scholar] [CrossRef] [PubMed]
- Tabet, F.; Cuesta Torres, L.F.; Ong, K.L.; Shrestha, S.; Choteau, S.A.; Barter, P.J.; Clifton, P.; Rye, K.-A. High-density lipoprotein-associated miR-223 is altered after diet-induced weight loss in overweight and obese males. PLoS ONE 2016, 11, e0151061. [Google Scholar] [CrossRef]
- Chamorro-Jorganes, A.; Anwar, M.; Emanueli, C. Changes in high-density lipoprotein microRNA might create a lasting memory of high-fat diet. Cardiovasc. Res. 2020, 116, 1237–1239. [Google Scholar] [CrossRef]
- Wang, J.; Calvert, A.E.; Kaplan, N.; McMahon, K.M.; Yang, W.; Lu, K.Q.; Peng, H.; Thaxton, C.S.; Lavker, R.M. HDL nanoparticles have wound healing and anti-inflammatory properties and can topically deliver miRNAs. Adv. Ther. 2020, 3, 2000138. [Google Scholar] [CrossRef]
- Li, M.; Su, Y.; Zhang, F.; Chen, K.; Xu, X.; Xu, L.; Zhou, J.; Wang, W. A dual–targeting reconstituted high density lipoprotein leveraging the synergy of sorafenib and anti-mirRNA21 for enhanced hepatocellular carcinoma therapy. Acta Biomater. 2018, 75, 413–426. [Google Scholar] [CrossRef]
- Rui, M.; Qu, Y.; Gao, T.; Ge, Y.; Feng, C.; Xu, X. Simultaneous delivery of anti-miR21 with doxorubicin pro-drug by mimetic lipoprotein nanoparticles for synergistic effect against drug resistance in cancer cells. Int. J. Nanomed. 2016, 12, 217–237. [Google Scholar] [CrossRef] [PubMed]
- Dehghankelishadi, P.; Maritz, M.F.; Badiee, P.; Thierry, B. High density lipoprotein nanoparticle as delivery system for radio-sensitising miRNA: An investigation in 2D/3D head and neck cancer models. Int. J. Pharm. 2022, 617, 121585. [Google Scholar] [CrossRef] [PubMed]
- Tarlton, J.M.R.; Lightbody, R.J.; Patterson, S.; Graham, A. Protection against glucolipotoxicity by high density lipoprotein in human PANC-1 hybrid 1.1B4 pancreatic beta cells: The role of microRNA. Biology 2021, 10, 218. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Xu, Y.; Zhu, Y.; Sun, H.; Jujuilon, C.; Li, F.; Fan, D.; Yin, L.; Zhang, Y. Macrophage miR-34a is a key regulator of cholesterol efflux and atherosclerosis. Mol. Ther. 2020, 28, 202–216. [Google Scholar] [CrossRef]
- Xie, Q.; Peng, J.; Guo, Y.; Li, F. MicroRNA-33-5p inhibits cholesterol efflux in vascular endothelial cells by regulating citrate synthase and ATP-binding cassette transporter A1. BMC Cardiovasc. Disord. 2021, 21, 433. [Google Scholar] [CrossRef]
- Tastsoglou, S.; Miliotis, M.; Kavakiotis, I.; Alexiou, A.; Gkotsi, E.C.; Lambropoulou, A.; Lygnos, V.; Kotsira, V.; Maroulis, V.; Zisis, D.; et al. PlasmiR: A manual collection of circulating microRNAs of prognostic and diagnostic value. Cancers 2021, 13, 3680. [Google Scholar] [CrossRef]
- Hourigan, S.T.; Solly, E.L.; Nankivell, V.A.; Ridiandries, A.; Weimann, B.M.; Henriquez, R.; Tepper, E.R.; Zhang, Q.J.; Tsatralis, T.; Clayton, Z.E.; et al. The regulation of miRNA by reconstituted high-density lipoproteins in diabetes-impaired angiogenesis. Sci. Rep. 2018, 8, 13596. [Google Scholar] [CrossRef]
- Li, H.-M.; Mo, Z.-W.; Peng, Y.-M.; Li, Y.; Dai, W.-P.; Yuan, H.-Y.; Chang, F.-J.; Wang, T.-T.; Wang, M.; Hu, K.-H.; et al. Angiogenic and antiangiogenic mechanisms of high density lipoprotein from healthy subjects and coronary artery disease patients. Redox Biol. 2020, 36, 101642. [Google Scholar] [CrossRef]
- Bastaki, K.M.; Tarlton, J.M.R.; Lightbody, R.J.; Graham, A.; Martin, P.E. Homo Sapiens (Hsa)-microRNA (miR)-6727-5p contributes to the impact of high-density lipoproteins on fibroblast wound healing in vitro. Membranes 2022, 12, 154. [Google Scholar] [CrossRef]
- Tall, A.J.; Rader, D.J. Trials and tribulations of CETP inhibitors. Circ. Res. 2018, 122, 106–112. [Google Scholar] [CrossRef]
- Gibson, C.M.; Kazmi, S.H.A.; Korjian, S.; Chi, G.; Phillips, A.T.; Montazerin, S.M.; Duffy, D.; Zheng, B.; Heise, M.; Liss, C.; et al. CSL112 (Apolipoprotein A-I [Human]) strongly enhances plasma Apoa-I and cholesterol efflux capacity in post-acute myocardial infarction patients: A PK/PD substudy of the AEGIS-1 trial. Cardiovasc Pharmacol Ther 2022, 27, 10742484221121507. [Google Scholar] [CrossRef] [PubMed]
- Anthony, K. RNA-based therapeutics for neurological diseases. RNA Biol. 2022, 18, 176–190. [Google Scholar] [CrossRef] [PubMed]
- Traber, G.M.; Yu, A.-M. RNAi-based therapeutics and novel RNA bioengineering technologies. J. Pharmacol. Exp. Ther. 2023, 384, 133–154. [Google Scholar] [CrossRef]
- Paunovska, K.; Loughrey, D.; Dahlman, J.E. Drug delivery systems for RNA therapeutics. Nat. Rev. Genet. 2022, 23, 265–280. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.-Y.; Qin, Z.; Zhu, Y.-H.; He, Z.-Y.; Xu, T. Current RNA-based therapeutics in clinical trials. Curr. Gene Ther. 2019, 19, 172–196. [Google Scholar] [CrossRef]
- Zhu, Y.; Zhu, L.; Wang, X.; Jin, H. RNA-based therapeutics: An overview and prospectus. Cell Death Dis. 2022, 13, 644. [Google Scholar] [CrossRef]
- Winkle, M.; El-Daly, S.M.; Fabbri, M.; Calin, G.A. Noncoding RNA therapeutics- challenges and potential solutions. Nat. Rev. Drug Discov. 2021, 20, 629–651. [Google Scholar] [CrossRef]
- Zhang, M.M.; Bahal, R.; Rasmussen, T.P.; Manautou, J.E.; Zhong, X.-B. The growth of siRNA-based therapeutics: Updated clinical studies. Biochem. Pharmacol. 2021, 189, 114432. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Graham, A. Modulation of the Cellular microRNA Landscape: Contribution to the Protective Effects of High-Density Lipoproteins (HDL). Biology 2023, 12, 1232. https://doi.org/10.3390/biology12091232
Graham A. Modulation of the Cellular microRNA Landscape: Contribution to the Protective Effects of High-Density Lipoproteins (HDL). Biology. 2023; 12(9):1232. https://doi.org/10.3390/biology12091232
Chicago/Turabian StyleGraham, Annette. 2023. "Modulation of the Cellular microRNA Landscape: Contribution to the Protective Effects of High-Density Lipoproteins (HDL)" Biology 12, no. 9: 1232. https://doi.org/10.3390/biology12091232
APA StyleGraham, A. (2023). Modulation of the Cellular microRNA Landscape: Contribution to the Protective Effects of High-Density Lipoproteins (HDL). Biology, 12(9), 1232. https://doi.org/10.3390/biology12091232





