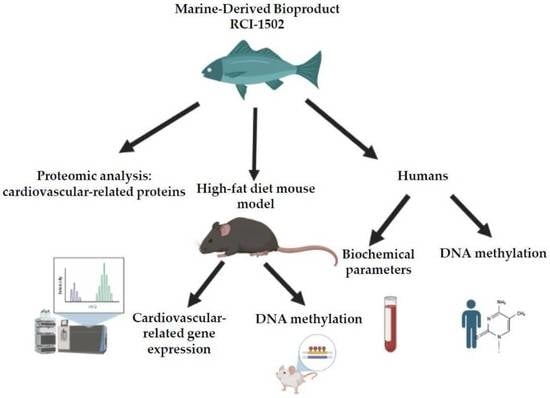
Proteomic and Global DNA Methylation Modulation in Lipid Metabolism Disorders with a Marine-Derived Bioproduct
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethical Approval
2.2. Human Blood Collection
2.3. Experimental Animals
2.4. Preparation of the Marine Lipoprotein Extract (RCI-1502)
2.5. HFD-induced Mouse Model of Obesity
2.6. Collection of Samples from Mice
2.7. DNA Extraction
2.8. RNA Extraction
2.9. Quantification of Global DNA Methylation (5 mC)
2.10. Quantitative Real Time RT-PCR
2.11. Preparation of RCI-1502 for Liquid Chromatography-Tandem Mass Spectrometry (LC-MS/MS) Analysis
2.12. LC-MS/MS Analysis
2.13. Proteomic Data Analysis
2.14. Statistical Analysis
3. Results
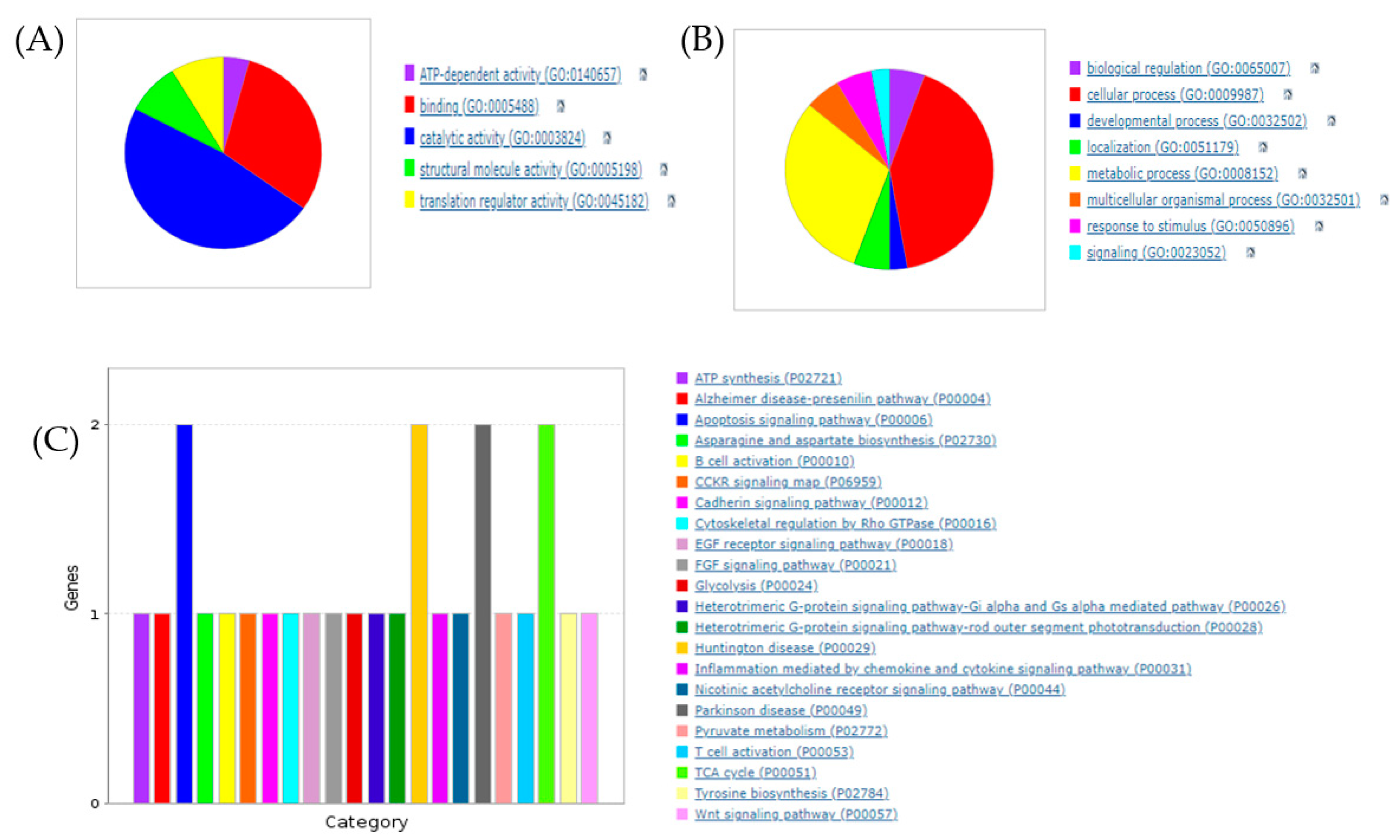
3.1. Proteomic Profiling of RCI-1502
3.2. RCI-1502 Reduces Cardiovascular Disease-Related Gene Expression in an HFD Mouse Model
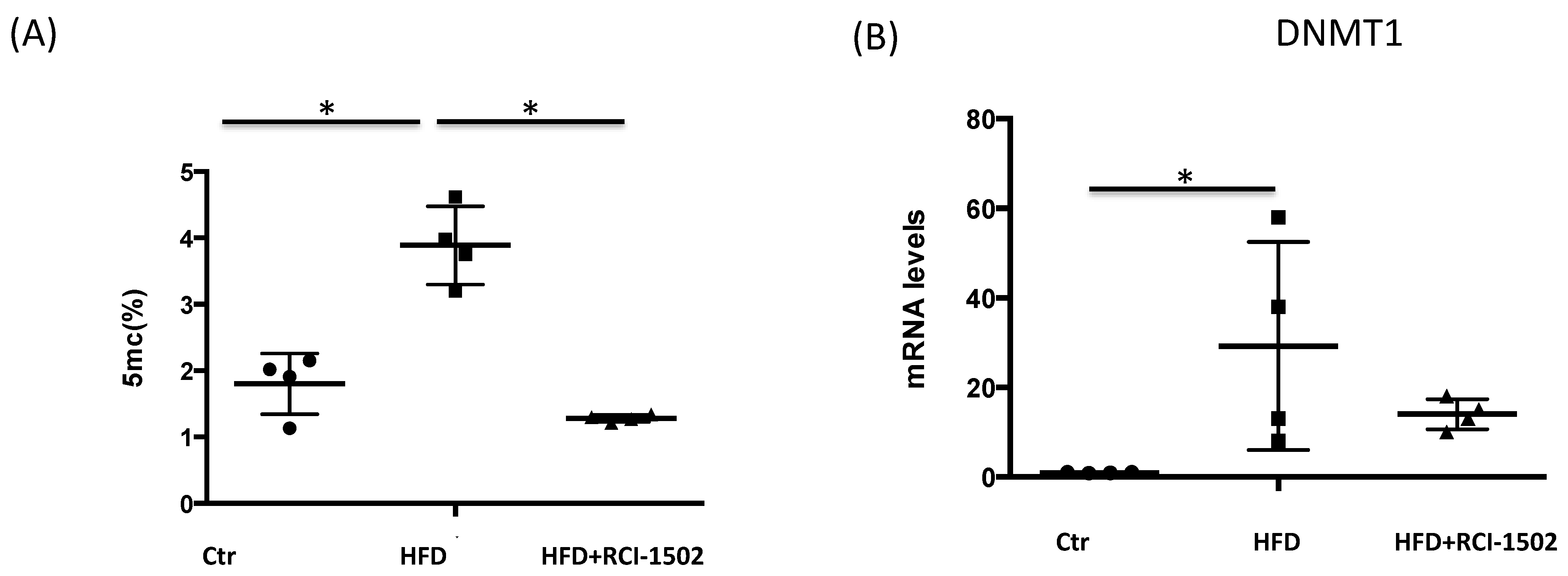
3.3. RCI-1502 Reduces DNA Methylation in an HFD Mouse Model
3.4. Patients with Dyslipidemia Exhibit Higher DNA Methylation Levels
3.5. RCI-1502 Reduces DNA Methylation and Regulates Cholesterol and Triglyceride Levels in Patients with Dyslipidemia
4. Discussion
5. Limitations of the Study
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Pappan, N. Dyslipidemia; StatPearls: Tampa, FL, USA, 2022. [Google Scholar]
- Porillo, A.; Casula, M.; Olmastroni, E.; Norata, G.; Catapano, A. Global epidemiology of dyslipidaemias. Nat. Rev. Cardiol. 2021, 18, 689–700. [Google Scholar] [CrossRef]
- Kandi, V.; Vadakedath, S. Effect of DNA methylation in various diseases and the probable protective role of nutrition: A mini-review. Cureus 2015, 7, e309. [Google Scholar] [CrossRef]
- Bird, A. The essentials of DNA methylation. Cell 1992, 70, 5–8. [Google Scholar] [CrossRef] [PubMed]
- Hermann, A.; Gowher, H.; Jeltsch, A. Biochemistry and biology of mammal DNA methyltransferases. Cell Mol. Life Sci. 2004, 61, 2571–2587. [Google Scholar] [CrossRef]
- Hermann, A.; Goyal, R.; Jeltsch, A. The Dnmt1 DNA-cytosine-c5-methyltransferase methylates DNA processively with high preference for hemimethylated target sites. J. Boil. Chem. 2004, 279, 48350–48359. [Google Scholar] [CrossRef]
- Okano, M.; Xie, S.; Li, E. Cloning and characterization of a family of novel mammalian DNMA (cytosine-5) methyltransferases. Nat. Genet. 1998, 19, 219–220. [Google Scholar] [CrossRef]
- Nan, X.; Campoy, F.J.; Bird, A. MeCP2 is a transcriptional repressor with abundant binding sites in genomic chromatin. Cell 1997, 88, 471–481. [Google Scholar] [CrossRef]
- Morofuji, Y.; Nakagawa, S.; Ujifuku, K.; Fujimoto, T.; Otsuka, K.; Niwa, M.; Tsutsumi, K. Beyond lipid-lowering: Effects of statins on cardiovascular and cerebrovascular diseases and cancer. Pharmaceuticals 2016, 15, 151. [Google Scholar] [CrossRef] [PubMed]
- Yao, C.M.; Hwang, S.-J.; Mendelson, M.; Huan, T.; Liu, C.; Levy, D. DNA methylation signature of statin therapy. Circulation 2017, 136, A20759. [Google Scholar]
- Gorica, E.; Mohammed, S.; Ambrosini, S.; Celderone, V.; Costantino, S.; Peneni, F. Epi-Drugs in heart failure. Front. Cardionasc. Med. 2022, 9, 923014. [Google Scholar] [CrossRef]
- Johnston, T.P.; Korolenko, T.A.; Pirro, M.; Sahebkar, A. preventing cardiovascular heart disease: Promising mutraceutical an non-nutracutical treatments for cholesterol management. Pharmacol. Res. 2017, 120, 219–225. [Google Scholar] [CrossRef]
- Szczpanska, E.; Bialek-Dratwa, A.; Janota, B.; Kowalski, O. Dietary therapy in prevention of Cardiovascular Disease (CVD)-Tradition or modernity? A review of the latest approaches to nutrition in CVD. Nutrients 2022, 14, 2649. [Google Scholar] [CrossRef] [PubMed]
- Maugeri, A.; Barchitta, M. How dietary factors affect DNA methylation: Lessons from epidemiological studies. Medicina 2020, 56, 374. [Google Scholar] [CrossRef] [PubMed]
- Kohil, A.; Al-Asmakh, M.; Al-Shafai, M.; Terranegra, A. The interpaly between diet and the epigenome in the apthogenesis of Type-q Diabetes. Froint. Nutr. 2021, 7, 612115. [Google Scholar] [CrossRef] [PubMed]
- Maugeri, A. the effect of dietary interventions on DNA methylation: Implications for obesity management. Int. J. Mol. Sci. 2020, 21, 8670. [Google Scholar] [CrossRef]
- Ma, J.; Rebholz, C.; Braun, K.; Reynolds, L.; Aslibekyan, S.; Xia, R.; Biligowda, N.G.; Huan, T.; Liu, C.; Mendelson, M.M.; et al. Whole blood DNA methylation signatures of diet are associated with cardiovascular risk factors and all-cause mortality. Circ. Genom. Pecicion Med. 2020, 13, e002766. [Google Scholar] [CrossRef]
- Lombardi, V.R.M.; Cacabelos, R. E-SAR-94010: A marine fish extract obtained by advanced biotechnological methods. Drugs Future 1999, 24, 167–176. [Google Scholar] [CrossRef]
- Carrera, I.; Corzo, L.; Naidoo, V.; Martínez-Iglesias, O.; Cacabelos, R. Cardiovascular and lipid-lowering effects of a marine lipoprotein extract in a high-fat diet-induced obesity mouse model. Int. J. Med. Sci. 2023, 20, 292–306. [Google Scholar] [CrossRef]
- Cacabelos, R. Application of Nutrigenomics to Alzheimer’s Disease. Agro Food Ind. Hi-Tech 2008, 19, 37. [Google Scholar]
- Lombardi, V.R.M.; Cagiao, A.; Fernández-Novoa, L.; Álvarez, X.A.; Corzo, M.D.; Zas, R.; Sampedro, C.; Cacabelos, R. Short-term food supplementation effects of a fish-derived extract on the immunological status of pregnant rats and their sucking pups. Nutr. Res. 2001, 21, 1425–1434. [Google Scholar] [CrossRef]
- Cacabelos, R. Pharmacogenomics, nutrigenomics and therapeutic optimization in Alzheimer’s disease. Aging Health 2005, 1, 303–348. [Google Scholar] [CrossRef]
- Lombardi, V.R.; Fernández-Novoa, L.; Etcheverría, I.; Seoane, S.; Cacabelos, R. Effects of fish-derived lipoprotein extracts on activation markers, Fas expression and apoptosis in peripheral blood lymphocytes. Int. Immunopharmacol. 2005, 5, 253–262. [Google Scholar] [CrossRef] [PubMed]
- Affane, F.; Louala, S.; El Imane Harrat, N.; Bensalah, F.; Chekkal, H.; Allaoui, A.; Lamri-Senhadji, M. Hypolipidemic, antioxidant and antiatherogenic property of sardine by-products proteins in high-fat diet induced obese rats. Life Sci. 2018, 199, 16–22. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.D.; Hu, F.B. Dietary fat and risk of cardiovascular disease: Recent controversies and advances. Annu. Rev. Nutr. 2017, 37, 423–446. [Google Scholar] [CrossRef] [PubMed]
- Aydis, L.R.; do Amaral, L.; de Souza, R.; Jacobowski, A.; Dos Santos, E.; Rodrigues Macedo, M.L. Nonalcoholic Fatty Liver Disease Induced by High-Fat Diet in C57bl/6 Models. Nutrients 2019, 11, 3067. [Google Scholar] [CrossRef] [PubMed]
- Wong, M.L.; Medrano, J.F. Real-time PCR for mRNA quantitation. BioTechniques 2005, 39, 75–85. [Google Scholar] [CrossRef] [PubMed]
- Malik, F.; Hartman, J.; Elias, K.; Morgan, B.; Rodriguez, H.; Brejc, K.; Anderson, R.L.; Sueoka, S.H.; Lee, K.H.; Finer, J.T.; et al. Cardiac myosin activation: A potential therapeutic approach for systolic heart failure. Science 2011, 331, 1439–1443. [Google Scholar] [CrossRef]
- Bai, F.; Wang, L.; Kawai, M. A study of tropomyosin’s role in cardiac function and disease using thin-filamnet reconstituted myocardium. J. Muscle Res. Cell Motil. 2013, 34, 295–310. [Google Scholar] [CrossRef]
- Long, Q.; Yang, K.; Yang, Q. Regulation of mitochondrial ATP synthase in cardiac pathophysiology. Am. J. Cardiovasc. Dis. 2015, 5, 19–32. [Google Scholar]
- Martinov, M.V.; Plotnikov, A.G.; Vitvitsky, V.; Ataullakhanov, F. deficiencies of glycolytic enzymes as a possible cause of hemolytic anemia. Biochim. Biophys. Acta 2000, 1474, 75–87. [Google Scholar] [CrossRef]
- Mizukami, Y.; Iwamatsu, A.; Aki, T.; Kimura, M.; Nakamura, K.; Nao, T.; Okusa, T.; Matsuzaki, M.; Yoshida, K.I.; Kobayashi, S. ERK1/2 intracellular ATP levels through a-enolase expression in cardiomyocites exposed to ischemic hypoxia and reoxygenation. J. Biol. Chem. 2004, 279, 50120–50131. [Google Scholar] [CrossRef]
- Distelmaier, K.; Muqaku, B.; Wurm, R.; Arfsten, H.; Seidel, S.; Kovacs, G.G.; Mayer, R.L.; Szekeres, T.; Wallisch, C.; Hubner, P.; et al. Proteomics-enriched prediction model for poor neurologic outcome in cardiac arrest survivors. Med. Care Med. 2020, 48, 167–175. [Google Scholar] [CrossRef] [PubMed]
- Kawase, Y.; Hajjar, R. The cardiac sarcoplasmic/endoplasmic reticulum carcium ATPase: A potent target for cardiovascular disease. Nat. Rev. Cardiol. 2008, 5, 554–565. [Google Scholar] [CrossRef]
- Qiu, Y.; Zhao, D.; Butenschön, V.M.; Bauer, A.T.; Schneider, S.W.; Skolnik, E.Y.; Hammes, H.P.; Wieland, T.; Feng, Y. Nucleoside diphospate kinase B deficiency causes a diabetes-like vascular pathology via up-regulation of endothelial angiopoietin-2 in the retina. Acta Diabet. 2015, 53, 81–89. [Google Scholar] [CrossRef]
- Sorensen, A.; Sondergaard, M.; Overgaard, M. Calmodulin in heartbeat. FEBS Lett. 2013, 280, 5511–5532. [Google Scholar] [CrossRef]
- Vogt, S.; Ruppert, V.; Pankuweit, S.; Paletta, J.; Rhiel, A.; Weber, P.; Irqsusi, M.; Cybulski, P.; Ramzan, R. Myocardial insuffieniency is related to reduced subunit 4 content of cytochrome c oxidase. J. Cardiothorac. Surg. 2018, 13, 95. [Google Scholar] [CrossRef] [PubMed]
- London, B.; Michalec, M.; Mehdi, H.; Zhu, X.; Kerchner, L.; Sanyal, S.; Viswanathan, P.C.; Pfahnl, A.E.; Shang, L.L.; Madhusudanan, M.; et al. Mutation in glycerol-3-phosphate dehydrogenase 1 like gene (GPD1-L) decreases cardiac Na+ current and causes inherited arrythmias. Circulation 2007, 116, 2260–2268. [Google Scholar] [CrossRef] [PubMed]
- Lei, I.; Tian, S.; Gao, W.; Liu, L.; Guo, Y.; Tang, P.; Chen, E.; Wang, Z. Acetyl-CoA production by specific metabolites promotes cardiac repair after myocardial infarction via histone acetylation. eLife 2021, 10, e60311. [Google Scholar] [CrossRef]
- Ashrafian, H.; Czibik, G.; Bellahcene, M.; Aksentjeviv, D.; Smith, A.; Mitchell, S.J.; Dodd, M.S.; Kirwan, J.; Byrne, J.J.; Ludwig, C.; et al. Fumarate is cardioprotective via activation of the Nrf2 antioxidant pathway. Cell Metab. 2012, 15, 161–371. [Google Scholar] [CrossRef]
- Alves-Bezerra, M.; Cohen, D.E. Triglyceride Metabolism in the Liver. Compr. Physiol. 2017, 8, 1–8. [Google Scholar]
- Mardinoglu, A.; Wu, H.; Bjornson, E.; Zhang, C.; Hakkarainen, A.; Räsänen, S.M.; Lee, S.; Mancina, R.M.; Bergentall, M.; Pietiläinen, K.H.; et al. An Integrated Understanding of the Rapid Metabolic Benefits of a Carbohydrate-Restricted Diet on Hepatic Steatosis in Humans. Cell Metab. 2018, 27, 559–571.e5. [Google Scholar] [CrossRef]
- Mahley, R.M. Apolipoprotein E: From cardiovascular disease to neurodegenrative disorders. J. Mol. Med. 2016, 94, 739–746. [Google Scholar] [CrossRef] [PubMed]
- Carr, R.M. VCA;-1: Closing the gap between lipotocicity and endothelial dysfunction in nonalcoholic steatohepatitis. J. Clin. Invest 2021, 131, e147556. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Rong, J.; Zhang, Z. The emerging role of angiotensinogen in cardiovascular diseases. J. Cell Physiol. 2021, 236, 68–78. [Google Scholar] [CrossRef] [PubMed]
- Nasution, S.A. The use of ACE inhibitor in cardiovascular disease. Acta Med. Indones 2006, 38, 60–64. [Google Scholar]
- Kumar, V.; Kumar, A.; Sanawar, R.; Jaleel, A.; Kumar, T.R.S.; Kartha, C. Chronic pressure overload results in deficiency of mitochondrial membrane transporter ABCB7 which contributes to iron overload, mitochondrial dysfunction, metabolic shift and worsens cardiac function. Sci. Rep. 2019, 9, 13170. [Google Scholar] [CrossRef]
- Zhang, Y.; Zeng, C. Role of DNA methylation in cardiovascular diseases. Clin. Exp. Hypertens. N. Y. 2016, 38, 261–267. [Google Scholar] [CrossRef]
- Ramos, R.B.; Fabris, V.; Lecke, S.; Maturana, M.A.; Spritzer, P. Association between global leukocyte DNA methylation and cardiovascular risk in postmenopausal women. MBMC Med. Genet. 2016, 17, 71. [Google Scholar] [CrossRef]
- Martins, J.; Steyn, N.; Rossouw, M.; Pillay, T. Best practice for LDL-cholesterol: When and how to calculate. J. Clin. Pathol. 2023, 76, 145–152. [Google Scholar] [CrossRef]
- Navas-Acien, A.; Domingo-Rellosos, A.; Subedi, P.; Riffo-Campos, A.; Xia, R.; Gomez, L.; Haack, K.; Goldsmith, J.; Howard, B.V.; Best, L.G.; et al. Blood DNA methylation and incident coronary heart disease: Evidence from the strong heart study. JAMA Cardiol. 2020, 6, 1237–1246. [Google Scholar] [CrossRef]
- Palou-marquez, G.; Subirana, I.; Nonell, L.; Fernández-Sanlés, A.; Elosua, R. DNA methylation and gene expression integration in cardiovascular disease. Clin. Epigenetics 2021, 13, 75. [Google Scholar] [CrossRef] [PubMed]
- Luo, X.; Hu, Y.; Shen, J.; Liu, X.; Wang, T.; Li, L.; Li, J. Integrative analysis of DNA methylation and gene expression reveals key molecular signatures in acute myocardial infarction. Clin. Epigenetics 2022, 14, 46. [Google Scholar] [CrossRef] [PubMed]
- Xiao, M.; Yang, H.; Xu, W.; Ma, S.; Lin, H.; Zhu, H.; Liu, L.; Liu, Y.; Yang, C.; Xu, Y.; et al. Inhibition of alpha-KG-dependent histone and DNA demethylases by fu, arate and succinate thata are accumulated in mutations of FH and SDH tumor suppressors. Genes Dev. 2012, 26, 1326–1338. [Google Scholar] [CrossRef] [PubMed]
- Karimi, M.; Vedin, J.; Levi, Y.; Basun, H.; Irving, G.; Eriksdotter, M.; Wahlund, L.O.; Schultzberg, M.; Hjorth, E.; Cederholm, T.; et al. DNA-rich n-3 fatty acid supplementation decreases DNA methylation in blood leukocytes: The omegAD study. Am. J. Clin. Nutr. 2017, 106, 1157–1165. [Google Scholar] [CrossRef]
- Donogran, R.; Wang, K.-H.; Lin, T.-J.; Yuan, T.-C.; Liu, C.-H. Anti-proliferative effects of statins is mediated by DNMT1 inhibition and p21 expression in OSCC cells. Cancers 2020, 12, 2084. [Google Scholar] [CrossRef]
- Kodach, L.; Jacobs, R.; Voorneveld, P.; Wildenberg, M.; Verspaget, H.; van Wezel, T.; Morreau, H.; Hommes, D.W.; Peppelenbosch, M.P.; van den Brink, G.R.; et al. Statins augment the chemosensitivity of colorectal cancer cells inducing epigenetic reprogramming and reducing colorectal cancer cell stemness via the bone morphogenetic protein pathway. Gut 2011, 60, 1544–1553. [Google Scholar] [CrossRef]
- Beltowski, J.; Wójcicka, G.; Jamroz-Wisniewska, A. Adverse effects of statins-mechanisms and consequences. Curr. Drug Saf. 2009, 4, 209–228. [Google Scholar] [CrossRef]
- Marais, A.D. Apolipoprotein E in lipoprotein metabolism, health and cardiovascular disease. Pathology 2019, 51, 165–176. [Google Scholar] [CrossRef]
- Cerda, A.; Genvigir, F.; Willrich, M.; Arazi, S.; Bernik, M.; Dorea, E.L.; Bertolami, M.C.; Faludi, A.A.; Hirata, M.H.; Hirata, R.D. Apolipoprotein E mRNA expression in monocclear cells from normolipidemic and hypercholesterolemic individuals treated with atorvastatin. Lipids Heakth Dis. 2011, 10, 206. [Google Scholar] [CrossRef]
- Troncoso, M.; Ortiz-Quintero, J.; Garrido-Moreno, V.; Sanhueza-Olivares, F.; Guerrero-Moncayo, A.; Sanhueza-Olivares, F.; Guerrero-Moncayo, A.; Chiong, M.; Castro, P.; García, L.; et al. VCAM-1as a predictor biomraker in cardiovascular disease. Biochim. Biophys. Acta Mol. Basis Dis. 2021, 1867, 166170. [Google Scholar] [CrossRef]
- Yin, L.; Bai, J.; Yu, W.-J.; Liu, Y.; Li, H.-H.; Lin, Q.-Y. Blocking VCAM-1 prevents Angiotensin II-induced hypertension and vascular remodeling in mice. Front. Pharm. 2022, 13, 2022. [Google Scholar] [CrossRef] [PubMed]
- Distasio, N.; Salmon, H.; Dierick, F.; Ebrahimian, T.; Tabrizian, M.; Lehoux, S. VCAM-1 targeted gene delivery nanoparticles localize to inflamed endothelial cells and atherosclerotic plaques. Adv. Ther. 2020, 4, 20000196. [Google Scholar] [CrossRef]
- Kheirolomoom, A.; Kim, C.; Seo, J.; Kumar, S.; Son, D.; Gagnon, M.K.; Ingham, E.; Ferrara, K.; Jo, H. Multifunctional nanoparticles facilitate molecular targeting and miRNA delivery to inhibit athesrosclerosis in ApoE-/- mice. ACS Nano 2015, 9, 8885–8897. [Google Scholar] [CrossRef] [PubMed]
- Morgan, E.S.; Tami, Y.; Hu, K.; Brambatti, M.; Mullick, A.E.; Geary, R.S.; Bakris, G.L.; Tsimikas, S. Antisense inhibition of Angiotensinogen with IONIS-AGT-LRX: Results of phase 1 and phase 2 studies. J. Am. Coll. Cardiol. Basic Trans. Sci. 2021, 6, 485–496. [Google Scholar]
- Gums, J.G. Use of ACW inhibitors in the treatment of cardiovascular disease. Am. Pharm. 1992, NS32, 62.70. [Google Scholar]
- Huang, C.W.; Chien, Y.S.; Chen, Y.J.; Ajuwon, K.M.; Mersmann, H.M.; Ding, S.T. Role of n-3 polyunsaturated fatty acids in ameliorating the obesity-induced metabolic syndrome in animal models and humans. Int. J. Mol. Sci. 2016, 17, 1689. [Google Scholar] [CrossRef]
- Brown, A.L.; Zhu, X.; Rong, S.; Shewale, S.; Seo, J.; Boudyguina, E.; Gebre, A.K.; Alexander-Miller, M.A.; Parks, J.S. Omega-3 fatty acids ameliorate atherosclerosis by favorably altering monocyte subsets and limiting monocyte recruitment to aortic lesions. Arter. Thromb. Vasc. Biol. 2012, 32, 2122–2130. [Google Scholar] [CrossRef]



| Week 1 | Week 2 | Week 3 | Week 4 | |
|---|---|---|---|---|
| Group A | Normal diet | Normal diet | Normal diet | Normal diet |
| Group B | HFD | HFD | HFD | HFD |
| Group C | HFD | HFD | HFD | HFD + RCI-1502 |
| GENE | ID |
|---|---|
| VCAM | Mm01320970_m1 |
| ACE | Mm00802048_m1 |
| AGT | Mm005996620_m1 |
| ABCB7 | Mm01235358_m1 |
| DNMT1 | Mm151854_m1 |
| Protein | Function | Reference |
|---|---|---|
| Myosin | Powers heart muscle contraction and increases cardiac function. | [28] |
| Tropomyosin | Plays a crucial role in cardiac muscle activation; mutations in this protein are associated with cardiomyopathy. | [29] |
| ATP synthase | Is the primary generator of cellular ATP and is a key regulator of mitochondrial function; ATP synthase dysfunction causes cardiomyopathy and congestive heart failure. | [30] |
| Triosephosphate isomerase | Deficiency in this glycolytic enzyme causes hemolitic anemia. | [31] |
| Alpha-enolase | Decreases in the aging heart and may be involved in the cardiomyopathy of aging; improves hypoxia-impaired cardiomyocyte contractility. | [32] |
| Heat shock cognate 71 KDa | Is a biomarker for poor neurological outcomes in survivors of cardiac arrest. | [33] |
| Sarcoplasmin/endoplasmic reticulum calcium ATPase | Has decreased expression in congestive heart failure. | [34] |
| Nucleoside diphosphate kinase B | Deficiency in this protein causes diabetes-like vascular pathology. | [35] |
| Calmodulin | Calmodulin dysfunction impairs critical cardiac calcium signaling processes. | [36] |
| Cytochrome C | Myocardial ischemia is related to reduced cytochrome C content. | [37] |
| Glycerol-3-phosphate dehydrogenase 1-like protein | Regulates cardiac sodium current; a novel mutation in the encoding gene is associated with early repolarization syndrome. | [38] |
| Acetyl-CoA acetyltransferase | Promotes cardiac repair after myocardial infarction via histone acetylation. | [39] |
| Fumarate hydrolase | Cardioprotective through the Nrf2 antioxidant pathway. | [40] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Martínez-Iglesias, O.; Naidoo, V.; Corzo, L.; Carrera, I.; Seoane, S.; Rodríguez, S.; Alcaraz, M.; Muñiz, A.; Cacabelos, N.; Cacabelos, R. Proteomic and Global DNA Methylation Modulation in Lipid Metabolism Disorders with a Marine-Derived Bioproduct. Biology 2023, 12, 806. https://doi.org/10.3390/biology12060806
Martínez-Iglesias O, Naidoo V, Corzo L, Carrera I, Seoane S, Rodríguez S, Alcaraz M, Muñiz A, Cacabelos N, Cacabelos R. Proteomic and Global DNA Methylation Modulation in Lipid Metabolism Disorders with a Marine-Derived Bioproduct. Biology. 2023; 12(6):806. https://doi.org/10.3390/biology12060806
Chicago/Turabian StyleMartínez-Iglesias, Olaia, Vinogran Naidoo, Lola Corzo, Iván Carrera, Silvia Seoane, Susana Rodríguez, Margarita Alcaraz, Adriana Muñiz, Natalia Cacabelos, and Ramón Cacabelos. 2023. "Proteomic and Global DNA Methylation Modulation in Lipid Metabolism Disorders with a Marine-Derived Bioproduct" Biology 12, no. 6: 806. https://doi.org/10.3390/biology12060806
APA StyleMartínez-Iglesias, O., Naidoo, V., Corzo, L., Carrera, I., Seoane, S., Rodríguez, S., Alcaraz, M., Muñiz, A., Cacabelos, N., & Cacabelos, R. (2023). Proteomic and Global DNA Methylation Modulation in Lipid Metabolism Disorders with a Marine-Derived Bioproduct. Biology, 12(6), 806. https://doi.org/10.3390/biology12060806