A Novel ENU-Induced Mfn2 Mutation Causes Motor Deficits in Mice without Causing Peripheral Neuropathy
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Generation and Maintenance of the MFN2 Mutant Line
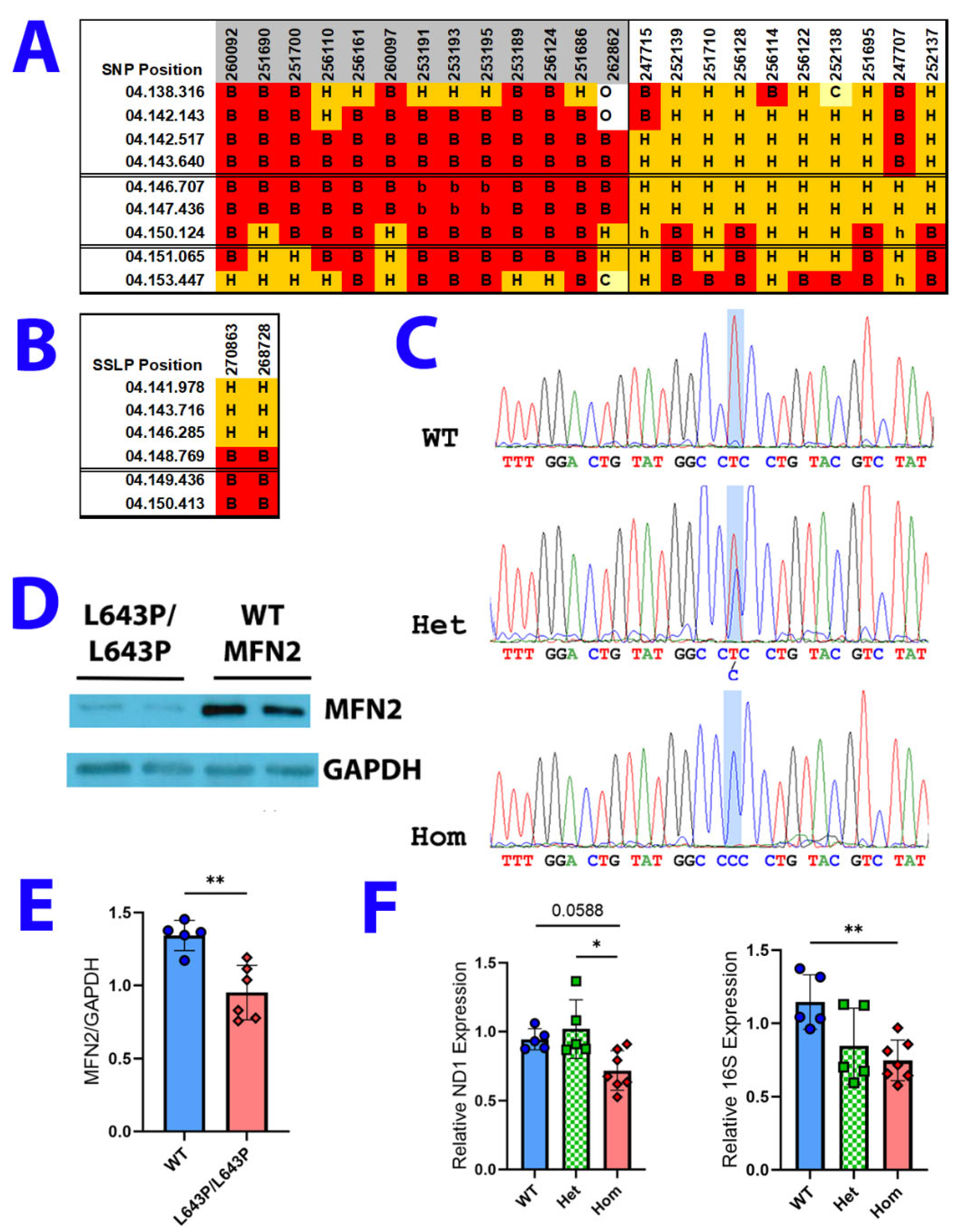
2.2. Single Nucleotide Polymorphism (SNP) and Microsatellite Mapping
2.3. Tissue Preparation for Histology and Electron Microscopy
2.4. Immunofluorescence
2.4.1. Spinal Cord Motor Neurons
2.4.2. Muscle Fiber Typing
2.4.3. Neuromuscular Junction Staining
2.5. Western Blotting
2.6. Analysis of mtDNA/nDNA Ratio
2.7. Open-Field Exploration
2.8. Inverted Wire Hang Test
2.9. Rotarod Assay
2.10. Mechanosensitivity and Thermosensitivity Test
2.11. Electromyography
2.12. In Vivo Contractile Function (Ankle Dorsiflexion)
2.13. MicroCT Analysis
3. Results
3.1. ENU Mutagenesis, Mapping, and Expression
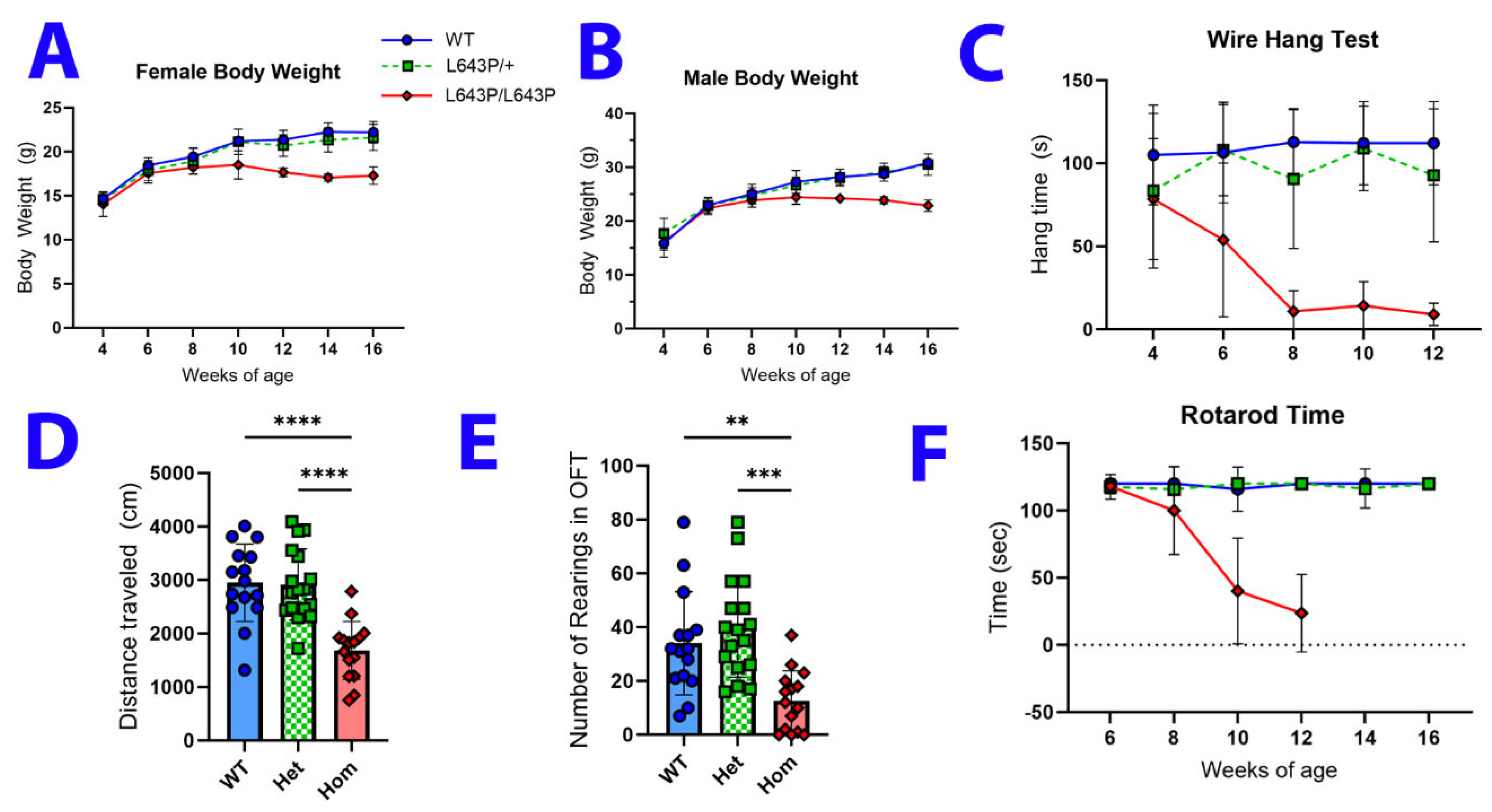
3.2. Changes in Body Weight and Behavior
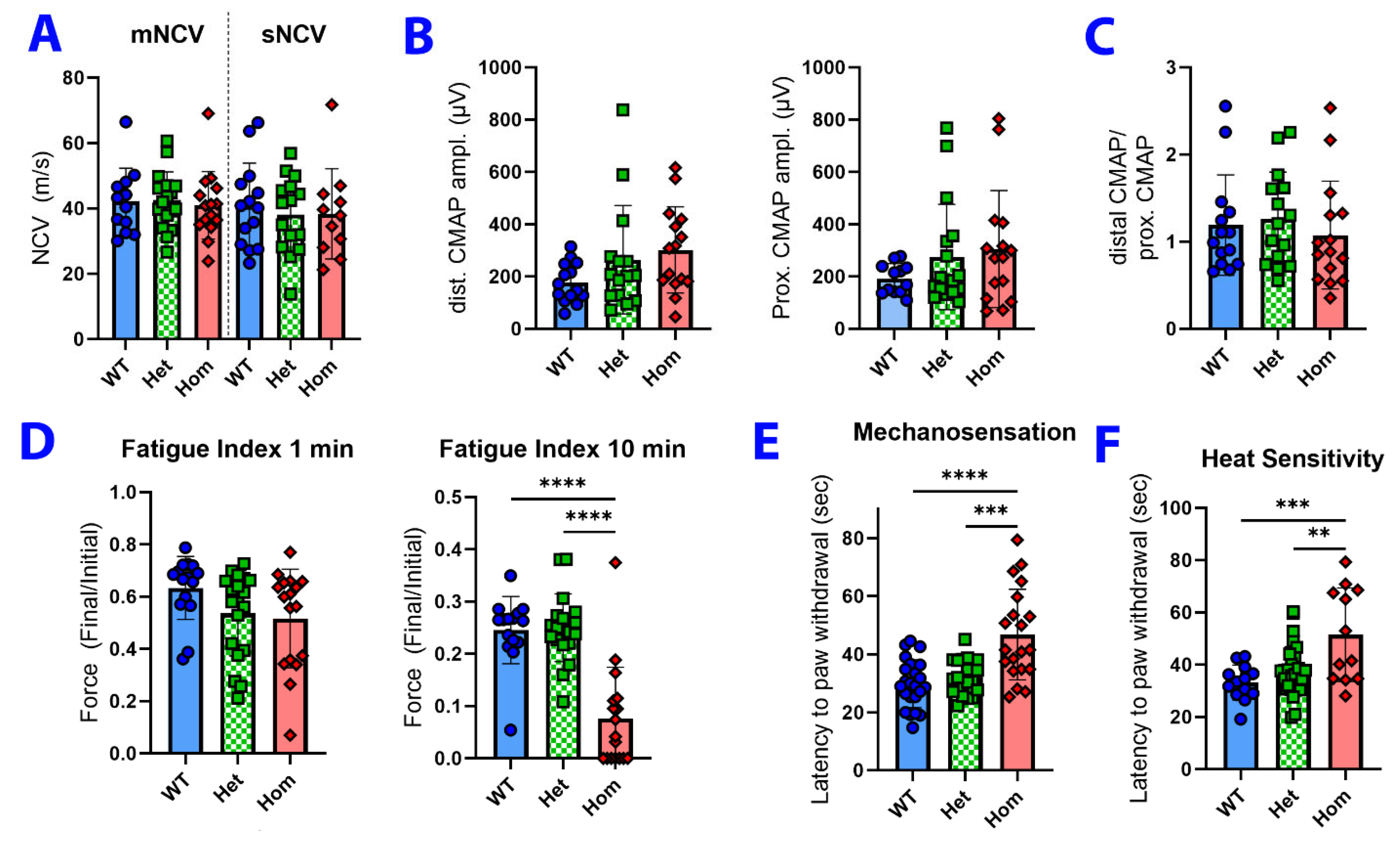
3.3. Neuromuscular Physiology and Sensory Testing
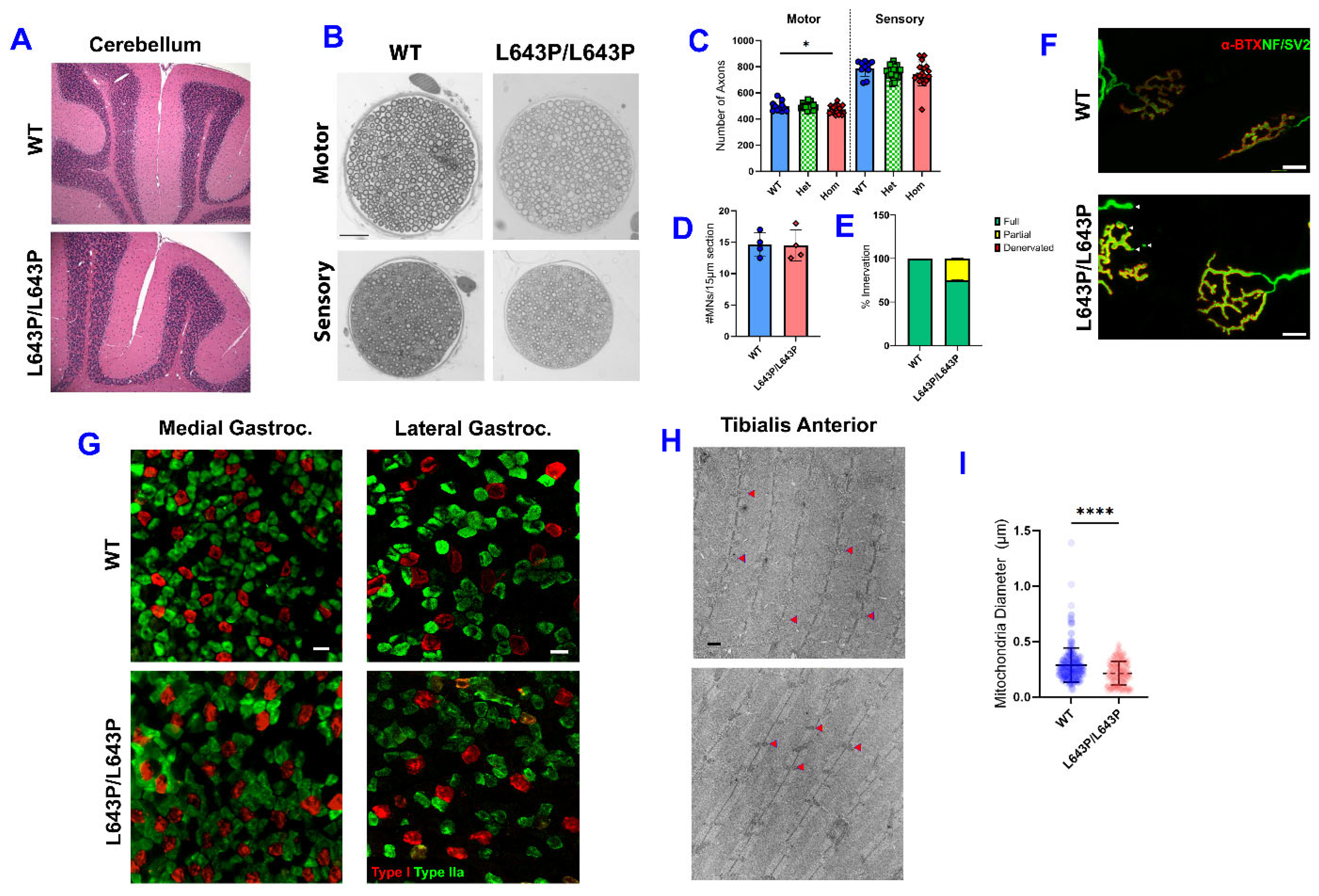
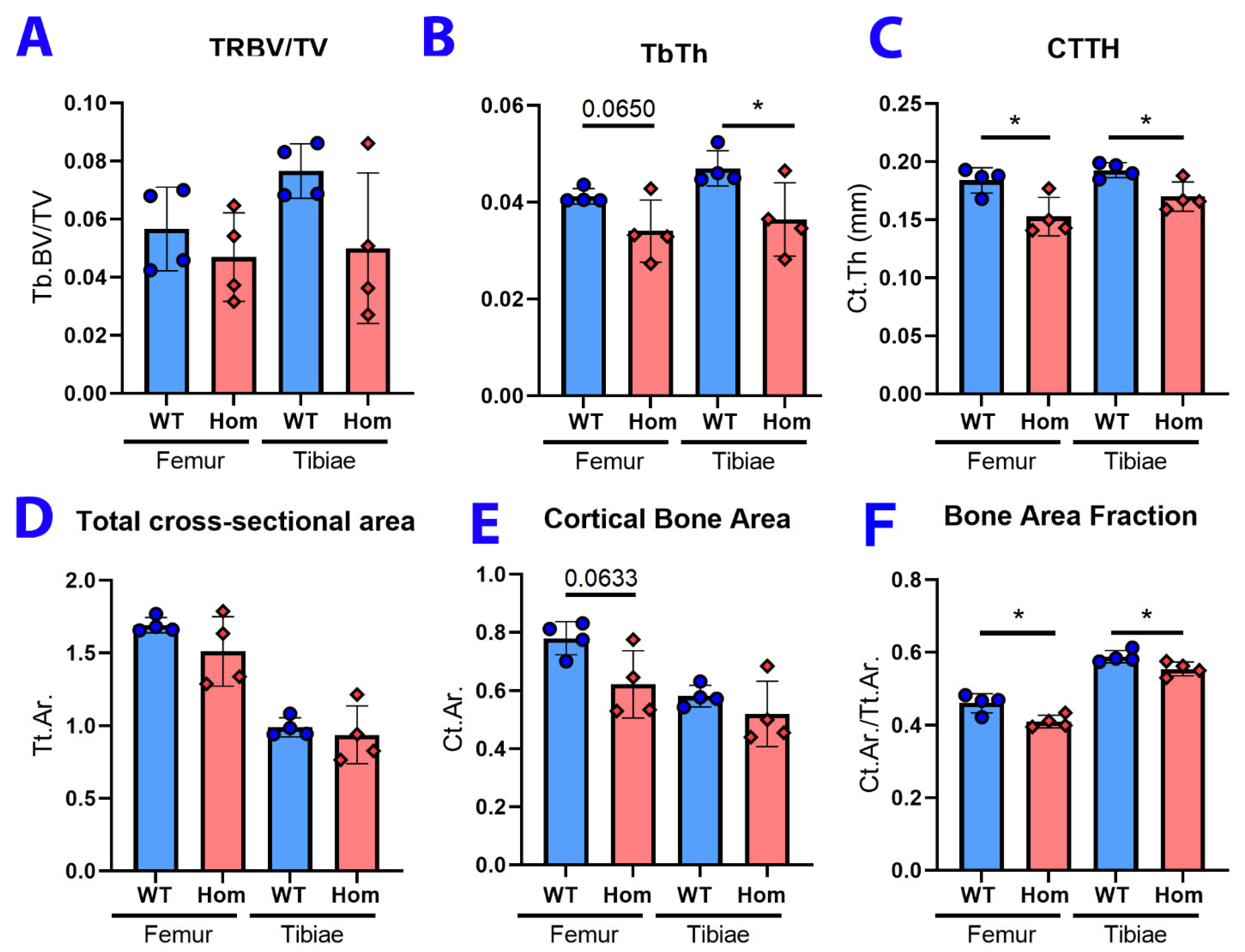
3.4. Histopathology
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interests
References
- McBride, H.M.; Neuspiel, M.; Wasiak, S. Mitochondria: More Than Just a Powerhouse. Curr. Biol. 2006, 16, R551–R560. [Google Scholar] [CrossRef] [PubMed]
- Dorn, G.W., 2nd. Mitochondrial dynamism and cardiac fate—A personal perspective. Circ. J. 2013, 77, 1370–1379. [Google Scholar] [CrossRef] [PubMed]
- Cohen, S.; Valm, A.M.; Lippincott-Schwartz, J. Interacting organelles. Curr. Opin. Cell Biol. 2018, 53, 84–91. [Google Scholar] [CrossRef]
- Yu, R.; Lendahl, U.; Nistér, M.; Zhao, J. Regulation of Mammalian Mitochondrial Dynamics: Opportunities and Challenges. Front. Endocrinol. 2020, 11, 374. [Google Scholar] [CrossRef] [PubMed]
- Tilokani, L.; Nagashima, S.; Paupe, V.; Prudent, J. Mitochondrial dynamics: Overview of molecular mechanisms. Essays Biochem. 2018, 62, 341–360. [Google Scholar] [CrossRef] [PubMed]
- Smirnova, E.; Griparic, L.; Shurland, D.-L.; van der Bliek, A.M.; Schiavon, C.R.; Turn, R.E.; Newman, L.E.; Kahn, R.A.; Spang, M.E.A.; Ganesan, V.; et al. Dynamin-related Protein Drp1 Is Required for Mitochondrial Division in Mammalian Cells. Mol. Biol. Cell 2001, 12, 2245–2256. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.E.; Westrate, L.M.; Wu, H.; Page, C.; Voeltz, G.K. Multiple dynamin family members collaborate to drive mitochondrial division. Nature 2016, 540, 139–143. [Google Scholar] [CrossRef]
- Eura, Y.; Ishihara, N.; Yokota, S.; Mihara, K. Two Mitofusin Proteins, Mammalian Homologues of FZO, with Distinct Functions Are Both Required for Mitochondrial Fusion. J. Biochem. 2003, 134, 333–344. [Google Scholar] [CrossRef]
- DeVay, R.M.; Dominguez-Ramirez, L.; Lackner, L.L.; Hoppins, S.; Stahlberg, H.; Nunnari, J. Coassembly of Mgm1 isoforms requires cardiolipin and mediates mitochondrial inner membrane fusion. J. Cell Biol. 2009, 186, 793–803. [Google Scholar] [CrossRef]
- Santel, A.; Fuller, M.T. Control of mitochondrial morphology by a human mitofusin. J. Cell Sci. 2001, 114, 867–874. [Google Scholar] [CrossRef]
- Silva Ramos, E.; Motori, E.; Brüser, C.; Kühl, I.; Yeroslaviz, A.; Ruzzenente, B.; Kauppila, J.H.K.; Busch, J.D.; Hultenby, K.; Habermann, B.H.; et al. Mitochondrial fusion is required for regulation of mitochondrial DNA replication. PLoS Genet. 2019, 15, e1008085. [Google Scholar] [CrossRef] [PubMed]
- Rooney, J.P.; Ryde, I.T.; Sanders, L.H.; Howlett, E.H.; Colton, M.D.; Germ, K.E.; Mayer, G.D.; Greenamyre, J.T.; Meyer, J.N. PCR Based Determination of Mitochondrial DNA Copy Number in Multiple Species. Mitochondrial Regul. Methods Protoc. 2014, 1241, 23–38. [Google Scholar] [CrossRef]
- Mattie, S.; Riemer, J.; Wideman, J.G.; McBride, H.M. A new mitofusin topology places the redox-regulated C terminus in the mitochondrial intermembrane space. J. Cell Biol. 2017, 217, 507–515. [Google Scholar] [CrossRef]
- Bertholet, A.; Delerue, T.; Millet, A.; Moulis, M.; David, C.; Daloyau, M.; Arnauné-Pelloquin, L.; Davezac, N.; Mils, V.; Miquel, M.; et al. Mitochondrial fusion/fission dynamics in neurodegeneration and neuronal plasticity. Neurobiol. Dis. 2016, 90, 3–19. [Google Scholar] [CrossRef] [PubMed]
- Westermann, B. Mitochondrial fusion and fission in cell life and death. Nat. Rev. Mol. Cell Biol. 2010, 11, 872–884. [Google Scholar] [CrossRef]
- Filadi, R.; Pendin, D.; Pizzo, P. Mitofusin 2: From functions to disease. Cell Death Dis. 2018, 9, 330. [Google Scholar] [CrossRef]
- Chandhok, G.; Lazarou, M.; Neumann, B. Structure, function, and regulation of mitofusin-2 in health and disease. Biol. Rev. 2017, 93, 933–949. [Google Scholar] [CrossRef]
- Züchner, S.; Mersiyanova, I.V.; Muglia, M.; Bissar-Tadmouri, N.; Rochelle, J.; Dadali, E.L.; Zappia, M.; Nelis, E.; Patitucci, A.; Senderek, J.; et al. Mutations in the mitochondrial GTPase mitofusin 2 cause Charcot-Marie-Tooth neuropathy type 2A. Nat. Genet. 2004, 36, 449–451. [Google Scholar] [CrossRef]
- Zaman, M.; Shutt, T.E. The Role of Impaired Mitochondrial Dynamics in MFN2-Mediated Pathology. Front. Cell Dev. Biol. 2022, 10, 858286. [Google Scholar] [CrossRef]
- Zuchner, S. MFN2 Hereditary Motor and Sensory Neuropathy. University of Washington, Seattle. Updated 14 May 2020. Available online: https://www.ncbi.nlm.nih.gov/books/NBK1511/ (accessed on 4 June 2023).
- Brožková, D.; Posadka, J.; Laššuthová, P.; Mazanec, R.; Haberlová, J.; Šišková, D.; Sakmaryová, I.; Neupauerová, J.; Seeman, P. Spectrum and frequencies of mutations in the MFN2 gene and its phenotypical expression in Czech hereditary motor and sensory neuropathy type II patients. Mol. Med. Rep. 2013, 8, 1779–1784. [Google Scholar] [CrossRef]
- Abati, E.; Manini, A.; Velardo, D.; Del Bo, R.; Napoli, L.; Rizzo, F.; Moggio, M.; Bresolin, N.; Bellone, E.; Bassi, M.T.; et al. Clinical and genetic features of a cohort of patients with MFN2-related neuropathy. Sci. Rep. 2022, 12, 6181. [Google Scholar] [CrossRef] [PubMed]
- Hsu, Y.; Lin, K.; Guo, Y.; Tsai, Y.; Liao, Y.; Lee, Y. Mutation spectrum of Charcot-Marie-Tooth disease among the Han Chinese in Taiwan. Ann. Clin. Transl. Neurol. 2019, 6, 1090–1101. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Sun, A.; Zhang, Y.; Fan, D.; Liu, X. The Genotype and Phenotype Features in a Large Chinese MFN2 Mutation Cohort. Front. Neurol. 2021, 12, 757518. [Google Scholar] [CrossRef] [PubMed]
- Stuppia, G.; Rizzo, F.; Riboldi, G.; Del Bo, R.; Nizzardo, M.; Simone, C.; Comi, G.P.; Bresolin, N.; Corti, S. MFN2-related neuropathies: Clinical features, molecular pathogenesis and therapeutic perspectives. J. Neurol. Sci. 2015, 356, 7–18. [Google Scholar] [CrossRef] [PubMed]
- Verhoeven, K.; Claeys, K.; Züchner, S.; Schröder, J.M.; Weis, J.; Ceuterick, C.; Jordanova, A.; Nelis, E.; De Vriendt, E.; Van Hul, M.; et al. MFN2 mutation distribution and genotype/phenotype correlation in Charcot-Marie-Tooth type 2. Brain 2006, 129, 2093–2102. [Google Scholar] [CrossRef]
- Detmer, S.A.; Chan, D.C. Complementation between mouse Mfn1 and Mfn2 protects mitochondrial fusion defects caused by CMT2A disease mutations. J. Cell Biol. 2007, 176, 405–414. [Google Scholar] [CrossRef]
- Zhou, Y.; Carmona, S.; Muhammad, A.; Bell, S.; Landeros, J.; Vazquez, M.; Ho, R.; Franco, A.; Lu, B.; Dorn, G.W.; et al. Restoring mitofusin balance prevents axonal degeneration in a Charcot-Marie-Tooth type 2A model. J. Clin. Investig. 2019, 129, 1756–1771. [Google Scholar] [CrossRef]
- Cipolat, S.; De Brito, O.M.; Dal Zilio, B.; Scorrano, L. OPA1 requires mitofusin 1 to promote mitochondrial fusion. Proc. Natl. Acad. Sci. USA 2004, 101, 15927–15932. [Google Scholar] [CrossRef]
- Ishihara, N.; Eura, Y.; Mihara, K. Mitofusin 1 and 2 play distinct roles in mitochondrial fusion reactions via GTPase activity. J. Cell Sci. 2004, 117, 6535–6546. [Google Scholar] [CrossRef]
- De Brito, O.M.; Scorrano, L. Mitofusin 2 tethers endoplasmic reticulum to mitochondria. Nature 2008, 456, 605–610. [Google Scholar] [CrossRef]
- Chen, H.C.; Detmer, S.A.; Ewald, A.J.; Griffin, E.E.; Fraser, S.E.; Chan, D.C. Mitofusins Mfn1 and Mfn2 coordinately regulate mitochondrial fusion and are essential for embryonic development. J. Cell Biol. 2003, 160, 189–200. [Google Scholar] [CrossRef]
- Misko, A.; Jiang, S.; Wegorzewska, I.; Milbrandt, J.; Baloh, R.H. Mitofusin 2 Is Necessary for Transport of Axonal Mitochondria and Interacts with the Miro/Milton Complex. J. Neurosci. 2010, 30, 4232–4240. [Google Scholar] [CrossRef] [PubMed]
- Misko, A.L.; Sasaki, Y.; Tuck, E.; Milbrandt, J.; Baloh, R.H. Mitofusin2 Mutations Disrupt Axonal Mitochondrial Positioning and Promote Axon Degeneration. J. Neurosci. 2012, 32, 4145–4155. [Google Scholar] [CrossRef] [PubMed]
- Deng, L.; Yi, S.; Yin, X.; Li, Y.; Luan, Q. MFN2 knockdown promotes osteogenic differentiation of iPSC-MSCs through aerobic glycolysis mediated by the Wnt/β-catenin signaling pathway. Stem Cell Res. Ther. 2022, 13, 162. [Google Scholar] [CrossRef] [PubMed]
- Zarei, A.; Ballard, A.; Cox, L.; Bayguinov, P.; Harris, T.; Davis, J.L.; Roper, P.; Fitzpatrick, J.; Faccio, R.; Veis, D.J. Osteolineage depletion of mitofusin2 enhances cortical bone formation in female mice. Bone 2021, 148, 115941. [Google Scholar] [CrossRef]
- De Gioia, R.; Citterio, G.; Abati, E.; Nizzardo, M.; Bresolin, N.; Comi, G.P.; Corti, S.; Rizzo, F. Animal Models of CMT2A: State-of-art and Therapeutic Implications. Mol. Neurobiol. 2020, 57, 5121–5129. [Google Scholar] [CrossRef]
- Feely, S.M.E.; Laura, M.; Siskind, C.E.; Sottile, S.; Davis, M.; Gibbons, V.S.; Reilly, M.M.; Shy, M.E. MFN2 mutations cause severe phenotypes in most patients with CMT2A. Neurology 2011, 76, 1690–1696. [Google Scholar] [CrossRef]
- Strickland, A.V.; Rebelo, A.P.; Zhang, F.; Price, J.; Bolon, B.; Silva, J.P.; Wen, R.; Züchner, S. Characterization of the mitofusin 2 R94W mutation in a knock-in mouse model. J. Peripher. Nerv. Syst. 2014, 19, 152–164. [Google Scholar] [CrossRef]
- Detmer, S.A.; Velde, C.V.; Cleveland, D.W.; Chan, D.C. Hindlimb gait defects due to motor axon loss and reduced distal muscles in a transgenic mouse model of Charcot–Marie–Tooth type 2A. Hum. Mol. Genet. 2007, 17, 367–375. [Google Scholar] [CrossRef]
- Cartoni, R.; Arnaud, E.; Médard, J.-J.; Poirot, O.; Courvoisier, D.S.; Chrast, R.; Martinou, J.-C. Expression of mitofusin 2R94Q in a transgenic mouse leads to Charcot–Marie–Tooth neuropathy type 2A. Brain 2010, 133, 1460–1469. [Google Scholar] [CrossRef]
- Bannerman, P.; Burns, T.; Xu, J.; Miers, L.; Pleasure, D. Mice Hemizygous for a Pathogenic Mitofusin-2 Allele Exhibit Hind Limb/Foot Gait Deficits and Phenotypic Perturbations in Nerve and Muscle. PLoS ONE 2016, 11, e0167573. [Google Scholar] [CrossRef] [PubMed]
- Cordes, S.P. N -Ethyl- N -Nitrosourea Mutagenesis: Boarding the Mouse Mutant Express. Microbiol. Mol. Biol. Rev. 2005, 69, 426–439. [Google Scholar] [CrossRef] [PubMed]
- Liesa, M.; Palacín, M.; Zorzano, A. Mitochondrial Dynamics in Mammalian Health and Disease. Physiol. Rev. 2009, 89, 799–845. [Google Scholar] [CrossRef] [PubMed]
- Hikiami, R.; Yamashita, H.; Koita, N.; Jingami, N.; Sawamoto, N.; Furukawa, K.; Kawai, H.; Terashima, T.; Oka, N.; Hashiguchi, A.; et al. Charcot–Marie–Tooth disease type 2A with an autosomal-recessive inheritance: The first report of an adult-onset disease. J. Hum. Genet. 2017, 63, 89–92. [Google Scholar] [CrossRef] [PubMed]
- FIBER TYPE STAINING (1, 2A, 2X) with Fiber Borders on Human Skeletal Muscle Cryosections. Available online: https://www.uky.edu/chs/sites/chs.uky.edu/files/fiber_type_staining_on_human_muscle_cryosections.doc (accessed on 4 June 2023).
- Quiros, P.M.; Goyal, A.; Jha, P.; Auwerx, J. Analysis of mtDNA/nDNA Ratio in Mice. Curr. Protoc. Mouse Biol. 2017, 7, 47–54. [Google Scholar] [CrossRef]
- Gomez, C.M.; Maselli, R.; Gundeck, J.E.; Chao, M.; Day, J.W.; Tamamizu, S.; Lasalde, J.A.; McNamee, M.; Wollmann, R.L. Slow-Channel Transgenic Mice: A Model of Postsynaptic Organellar Degeneration at the Neuromuscular Junction. J. Neurosci. 1997, 17, 4170–4179. [Google Scholar] [CrossRef]
- Rafael, J.A.; Nitta, Y.; Peters, J.; Davies, K.E. Testing of SHIRPA, a mouse phenotypic assessment protocol, on Dmd mdx and Dmd mdx3cv dystrophin-deficient mice. Mamm. Genome 2000, 11, 725–728. [Google Scholar] [CrossRef]
- Spaulding, E.L.; Sleigh, J.N.; Morelli, K.H.; Pinter, M.J.; Burgess, R.W.; Seburn, K.L. Synaptic Deficits at Neuromuscular Junctions in Two Mouse Models of Charcot–Marie–Tooth Type 2d. J. Neurosci. 2016, 36, 3254–3267. [Google Scholar] [CrossRef]
- Gerlinger-Romero, F.; Addinsall, A.B.; Lovering, R.M.; Foletta, V.C.; van der Poel, C.; Della-Gatta, P.A.; Russell, A.P. Non-invasive Assessment of Dorsiflexor Muscle Function in Mice. J. Vis. Exp. 2019, 143, e58696. [Google Scholar] [CrossRef]
- Bouxsein, M.L.; Boyd, S.K.; Christiansen, B.A.; Guldberg, R.E.; Jepsen, K.J.; Müller, R. Guidelines for assessment of bone microstructure in rodents using micro-computed tomography. J. Bone Miner. Res. 2010, 25, 1468–1486. [Google Scholar] [CrossRef]
- Zhao, C.; Takita, J.; Tanaka, Y.; Setou, M.; Nakagawa, T.; Takeda, S.; Yang, H.W.; Terada, S.; Nakata, T.; Takei, Y.; et al. Charcot-Marie-Tooth Disease Type 2A Caused by Mutation in a Microtubule Motor KIF1Bβ. Cell 2001, 105, 587–597. [Google Scholar] [CrossRef] [PubMed]
- Ballard, A.; Zeng, R.; Zarei, A.; Shao, C.; Cox, L.; Yan, H.; Franco, A.; Ii, G.W.D.; Faccio, R.; Veis, D.J. The tethering function of mitofusin2 controls osteoclast differentiation by modulating the Ca2+–NFATc1 axis. J. Biol. Chem. 2020, 295, 6629–6640. [Google Scholar] [CrossRef] [PubMed]
- Luo, N.; Yue, F.; Jia, Z.; Chen, J.; Deng, Q.; Zhao, Y.; Kuang, S. Reduced electron transport chain complex I protein abundance and function in Mfn2-deficient myogenic progenitors lead to oxidative stress and mitochondria swelling. FASEB J. 2021, 35, e21426. [Google Scholar] [CrossRef] [PubMed]
- Ainbinder, A.; Boncompagni, S.; Protasi, F.; Dirksen, R.T. Role of Mitofusin-2 in mitochondrial localization and calcium uptake in skeletal muscle. Cell Calcium 2015, 57, 14–24. [Google Scholar] [CrossRef] [PubMed]
- Vielhaber, S.; Debska-Vielhaber, G.; Peeva, V.; Schoeler, S.; Kudin, A.P.; Minin, I.; Schreiber, S.; Dengler, R.; Kollewe, K.; Zuschratter, W.; et al. Mitofusin 2 mutations affect mitochondrial function by mitochondrial DNA depletion. Acta Neuropathol. 2012, 125, 245–256. [Google Scholar] [CrossRef]
- Rouzier, C.; Bannwarth, S.; Chaussenot, A.; Chevrollier, A.; Verschueren, A.; Bonello-Palot, N.; Fragaki, K.; Cano, A.; Pouget, J.; Pellissier, J.-F.; et al. The MFN2 gene is responsible for mitochondrial DNA instability and optic atrophy ‘plus’ phenotype. Brain 2012, 135, 23–34. [Google Scholar] [CrossRef]
- Lewis, S.C.; Uchiyama, L.F.; Nunnari, J. ER-mitochondria contacts couple mtDNA synthesis with mitochondrial division in human cells. Science 2016, 353, aaf5549. [Google Scholar] [CrossRef]
- Bach, D.; Pich, S.; Soriano, F.X.; Vega, N.; Baumgartner, B.; Oriola, J.; Daugaard, J.R.; Lloberas, J.; Camps, M.; Zierath, J.R.; et al. Mitofusin-2 Determines Mitochondrial Network Architecture and Mitochondrial Metabolism: A novel regulatory mechanism altered in obesity. J. Biol. Chem. 2003, 278, 17190–17197. [Google Scholar] [CrossRef]
| Chromosome | Start | End | cM | Strand GRCm39 | MGI ID | Feature Type | Symbol | Name |
|---|---|---|---|---|---|---|---|---|
| 4 | 148085179 | 148086531 | 78.66 | + | MGI:97367 | protein coding gene | Nppa | natriuretic peptide type A |
| 4 | 148070264 | 148071662 | 78.57 | + | MGI:97368 | protein coding gene | Nppb | natriuretic peptide type B |
| 4 | 149234448 | 149251162 | 78.96 | − | MGI:97553 | protein coding gene | Pgd | phosphogluconate dehydrogenase |
| 4 | 149260776 | 149392150 | 79.05 | − | MGI:108426 | protein coding gene | Kif1b | kinesin family member 1B |
| 4 | 149209491 | 149211220 | 78.89 | − | MGI:109538 | protein coding gene | Cort | cortistatin |
| 4 | 149733625 | 149787023 | 80.15 | − | MGI:1098211 | protein coding gene | Pik3cd | phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit delta |
| 4 | 149188603 | 149205104 | 78.87 | + | MGI:1196227 | protein coding gene | Dffa | DNA fragmentation factor, alpha subunit |
| 4 | 148888886 | 149039346 | 78.87 | + | MGI:1196251 | protein coding gene | Casz1 | castor zinc finger 1 |
| 4 | 149799832 | 149822501 | 80.15 | − | MGI:1196277 | protein coding gene | Tmem201 | transmembrane protein 201 |
| 4 | 147945235 | 147953176 | 78.56 | − | MGI:106506 | protein coding gene | Miip | migration and invasion inhibitory protein |
| 4 | 148123534 | 148144008 | 78.67 | + | MGI:106639 | protein coding gene | Mthfr | methylenetetrahydrofolate reductase |
| 4 | 147953436 | 147954815 | 78.56 | + | MGI:95595 | protein coding gene | Fv1 | Friend virus susceptibility 1 |
| 4 | 149552029 | 149569659 | 79.47 | − | MGI:1913704 | protein coding gene | Nmnat1 | nicotinamide nucleotide adenylyltransferase 1 |
| 4 | 149602698 | 149650894 | 79.66 | + | MGI:1915756 | protein coding gene | Ctnnbip1 | catenin beta interacting protein 1 |
| 4 | 149569686 | 149581125 | 79.53 | + | MGI:1916401 | protein coding gene | Lzic | leucine zipper and CTNNBIP1 domain containing |
| 4 | 144144759 | 144165342 | 77.98 | − | MGI:1916614 | protein coding gene | Cfap107 | cilia and flagella associated protein 107 |
| 4 | 148025352 | 148031771 | 78.57 | + | MGI:1924284 | protein coding gene | 2510039O18Rik | RIKEN cDNA 2510039O18 gene |
| 4 | 144118244 | 144135034 | 77.98 | − | MGI:1924882 | protein coding gene | Pramel13 | PRAME like 13 |
| 4 | 148161518 | 148172488 | 78.67 | − | MGI:1339977 | protein coding gene | Agtrap | angiotensin II, type I receptor-associated protein |
| 4 | 148088716 | 148123270 | 78.67 | − | MGI:1347049 | protein coding gene | Clcn6 | chloride channel, voltage-sensitive 6 |
| 4 | 148696839 | 148711476 | 78.77 | − | MGI:2387629 | protein coding gene | Tardbp | TAR DNA binding protein |
| 4 | 147958056 | 147989161 | 78.56 | − | MGI:2442230 | protein coding gene | Mfn2 | mitofusin 2 |
| 4 | 148230173 | 148236592 | 78.67 | − | MGI:1354743 | protein coding gene | Fbxo6 | F-box protein 6 |
| 4 | 148237256 | 148244663 | 78.67 | − | MGI:1354744 | protein coding gene | Fbxo44 | F-box protein 44 |
| 4 | 148642886 | 148666858 | 78.76 | + | MGI:1355322 | protein coding gene | Exosc10 | exosome component 10 |
| 4 | 149412873 | 149511206 | 79.08 | − | MGI:1927086 | protein coding gene | Ube4b | ubiquitination factor E4B |
| 4 | 149534144 | 149539435 | 79.4 | − | MGI:1890409 | protein coding gene | Rbp7 | retinol binding protein 7, cellular |
| 4 | 148245078 | 148250881 | 78.68 | + | MGI:2446216 | protein coding gene | Fbxo2 | F-box protein 2 |
| 4 | 144180341 | 144190326 | 77.98 | − | MGI:2685281 | protein coding gene | Aadacl3 | arylacetamide deacetylase like 3 |
| 4 | 144396507 | 144412938 | 78.04 | − | MGI:2685282 | protein coding gene | Aadacl4fm4 | AADACL4 family member 4 |
| 4 | 144503774 | 144513153 | 78.08 | − | MGI:2685284 | protein coding gene | Aadacl4fm5 | AADACL4 family member 5 |
| 4 | 148727774 | 148756029 | 78.82 | + | MGI:2685418 | protein coding gene | Gm572 | predicted gene 572 |
| 4 | 144246392 | 144255923 | 77.98 | + | MGI:2685880 | protein coding gene | Aadacl4fm1 | AADACL4 family member 1 |
| 4 | 147757959 | 147787010 | 78.53 | − | MGI:3650650 | protein coding gene | Zfp534 | zinc finger protein 534 |
| 4 | 144429761 | 144447974 | 78.05 | − | MGI:3650721 | protein coding gene | AAdacl4fm3 | AADACL4 family member 3 |
| 4 | 145311770 | 145351915 | 78.22 | + | MGI:3651014 | protein coding gene | Zfp268 | zinc finger protein 268 |
| 4 | 147056433 | 147075212 | 78.41 | + | MGI:3651739 | protein coding gene | Zfp989 | zinc finger protein 989 |
| 4 | 147838431 | 147894245 | 78.54 | − | MGI:3651978 | protein coding gene | Zfp984 | zinc finger protein 984 |
| 4 | 147390131 | 147418191 | 78.47 | + | MGI:3651985 | protein coding gene | Zfp988 | zinc finger protein 988 |
| 4 | 147637734 | 147669655 | 78.51 | + | MGI:3651986 | protein coding gene | Zfp985 | zinc finger protein 985 |
| 4 | 145237329 | 145265751 | 78.2 | + | MGI:3652161 | protein coding gene | Zfp990 | zinc finger protein 990 |
| 4 | 144281570 | 144291704 | 77.99 | − | MGI:3652194 | protein coding gene | Aadacl4fm2 | AADACL4 family member 2 |
| 4 | 144699192 | 144921575 | 78.17 | − | MGI:2448530 | protein coding gene | Vps13d | vacuolar protein sorting 13D |
| 4 | 145595364 | 145626545 | 78.26 | + | MGI:3649925 | protein coding gene | Zfp986 | zinc finger protein 986 |
| 4 | 144340277 | 144349968 | 78.01 | + | MGI:3650257 | protein coding gene | Aadacl4 | arylacetamide deacetylase like 4 |
| 4 | 147907443 | 147932823 | 78.56 | − | MGI:1922865 | protein coding gene | Zfp933 | zinc finger protein 933 |
| 4 | 148579737 | 148584919 | 78.76 | − | MGI:3605801 | protein coding gene | Angptl7 | angiopoietin-like 7 |
| 4 | 146533487 | 146553897 | 78.31 | + | MGI:3700963 | protein coding gene | Zfp992 | zinc finger protein 992 |
| 4 | 146586484 | 146623852 | 78.3 | + | MGI:3700965 | protein coding gene | Zfp981 | zinc finger protein 981 |
| 4 | 147576874 | 147597943 | 78.5 | + | MGI:3701121 | protein coding gene | Zfp982 | zinc finger protein 982 |
| 4 | 147445760 | 147475461 | 78.48 | + | MGI:3701123 | protein coding gene | Zfp978 | zinc finger protein 978 |
| 4 | 149561883 | 149567796 | 79.51 | + | MGI:3701129 | protein coding gene | Tmem274 | transmembrame protein 274 |
| 4 | 147216495 | 147265036 | 78.44 | + | MGI:3701604 | protein coding gene | Zfp991 | zinc finger protein 991 |
| 4 | 146033882 | 146063194 | 78.37 | + | MGI:3702694 | protein coding gene | Zfp987 | zinc finger protein 987 |
| 4 | 146093397 | 146135326 | 78.38 | + | MGI:3705222 | protein coding gene | Zfp600 | zinc finger protein 600 |
| 4 | 145397267 | 145431007 | 78.23 | + | MGI:3712454 | protein coding gene | Zfp980 | zinc finger protein 980 |
| 4 | 144099330 | 144104503 | 77.98 | − | MGI:3712553 | protein coding gene | Pramel15 | PRAME like 15 |
| 4 | 146695418 | 146742617 | 78.28 | + | MGI:3713585 | protein coding gene | Zfp993 | zinc finger protein 993 |
| 4 | 148518952 | 148529217 | 78.76 | − | MGI:1918957 | protein coding gene | Ubiad1 | UbiA prenyltransferase domain containing 1 |
| 4 | 148214841 | 148230156 | 78.67 | + | MGI:1919140 | protein coding gene | Mad2l2 | MAD2 mitotic arrest deficient-like 2 |
| 4 | 144993707 | 145041734 | 78.17 | − | MGI:99908 | protein coding gene | Tnfrsf8 | tumor necrosis factor receptor superfamily, member 8 |
| 4 | 144940033 | 144973440 | 78.17 | − | MGI:1314883 | protein coding gene | Tnfrsf1b | tumor necrosis factor receptor superfamily, member 1b |
| 4 | 144619397 | 144654779 | 78.14 | + | MGI:1315215 | protein coding gene | Dhrs3 | dehydrogenase/reductase (SDR family) member 3 |
| 4 | 147106307 | 147145251 | 78.42 | + | MGI:1328322 | protein coding gene | Rex2 | reduced expression 2 |
| 4 | 148687011 | 148699956 | 78.76 | + | MGI:1330832 | protein coding gene | Masp2 | mannan-binding lectin serine peptidase 2 |
| 4 | 147994210 | 148021224 | 78.57 | − | MGI:99907 | protein coding gene | Plod1 | procollagen-lysine, 2-oxoglutarate 5-dioxygenase 1 |
| 4 | 149212806 | 149222057 | 78.89 | − | MGI:1917178 | protein coding gene | Cenps | centromere protein S |
| 4 | 148182894 | 148215155 | 78.67 | − | MGI:1917683 | protein coding gene | Draxin | dorsal inhibitory axon guidance protein |
| 4 | 149828493 | 149858734 | 80.15 | − | MGI:1917806 | protein coding gene | Slc25a33 | solute carrier family 25, member 33 |
| 4 | 148324721 | 148372422 | 78.73 | − | MGI:2444403 | protein coding gene | Disp3 | dispatched RND transporter family member 3 |
| 4 | 149044992 | 149184300 | 78.87 | − | MGI:1927868 | protein coding gene | Pex14 | peroxisomal biogenesis factor 14 |
| 4 | 148533068 | 148642140 | 78.76 | + | MGI:1928394 | protein coding gene | Mtor | mechanistic target of rapamycin kinase |
| 4 | 149670925 | 149733356 | 79.91 | + | MGI:1929895 | protein coding gene | Clstn1 | calsyntenin 1 |
| 4 | 148675970 | 148679076 | 78.76 | + | MGI:102690 | protein coding gene | Srm | spermidine synthase |
| 4 | 149980740 | 150039463 | 80.46 | − | MGI:1921896 | protein coding gene | Spsb1 | splA/ryanodine receptor domain and SOCS box containing 1 |
| 4 | 147696394 | 147726970 | − | MGI:2148252 | protein coding gene | Zfp979 | zinc finger protein 979 | |
| 4 | 146976154 | 146994032 | − | MGI:5434766 | protein coding gene | Gm21411 | predicted gene, 21411 | |
| 4 | 145130858 | 145178235 | + | MGI:5625231 | protein coding gene | Gm42346 | predicted gene, 42346 | |
| 4 | 146340962 | 146341306 | − | MGI:6721541 | protein coding gene | Gm53232 | predicted gene, 53232 | |
| 4 | 147303548 | 147311932 | + | MGI:6723559 | protein coding gene | Gm54268 | predicted gene, 54268 |
| IVFs performed | 13 |
| Successful IVFs (pups produced) | 5/13(38.4%) |
| Average oocyte fertilization rate | 5.7% (vs. 65% for wild type B6) |
| Presumptive 2-cel embryos transferred | 198 |
| Number of pups produced | 28 (14.1% vs. 50% for wild type B6) |
| Absolute Muscle Force | Normalized Muscle Force | ||||||
|---|---|---|---|---|---|---|---|
| GenoType | Muscle Weight (mg) | Pt (g) | PO 50 Hz (g) | PO 150 Hz (g) | Pt (g/g) | PO 50 Hz (g/g) | PO 150 Hz (g/g) |
| WT | 55.9 +/− 7.1 | 1.9 +/− 0.58 | 4.6 +/− 1.5 | 5.6 +/− 1.4 | 33.3 +/− 8 | 81.9 +/− 30.1 | 99.3 +/− 23.3 |
| Het | 56.5 +/− 10.2 | 1.9 +/− 0.66 | 4.4 +/− 1.7 | 5.3 +/− 1.8 | 34.2 +/− 12 | 79.1 +/− 29.8 | 96.4 +/− 29.8 |
| Hom | ** 42.9 +/− 6.8 | * 1.4 +/− 0.48 | 3.4 +/− 1.3 | * 3.8 +/− 1.4 | 33.1 +/− 11 | 88.1 +/− 30.2 | 89.4 +/− 31.1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hines, T.J.; Bailey, J.; Liu, H.; Guntur, A.R.; Seburn, K.L.; Pratt, S.L.; Funke, J.R.; Tarantino, L.M.; Burgess, R.W. A Novel ENU-Induced Mfn2 Mutation Causes Motor Deficits in Mice without Causing Peripheral Neuropathy. Biology 2023, 12, 953. https://doi.org/10.3390/biology12070953
Hines TJ, Bailey J, Liu H, Guntur AR, Seburn KL, Pratt SL, Funke JR, Tarantino LM, Burgess RW. A Novel ENU-Induced Mfn2 Mutation Causes Motor Deficits in Mice without Causing Peripheral Neuropathy. Biology. 2023; 12(7):953. https://doi.org/10.3390/biology12070953
Chicago/Turabian StyleHines, Timothy J., Janice Bailey, Hedi Liu, Anyonya R. Guntur, Kevin L. Seburn, Samia L. Pratt, Jonathan R. Funke, Lisa M. Tarantino, and Robert W. Burgess. 2023. "A Novel ENU-Induced Mfn2 Mutation Causes Motor Deficits in Mice without Causing Peripheral Neuropathy" Biology 12, no. 7: 953. https://doi.org/10.3390/biology12070953
APA StyleHines, T. J., Bailey, J., Liu, H., Guntur, A. R., Seburn, K. L., Pratt, S. L., Funke, J. R., Tarantino, L. M., & Burgess, R. W. (2023). A Novel ENU-Induced Mfn2 Mutation Causes Motor Deficits in Mice without Causing Peripheral Neuropathy. Biology, 12(7), 953. https://doi.org/10.3390/biology12070953





