VDR Gene Single Nucleotide Polymorphisms and Autoimmunity: A Narrative Review
Abstract
Simple Summary
Abstract
1. Introduction
1.1. Vitamin D and VDR
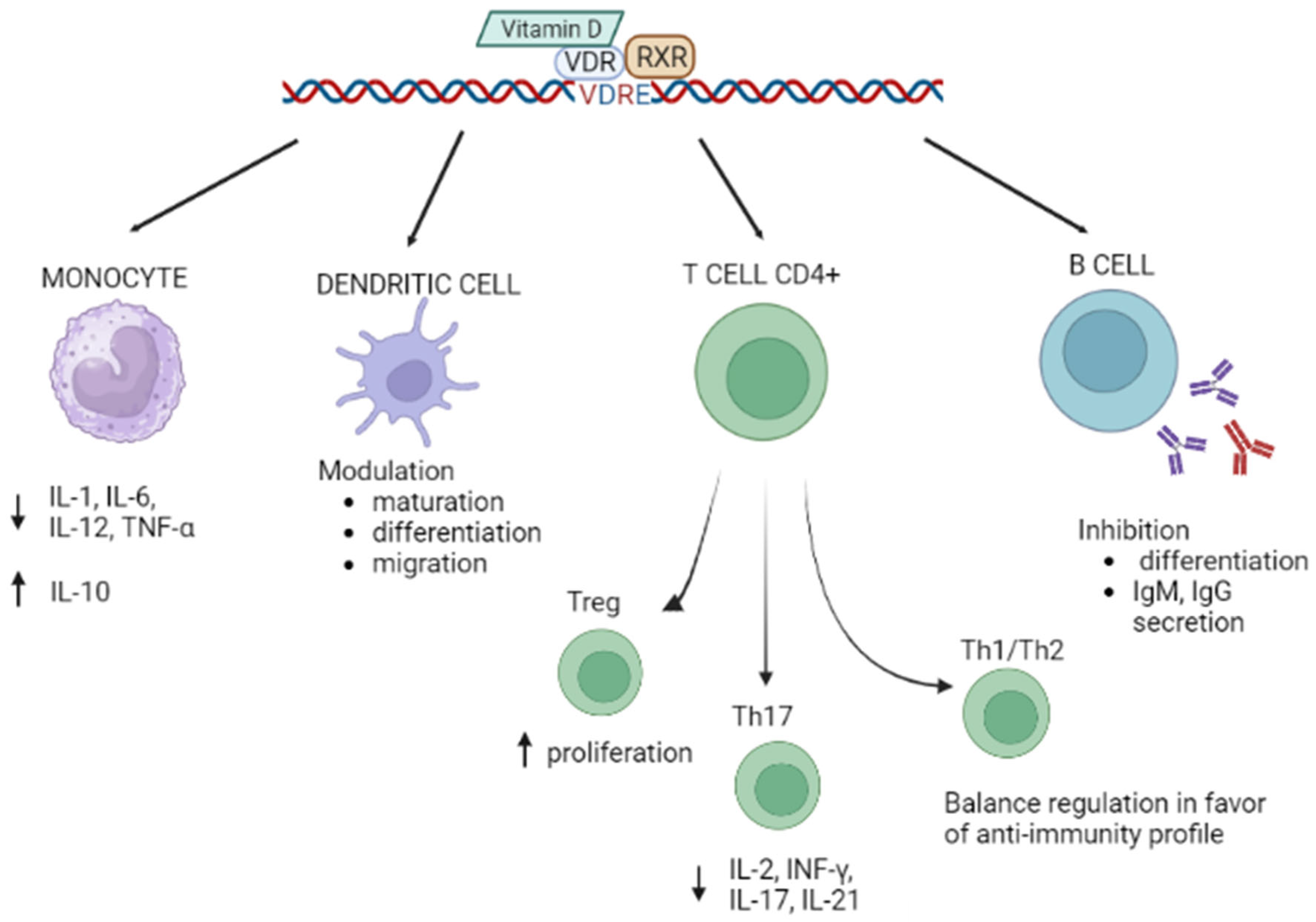
1.2. Vitamin D/VDR and Autoimmunity
1.3. VDR Gene Structure and Principal Polymorphisms
- VDRA: start site in exon 2, 427 amino acids, 48 kDa;
- VDRB1: start site in exon 1d, 477 amino acids, 54 kDa;
- a shorter isoform with higher transcriptional activity is created by a FokI single nucleotide polymorphism (SNP) that creates a translation initiation codon [28]: 424 amino acids, 47 kDa.
2. VDR Polymorphisms and Principal Autoimmune Diseases
2.1. Multiple Sclerosis
2.2. Behcet’s Disease
2.3. Systemic Lupus Erythematosus (SLE)
2.4. Type 1 Diabetes
2.5. Celiac Disease
2.6. Vitiligo
2.7. Psoriasis
2.8. Rheumatoid Arthritis (RA)
2.9. Systemic Sclerosis
2.10. Sjögren Syndrome
2.11. Hashimoto’s Thyroiditis (HT) and Grave’s Disease (GD)
| Disease | ApaI (Cases/Controls) Etnicity | BsmI (Cases/Controls) Etnicity | FokI (Cases/Controls), Etnicity | TaqI (Cases/Controls), Etnicity | Ref. | ||||
|---|---|---|---|---|---|---|---|---|---|
| risk | protection | risk | protection | risk | protection | risk | protection | ||
| Multiple sclerosis | A vs. C (596/731) Asians | [36] | |||||||
| A, AA vs. CC (721/696) Iranian | TT (721/696) Iranian | [38] | |||||||
| T, TT (in HLADRB1*15+) (641/558) Italians | [39] | ||||||||
| CT vs. TT (3758/3992) overall | [37] | ||||||||
| Behcet’s disease | A vs. C (478/666) overall AA vs. CC (237/230) Caucasians | C vs. T CC vs. CT + TT (200/200) Caucasians | T vs. C TT + CT vs. CC (176/197) Africans |
T vs. C (188/239) Africans | [42] | ||||
| Systemic lupus erythematosus | AA vs. CC (1320/1736) overall AA vs. CC females AA vs. CC (759/1019) Caucasians | C vs. T, CC + CT vs. TT (241/245) Africans | CC vs. CT + TT, C vs. T (913/1271) Caucasians | CC vs. CT + TT, C vs. T, CC vs. TT, CC + CT vs. TT (441/445) Africans | [45] | ||||
| Type 1 diabetes | TT + CT vs. CC TT vs. CT+CC T vs. C TT vs. CC (463/479) Americans | TT + CT vs. CC TT vs. CT + CC T vs. C TT vs. CC CT vs. CC (312/220) Africans | TT + CT vs. CC CT vs. CC (2077/3849) Europeans | [50] | |||||
| Celiac disease | T (176/402) | [53] | |||||||
| Vitiligo | CC + AC vs. AA CC vs. AC + AA C vs. A (1250/1400) | [55] | |||||||
| A vs. C (1048/1058) Asians | C vs. T (849/863) Asians | [56] | |||||||
| Psoriasis | TT (205/550) Caucasians | [58] | |||||||
| Rheumatoid arthritis |
AC vs. AA (1191/1451) | TT vs. CT + CC T vs. C TT vs. CC CT vs. CC (541/532) Africans | TT (2170/2452), overall | CC + CT vs. TT CC vs. CT + TT C vs. T CC vs. TT CT vs. TT (339/383) Africans CC vs. CT + TT CC vs. TT (560/623) Arabs | [62] | ||||
| CC (191/246) Italians | [64] | ||||||||
| Hashimoto’s thyroiditis | C vs. T CC vs. TT + CT (978/938) overall | [74] | |||||||
| CC (121/117) | [75] | ||||||||
| Grave’s disease | A vs. C (300/295) Asians | C vs. T (300/297) Asians | C vs. T (219/240) Asians | [77] | |||||
3. Discussion
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bikle, D.D. Vitamin D: Production, Metabolism and Mechanisms of Action. In Endotext; Feingold, K.R., Anawalt, B., Blackman, M.R., Boyce, A., Chrousos, G., Corpas, E., de Herder, W.W., Dhatariya, K., Dungan, K., Hofland, J., et al., Eds.; MDText.com, Inc.: South Dartmouth, MA, USA, 2021. [Google Scholar]
- Pike, J.W. Vitamin D3 receptors: Structure and function in transcription. Annu. Rev. Nutr. 1991, 11, 189–216. [Google Scholar] [CrossRef] [PubMed]
- Christakos, S.; Raval-Pandya, M.; Wernyj, R.P.; Yang, W. Genomic mechanisms involved in the pleiotropic actions of 1,25-dihydroxyvitamin D3. Biochem. J. 1996, 316 Pt 2, 361–371, Correction in Biochem. J. 1996, 318 Pt 3, 1079. [Google Scholar] [CrossRef] [PubMed]
- Brumbaugh, P.F.; Haussler, M.R. 1 Alpha,25-dihydroxycholecalciferol receptors in intestine. I. Association of 1 alpha,25-dihydroxycholecalciferol with intestinal mucosa chromatin. J. Biol. Chem. 1974, 249, 1251–1257. [Google Scholar] [CrossRef] [PubMed]
- Brumbaugh, P.F.; Haussler, M.R. 1 Alpha,25-dihydroxycholecalciferol receptors in intestine. II. Temperature-dependent transfer of the hormone to chromatin via a specific cytosol receptor. J. Biol. Chem. 1974, 249, 1258–1262. [Google Scholar] [CrossRef] [PubMed]
- Bizzaro, G.; Antico, A.; Fortunato, A.; Bizzaro, N. Vitamin D and Autoimmune Diseases: Is Vitamin D Receptor (VDR) Polymorphism the Culprit? Isr. Med. Assoc. J. IMAJ 2017, 19, 438–443. [Google Scholar]
- Haussler, M.R.; Jurutka, P.W.; Mizwicki, M.; Norman, A.W. Vitamin D receptor (VDR)-mediated actions of 1α,25(OH)2vitamin D₃: Genomic and non-genomic mechanisms. Best Pract. Res. Clin. Endocrinol. Metab. 2011, 25, 543–559. [Google Scholar] [CrossRef]
- Hii, C.S.; Ferrante, A. The Non-Genomic Actions of Vitamin D. Nutrients 2016, 8, 135. [Google Scholar] [CrossRef]
- Veldurthy, V.; Wei, R.; Oz, L.; Dhawan, P.; Jeon, Y.H.; Christakos, S. Vitamin D, calcium homeostasis and aging. Bone Res. 2016, 4, 16041. [Google Scholar] [CrossRef]
- Muller, V.; de Boer, R.J.; Bonhoeffer, S.; Szathmary, E. An evolutionary perspective on the systems of adaptive immunity. Biol. Rev. Camb. Philos. Soc. 2018, 93, 505–528. [Google Scholar] [CrossRef]
- Vanherwegen, A.S.; Gysemans, C.; Mathieu, C. Vitamin D endocrinology on the cross-road between immunity and metabolism. Mol. Cell. Endocrinol. 2017, 453, 52–67. [Google Scholar] [CrossRef]
- Cortes, M.; Chen, M.J.; Stachura, D.L.; Liu, S.Y.; Kwan, W.; Wright, F.; Vo, L.T.; Theodore, L.N.; Esain, V.; Frost, I.M.; et al. Developmental vitamin D availability impacts hematopoietic stem cell production. Cell Rep. 2016, 17, 458–468. [Google Scholar] [CrossRef]
- Zenata, O.; Vrzal, R. Fine tuning of vitamin D receptor (VDR) activity by post-transcriptional and post-translational modifications. Oncotarget 2017, 8, 35390–35402. [Google Scholar] [CrossRef]
- Agliardi, C.; Guerini, F.R.; Zanzottera, M.; Bolognesi, E.; Meloni, M.; Riboldazzi, G.; Zangaglia, R.; Sturchio, A.; Casali, C.; Di Lorenzo, C.; et al. The VDR FokI (rs2228570) polymorphism is involved in Parkinson’s disease. J. Neurol. Sci. 2021, 428, 117606. [Google Scholar] [CrossRef]
- Guerini, F.R.; Bolognesi, E.; Chiappedi, M.; Mensi, M.M.; Fumagalli, O.; Rogantini, C.; Zanzottera, M.; Ghezzo, A.; Zanette, M.; Agliardi, C.M.; et al. Vitamin D Receptor Polymorphisms Associated with Autism Spectrum Disorder. Autism Res. 2020, 13, 680–690. [Google Scholar] [CrossRef]
- Arosio, B.; Guerini, F.R.; Costa, A.S.; Dicitore, A.; Ferri, E.; Mari, D.; Torresani, E.; Clerici, M.; Cesari, M.; Vitale, G. Vitamin D Receptor Polymorphisms in Sex-Frailty Paradox. Nutrients 2020, 12, 2714. [Google Scholar] [CrossRef]
- Mazur, A.; Frączek, P.; Tabarkiewicz, J. Vitamin D as a Nutri-Epigenetic Factor in Autoimmunity-A Review of Current Research and Reports on Vitamin D Deficiency in Autoimmune Diseases. Nutrients 2022, 14, 4286. [Google Scholar] [CrossRef]
- Agmon-Levin, N.; Theodor, E.; Segal, R.M.; Shoenfeld, Y. Vitamin D in systemic and organ-specific autoimmune diseases. Clin. Rev. Allergy Immunol. 2013, 45, 256–266. [Google Scholar] [CrossRef]
- Almerighi, C.; Sinistro, A.; Cavazza, A.; Ciaprini, C.; Rocchi, G.; Bergamini, A. 1Alpha,25-dihydroxyvitamin D3 inhibits CD40L-induced pro-inflammatory and immunomodulatory activity in human monocytes. Cytokine 2009, 45, 190–197. [Google Scholar] [CrossRef]
- Piemonti, L.; Monti, P.; Sironi, M.; Fraticelli, P.; Leone, B.E.; Dal Cin, E.; Allavena, P.; Di Carlo, V. Vitamin D3 affects differentiation, maturation, and function of human monocyte-derived dendritic cells. J. Immunol. 2000, 164, 4443–4451. [Google Scholar] [CrossRef] [PubMed]
- Bscheider, M.; Butcher, E.C. Vitamin D immunoregulation through dendritic cells. Immunology 2016, 148, 227–236. [Google Scholar] [CrossRef]
- Gregori, S.; Casorati, M.; Amuchastegui, S.; Smiroldo, S.; Davalli, A.M.; Adorini, L. Regulatory T cells induced by 1 alpha,25-dihydroxyvitamin D3 and mycophenolate mofetil treatment mediate transplantation tolerance. J. Immunol. 2001, 167, 1945–1953. [Google Scholar] [CrossRef]
- Boonstra, A.; Barrat, F.J.; Crain, C.; Heath, V.L.; Savelkoul, H.F.; O’Garra, A. 1alpha,25-Dihydroxyvitamin d3 has a direct effect on naive CD4(+) T cells to enhance the development of Th2 cells. J. Immunol. 2001, 167, 4974–4980. [Google Scholar] [CrossRef] [PubMed]
- Tang, J.; Zhou, R.; Luger, D.; Zhu, W.; Silver, P.B.; Grajewski, R.S.; Su, S.B.; Chan, C.C.; Adorini, L.; Caspi, R.R. Calcitriol suppresses antiretinal autoimmunity through inhibitory effects on the Th17 effector response. J. Immunol. 2009, 182, 4624–4632. [Google Scholar] [CrossRef]
- Jeffery, L.E.; Burke, F.; Mura, M.; Zheng, Y.; Qureshi, O.S.; Hewison, M.; Walker, L.S.; Lammas, D.A.; Raza, K.; Sansom, D.M. 1,25-Dihydroxyvitamin D3 and IL-2 combine to inhibit T cell production of inflammatory cytokines and promote development of regulatory T cells expressing CTLA-4 and FoxP3. J. Immunol. 2009, 183, 5458–5467. [Google Scholar] [CrossRef]
- Rolf, L.; Muris, A.H.; Hupperts, R.; Damoiseaux, J. Vitamin D effects on B cell function in autoimmunity. Ann. N. Y. Acad. Sci. 2014, 1317, 84–91. [Google Scholar] [CrossRef]
- Jehan, F.; d’Alésio, A.; Garabédian, M. Exons and functional regions of the human vitamin D receptor gene around and within the main 1a promoter are well conserved among mammals. J. Steroid Biochem. Mol. Biol. 2007, 103, 361–367. [Google Scholar] [CrossRef]
- Jurutka, P.W.; Remus, L.S.; Whitfield, G.K.; Thompson, P.D.; Hsieh, J.C.; Zitzer, H.; Tavakkoli, P.; Galligan, M.A.; Dang, H.T.; Haussler, C.A.; et al. The polymorphic N terminus in human vitamin D receptor isoforms influences transcriptional activity by modulating interaction with transcription factor IIB. Mol. Endocrinol. 2000, 14, 401–420. [Google Scholar] [CrossRef]
- Usategui-Martín, R.; De Luis-Román, D.-A.; Fernández-Gómez, J.M.; Ruiz-Mambrilla, M.; Pérez-Castrillón, J.-L. Vitamin D Receptor (VDR) Gene Polymorphisms Modify the Response to Vitamin D Supplementation: A Systematic Review and Meta-Analysis. Nutrients 2022, 14, 360. [Google Scholar] [CrossRef]
- Hussain, T.; Naushad, S.M.; Ahmed, A.; Alamery, S.; Mohammed, A.A.; Abdelkader, M.O.; Alkhrm, N.A.N. Association of vitamin D receptor TaqI and ApaI genetic polymorphisms with nephrolithiasis and end stage renal disease: A meta-analysis. BMC Med. Genet. 2019, 20, 193. [Google Scholar] [CrossRef]
- Karussis, D. The diagnosis of multiple sclerosis and the various related demyelinating syndromes: A critical review. J. Autoimmun. 2014, 48–49, 134–142. [Google Scholar] [CrossRef]
- Browne, P.; Chandraratna, D.; Angood, C.; Tremlett, H.; Baker, C.; Taylor, B.V.; Thompson, A.J. Atlas of Multiple Sclerosis 2013: A growing global problem with widespread inequity. Neurology 2014, 83, 1022–1024. [Google Scholar] [CrossRef]
- Olerup, O.; Hillert, J. HLA class II-associated genetic susceptibility in multiple sclerosis: A critical evaluation. Tissue Antigens 1991, 38, 1–15. [Google Scholar] [CrossRef]
- Haines, J.L.; Terwedow, H.A.; Burgess, K.; Pericak-Vance, M.A.; Rimmler, J.B.; Martin, E.R.; Oksenberg, J.R.; Lincoln, R.; Zhang, D.Y.; Banatao, D.R.; et al. Linkage of the MHC to familial multiple sclerosis suggests genetic heterogeneity. The Multiple Sclerosis Genetics Group. Hum. Mol. Genet. 1998, 7, 1229–1234. [Google Scholar] [CrossRef]
- International Multiple Sclerosis Genetics Consortium. Multiple sclerosis genomic map implicates peripheral immune cells and microglia in susceptibility. Science 2019, 365, eaav7188. [Google Scholar] [CrossRef]
- Zhang, Y.J.; Zhang, L.; Chen, S.Y.; Yang, G.J.; Huang, X.L.; Duan, Y.; Yang, L.J.; Ye, D.Q.; Wang, J. Association between VDR polymorphisms and multiple sclerosis: Systematic review and updated meta-analysis of case-control studies. Neurol. Sci. 2018, 39, 225–234. [Google Scholar] [CrossRef]
- Imani, D.; Razi, B.; Motallebnezhad, M.; Rezaei, R. Association between vitamin D receptor (VDR) polymorphisms and the risk of multiple sclerosis (MS): An updated meta-analysis. BMC Neurol. 2019, 19, 339. [Google Scholar] [CrossRef] [PubMed]
- Mohammadi, A.; Azarnezhad, A.; Khanbabaei, H.; Izadpanah, E.; Abdollahzadeh, R.; Barreto, G.E.; Sahebkar, A. Vitamin D receptor genetic polymorphisms and the risk of multiple sclerosis: A systematic review and meta-analysis. Steroids 2020, 158, 108615. [Google Scholar] [CrossRef] [PubMed]
- Agliardi, C.; Guerini, F.R.; Saresella, M.; Caputo, D.; Leone, M.A.; Zanzottera, M.; Bolognesi, E.; Marventano, I.; Barizzone, N.; Fasano, M.E.; et al. Vitamin D receptor (VDR) gene SNPs influence VDR expression and modulate protection from multiple sclerosis in HLA-DRB1*15-positive individuals. Brain Behav. Immun. 2011, 25, 1460–1467. [Google Scholar] [CrossRef]
- Kiafar, M.; Faezi, S.T.; Kasaeian, A.; Baghdadi, A.; Kakaei, S.; Mousavi, S.A.; Nejadhosseinian, M.; Shahram, F.; Ghodsi, S.Z.; Shams, H.; et al. Diagnosis of Behçet’s disease: Clinical characteristics, diagnostic criteria, and differential diagnoses. BMC Rheumatol. 2021, 5, 2. [Google Scholar] [CrossRef]
- Davatchi, F.; Chams-Davatchi, C.; Shams, H.; Shahram, F.; Nadji, A.; Akhlaghi, M.; Faezi, T.; Ghodsi, Z.; Sadeghi Abdollahi, B.; Ashofteh, F.; et al. Behcet’s disease: Epidemiology, clinical manifestations, and diagnosis. Expert. Rev. Clin. Immunol. 2017, 13, 57–65. [Google Scholar] [CrossRef]
- Wu, M.; Li, L.; Tian, L.; Liu, D.; Jian, J.; Zhou, Y.; Xu, Y. 5Apal, Taql, Fokl, and Bsml polymorphisms and the susceptibility of Behcet’s disease: An updated meta-analysis. Immunol. Res. 2022, 70, 781–792. [Google Scholar] [CrossRef]
- Zandman-Goddard, G.; Solomon, M.; Rosman, Z.; Peeva, E.; Shoenfeld, Y. Environment and lupus-related diseases. Lupus 2012, 21, 241–250. [Google Scholar] [CrossRef] [PubMed]
- Rullo, O.J.; Tsao, B.P. Recent insights into the genetic basis of systemic lupus erythematosus. Ann. Rheum. Dis. 2013, 72 (Suppl. 2), ii56–ii61. [Google Scholar] [CrossRef]
- Yang, S.K.; Liu, N.; Zhang, W.J.; Song, N.; Yang, J.P.; Zhang, H.; Gui, M. Impact of Vitamin D Receptor Gene Polymorphism on Systemic Lupus Erythematosus Susceptibility: A Pooled Analysis. Genet. Test. Mol. Biomark. 2022, 26, 228–238. [Google Scholar] [CrossRef]
- van Belle, T.L.; Coppieters, K.T.; von Herrath, M.G. Type 1 diabetes: Etiology, immunology, and therapeutic strategies. Physiol. Rev. 2011, 91, 79–118. [Google Scholar] [CrossRef] [PubMed]
- Diaz-Valencia, P.A.; Bougnères, P.; Valleron, A.J. Global epidemiology of type 1 diabetes in young adults and adults: A systematic review. BMC Public Health 2015, 15, 255. [Google Scholar] [CrossRef] [PubMed]
- Pociot, F.; McDermott, M.F. Genetics of type 1 diabetes mellitus. Genes. Immun. 2002, 3, 235–249. [Google Scholar] [CrossRef]
- Tizaoui, K.; Kaabachi, W.; Hamzaoui, A.; Hamzaoui, K. Contribution of VDR polymorphisms to type 1 diabetes susceptibility: Systematic review of case-control studies and meta-analysis. J. Steroid Biochem. Mol. Biol. 2014, 143, 240–249. [Google Scholar] [CrossRef]
- Zhai, N.; Bidares, R.; Makoui, M.H.; Aslani, S.; Mohammadi, P.; Razi, B.; Imani, D.; Yazdchi, M.; Mikaeili, H. Vitamin D receptor gene polymorphisms and the risk of the type 1 diabetes: A meta-regression and updated meta-analysis. BMC Endocr. Disord. 2020, 20, 121. [Google Scholar] [CrossRef]
- Taylor, A.K.; Lebwohl, B.; Snyder, C.L.; Green, P.H.R. Celiac Disease. In GeneReviews®; Adam, M.P., Mirzaa, G.M., Pagon, R.A., Wallace, S.E., Bean, L.J.H., Gripp, K.W., Amemiya, A., Eds.; University of Washington: Seattle, DC, USA, 2008. [Google Scholar]
- Corazza, G.R.; Di Sario, A.; Cecchetti, L.; Tarozzi, C.; Corrao, G.; Bernardi, M.; Gasbarrini, G. Bone mass and metabolism in patients with celiac disease. Gastroenterology 1995, 109, 122–128. [Google Scholar] [CrossRef]
- Shree, T.; Banerjee, P.; Senapati, S. A meta-analysis suggests the association of reduced serum level of vitamin D and T-allele of Fok1 (rs2228570) polymorphism in the vitamin D receptor gene with celiac disease. Front. Nutr. 2023, 9, 996450. [Google Scholar] [CrossRef] [PubMed]
- Picardo, M.; Dell’Anna, M.L.; Ezzedine, K.; Hamzavi, I.; Harris, J.E.; Parsad, D.; Taieb, A. Vitiligo. Nat. Rev. Dis. Prim. 2015, 1, 15011. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.Z.; Wang, M.; Ding, Y.; Gao, F.; Feng, Y.Y.; Yakeya, B.; Wang, P.; Wu, X.J.; Hu, F.X.; Xian, J.; et al. Vitamin D receptor gene polymorphism, serum 25-hydroxyvitamin D levels, and risk of vitiligo: A meta-analysis. Medicine 2018, 97, e11506. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.H.; Song, G.G. Association between vitamin D receptor polymorphisms and vitiligo susceptibility: An updated meta-analysis. J. Cosmet. Dermatol. 2023, 22, 969–979. [Google Scholar] [CrossRef] [PubMed]
- Menter, A.; Gottlieb, A.; Feldman, S.R.; Van Voorhees, A.S.; Leonardi, C.L.; Gordon, K.B.; Lebwohl, M.; Koo, J.Y.; Elmets, C.A.; Korman, N.J.; et al. Guidelines of care for the management of psoriasis and psoriatic arthritis: Section 1. Overview of psoriasis and guidelines of care for the treatment of psoriasis with biologics. J. Am. Acad. Dermatol. 2008, 58, 826–850. [Google Scholar] [CrossRef]
- Li, J.; Sun, L.; Sun, J.; Yan, M. Pooling analysis regarding the impact of human vitamin D receptor variants on the odds of psoriasis. BMC Med. Genet. 2019, 20, 161. [Google Scholar] [CrossRef]
- Lee, Y.H. Vitamin D receptor ApaI, TaqI, BsmI, and FokI polymorphisms and psoriasis susceptibility: An updated meta-analysis. Clin. Exp. Dermatol. 2019, 44, 498–505. [Google Scholar] [CrossRef]
- McInnes, I.B.; Schett, G. The pathogenesis of rheumatoid arthritis. N. Engl. J. Med. 2011, 365, 2205–2219. [Google Scholar] [CrossRef]
- Orozco, G.; Barton, A. Update on the genetic risk factors for rheumatoid arthritis. Expert. Rev. Clin. Immunol. 2010, 6, 61–75. [Google Scholar] [CrossRef]
- Bagheri-Hosseinabadi, Z.; Imani, D.; Yousefi, H.; Abbasifard, M. Vitamin D receptor (VDR) gene polymorphism and risk of rheumatoid arthritis (RA): Systematic review and meta-analysis. Clin. Rheumatol. 2020, 39, 3555–3569. [Google Scholar] [CrossRef]
- Despotović, M.; Jevtović Stoimenov, T.; Stojanović, S.; Bašić, J.; Kundalić, J.; Đorđević, B.; Ranđelović, M.; Pavlović, D. Association of vitamin D receptor genetic variants with bone mineral density and inflammatory markers in rheumatoid arthritis. Clin. Biochem. 2021, 87, 26–31. [Google Scholar] [CrossRef]
- Latini, A.; De Benedittis, G.; Perricone, C.; Colafrancesco, S.; Conigliaro, P.; Ceccarelli, F.; Chimenti, M.S.; Novelli, L.; Priori, R.; Conti, F.; et al. VDR Polymorphisms in Autoimmune Connective Tissue Diseases: Focus on Italian Population. J. Immunol. Res. 2021, 2021, 5812136. [Google Scholar] [CrossRef] [PubMed]
- Bairkdar, M.; Rossides, M.; Westerlind, H.; Hesselstrand, R.; Arkema, E.V.; Holmqvist, M. Incidence and prevalence of systemic sclerosis globally: A comprehensive systematic review and meta-analysis. Rheumatology 2021, 60, 3121–3133. [Google Scholar] [CrossRef] [PubMed]
- Kamal, A.; Gamal, S.M.; Elgengehy, F.T.; Alkemary, A.K.; Siam, I. Association of VDR ApaI and TaqI Gene Polymorphisms with the Risk of Scleroderma and Behçet’s Disease. Immunol. Investig. 2016, 45, 531–542. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Chen, S.-Y.; Liu, H.-H.; Yin, X.-D.; Cao, L.-T.; Xu, J.-H.; Li, X.M.; Ye, D.Q.; Wang, J. Associations of Vitamin D Receptor Single Nucleotide Polymorphisms with Susceptibility to Systemic Sclerosis. Arch. Med. Res. 2019, 50, 368–376. [Google Scholar] [CrossRef]
- Fox, R.I.; Howell, F.V.; Bone, R.C.; Michelson, P. Primary Sjogren syndrome: Clinical and immunopathologic features. Semin. Arthritis Rheum. 1984, 14, 77–105. [Google Scholar] [CrossRef]
- Shiboski, C.H.; Baer, A.N.; Shiboski, S.C.; Lam, M.; Challacombe, S.; Lanfranchi, H.E.; Schiødt, M.; Shirlaw, P.; Srinivasan, M.; Umehara, H.; et al. Natural History and Predictors of Progression to Sjögren’s Syndrome Among Participants of the Sjögren’s International Collaborative Clinical Alliance Registry. Arthritis Care Res. 2018, 70, 284–294. [Google Scholar] [CrossRef]
- Nikolov, N.P.; Illei, G.G. Pathogenesis of Sjögren’s syndrome. Curr. Opin. Rheumatol. 2009, 21, 465–470. [Google Scholar] [CrossRef]
- Zilahi, E.; Chen, J.Q.; Papp, G.; Szántó, A.; Zeher, M. Lack of association of vitamin D receptor gene polymorphisms/haplotypes in Sjögren’s syndrome. Clin. Rheumatol. 2015, 34, 247–253. [Google Scholar] [CrossRef]
- Ragusa, F.; Fallahi, P.; Elia, G.; Gonnella, D.; Paparo, S.R.; Giusti, C.; Churilov, L.P.; Ferrari, S.M.; Antonelli, A. Hashimotos’ thyroiditis: Epidemiology, pathogenesis, clinic and therapy. Best. Pract. Res. Clin. Endocrinol. Metab. 2019, 33, 101367. [Google Scholar] [CrossRef]
- Kamyshna, I.I.; Pavlovych, L.B.; Malyk, I.V.; Kamyshnyi, A.M. 25-OH Vitamin D blood serum linkage with VDR gene polymorphism (rs2228570) in thyroid pathology patients in the West-Ukrainian population. J. Med. Life 2021, 14, 549–556. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Cheng, W.; Ma, Y.; Zhu, J. Vitamin D receptor gene FokI but not TaqI, ApaI, BsmI polymorphism is associated with Hashimoto’s thyroiditis: A meta-analysis. Sci. Rep. 2017, 7, 41540. [Google Scholar] [CrossRef] [PubMed]
- Zarrin, R.; Bagheri, M.; Mehdizadeh, A.; Ayremlou, P.; Faghfouri, A.H. The association of FokI and ApaI polymorphisms in vitamin D receptor gene with autoimmune thyroid diseases in the northwest of Iran. Med. J. Islam. Repub. Iran. 2018, 32, 4. [Google Scholar] [CrossRef] [PubMed]
- Prabhakar, B.S.; Bahn, R.S.; Smith, T.J. Current perspective on the pathogenesis of Graves’ disease and ophthalmopathy. Endocr. Rev. 2003, 24, 802–835. [Google Scholar] [CrossRef]
- Zhou, H.; Xu, C.; Gu, M. Vitamin D receptor (VDR) gene polymorphisms and Graves’ disease: A meta-analysis. Clin. Endocrinol. 2009, 70, 938–945. [Google Scholar] [CrossRef]
- Gregersen, P.K.; Olsson, L.M. Recent advances in the genetics of autoimmune disease. Annu. Rev. Immunol. 2009, 27, 363–391. [Google Scholar] [CrossRef]
- Li, K.; Shi, Q.; Yang, L.; Li, X.; Liu, L.; Wang, L.; Li, Q.; Wang, G.; Li, C.Y.; Gao, T.W. The association of vitamin D receptor gene polymorphisms and serum 25-hydroxyvitamin D levels with generalized vitiligo. Br. J. Dermatol. 2012, 167, 815–821. [Google Scholar] [CrossRef]
- Shimada, A.; Kanazawa, Y.; Motohashi, Y.; Yamada, S.; Maruyama, T.; Ikegami, H.; Awata, T.; Kawasaki, E.; Kobayashi, T.; Nakanishi, K.; et al. Evidence for association between vitamin D receptor BsmI polymorphism and type 1 diabetes in Japanese. J. Autoimmun. 2008, 30, 207–211. [Google Scholar] [CrossRef]
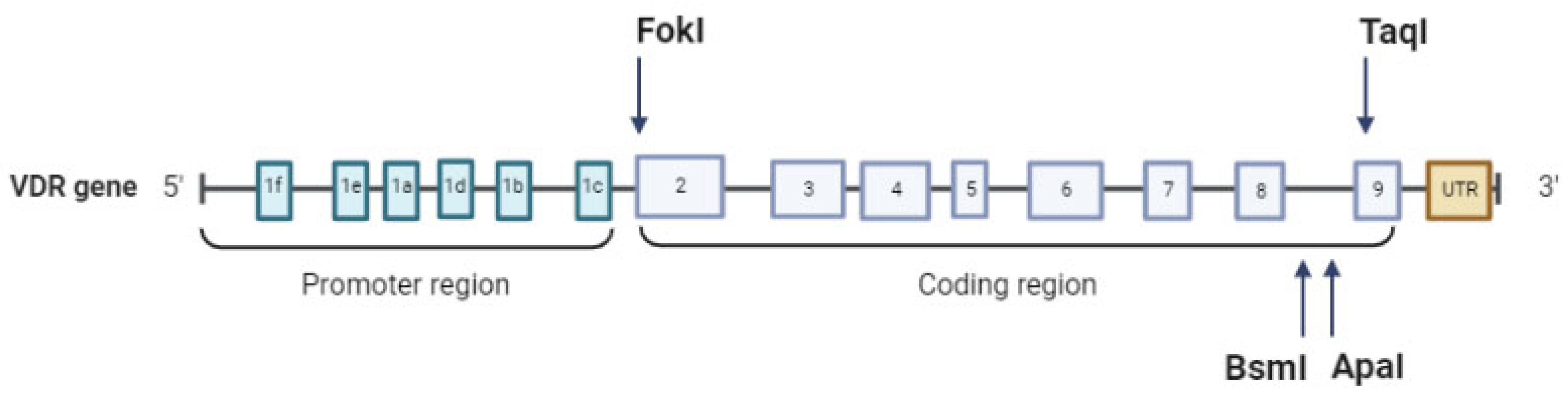
| VDR SNP | Position | Description | Effect |
|---|---|---|---|
| TaqI (rs731236) | +65,058, intronic | T > C (T < t) | Affects splicing and VDR translation |
| BsmI (rs1544410) | +63,980, intronic | C < T (B > b) | Decreases mRNA stability |
| ApaI (rs7975232) | +64,978, intronic | A > C (A > a) | Decreases mRNA stability |
| FokI (rs2228570) | +30,920, exon 2 | C > T (F < f) | Start loss |
| Genetic Model | ApaI | BsmI | FokI | TaqI |
|---|---|---|---|---|
| Dominant | CC + AC vs. AA | TT + CT vs. CC | TT + CT vs. CC | CC + CT vs. TT |
| Recessive | CC vs. AC + AA | TT vs. CT + CC | TT vs. CT + CC | CC vs. CT + TT |
| Heterozygote | AC vs. AA | CT vs. CC | CT vs. CC | CT vs. TT |
| Homozygote | CC vs. AA/AA vs. CC | TT vs. CC/CC vs. TT | TT vs. CC/CC vs. TT | CC vs. TT/TT vs. CC |
| Allelic | C vs. A/A vs. C | T vs. C/C vs. T | T vs. C/C vs. T | C vs. T/T vs. C |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Agliardi, C.; Guerini, F.R.; Bolognesi, E.; Zanzottera, M.; Clerici, M. VDR Gene Single Nucleotide Polymorphisms and Autoimmunity: A Narrative Review. Biology 2023, 12, 916. https://doi.org/10.3390/biology12070916
Agliardi C, Guerini FR, Bolognesi E, Zanzottera M, Clerici M. VDR Gene Single Nucleotide Polymorphisms and Autoimmunity: A Narrative Review. Biology. 2023; 12(7):916. https://doi.org/10.3390/biology12070916
Chicago/Turabian StyleAgliardi, Cristina, Franca Rosa Guerini, Elisabetta Bolognesi, Milena Zanzottera, and Mario Clerici. 2023. "VDR Gene Single Nucleotide Polymorphisms and Autoimmunity: A Narrative Review" Biology 12, no. 7: 916. https://doi.org/10.3390/biology12070916
APA StyleAgliardi, C., Guerini, F. R., Bolognesi, E., Zanzottera, M., & Clerici, M. (2023). VDR Gene Single Nucleotide Polymorphisms and Autoimmunity: A Narrative Review. Biology, 12(7), 916. https://doi.org/10.3390/biology12070916







