Screening of Comprehensive Panel of Cultivated and Wild Vigna Species for Resistance to Pulse Beetle, Callosobruchus chinensis L.
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material and Insect Culturing
2.2. Screening of Test Genotypes
2.2.1. Percent Adult Emergence
2.2.2. Mean Development Period
2.2.3. Growth Index
2.2.4. Seed Weight Loss (%)
2.2.5. Grouping of Test Genotypes for Pulse Beetle Reactions
2.3. Basal Expression Profiling Antioxidant
2.4. Molecular Genotyping
2.5. Statistical Analyses
3. Results
3.1. Screening of Vigna Genotypes
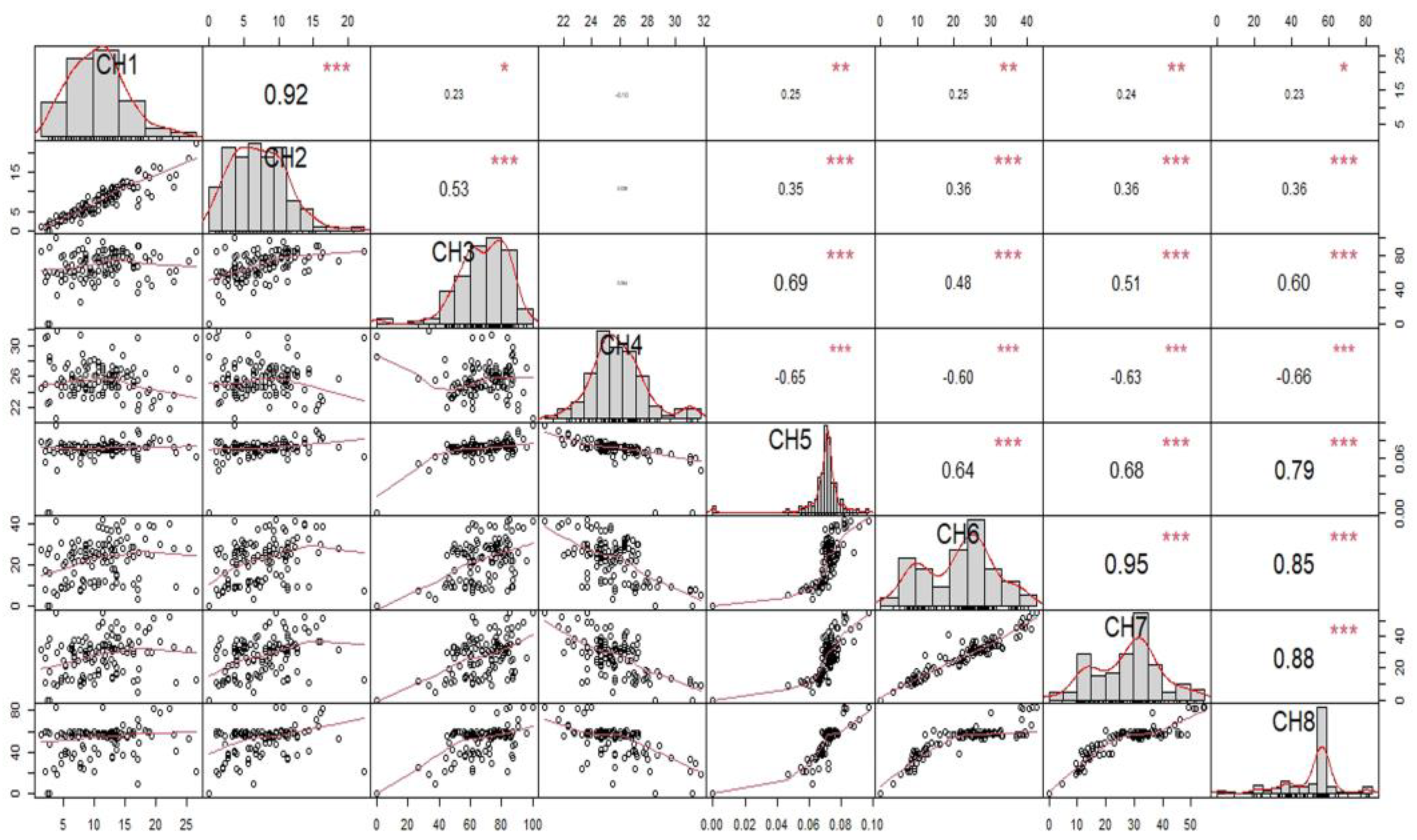
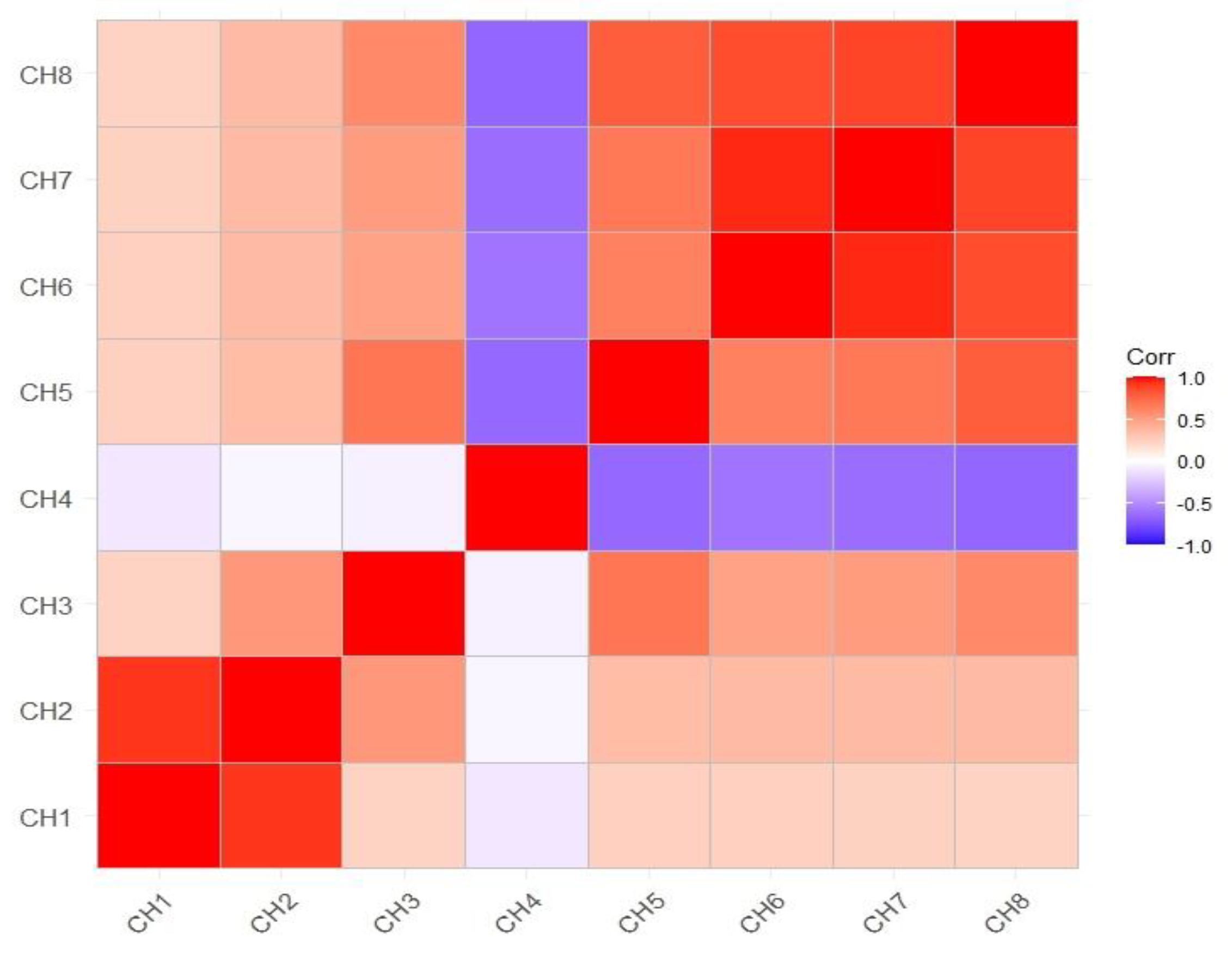
3.2. Correlation of Growth Index with Various Parameters
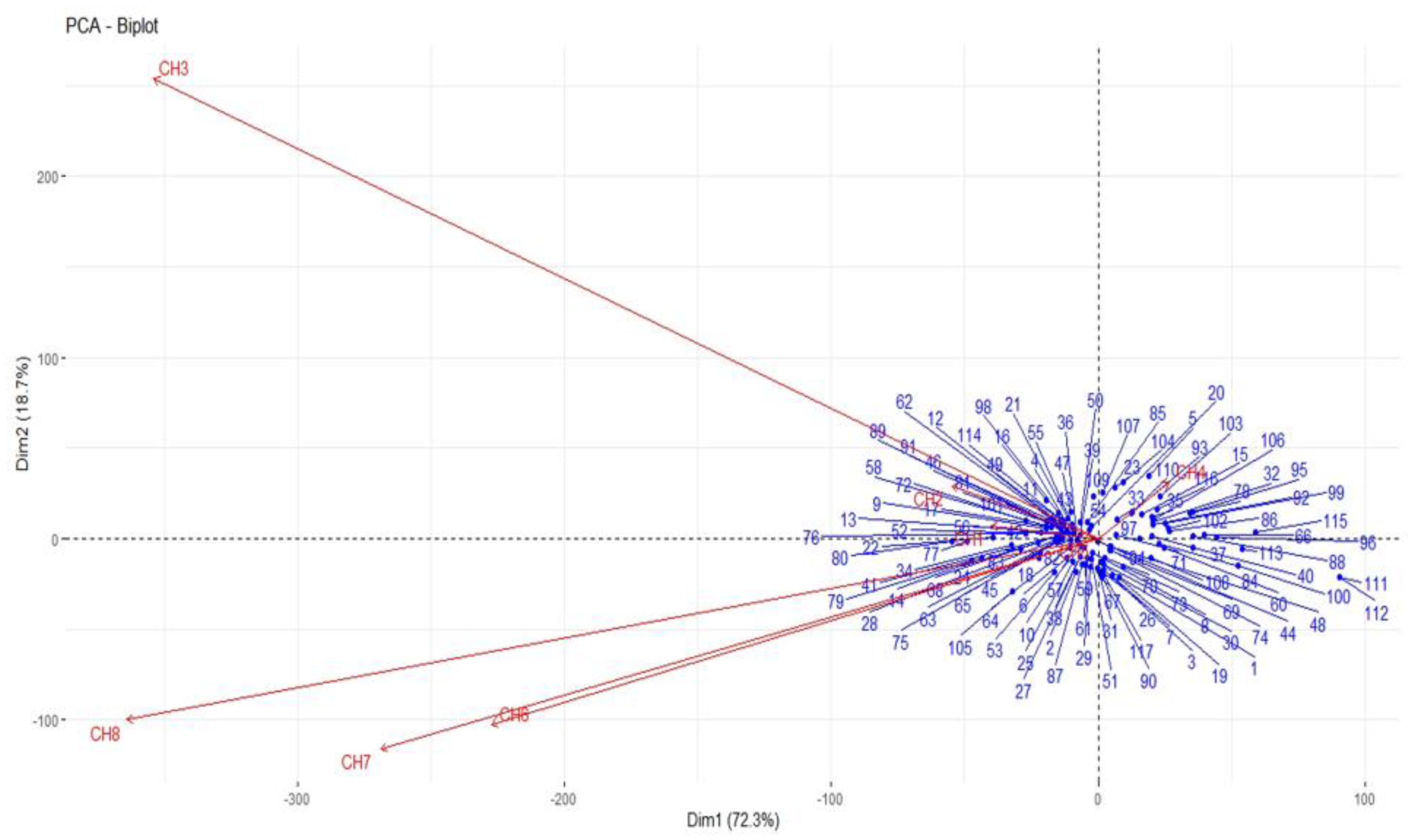
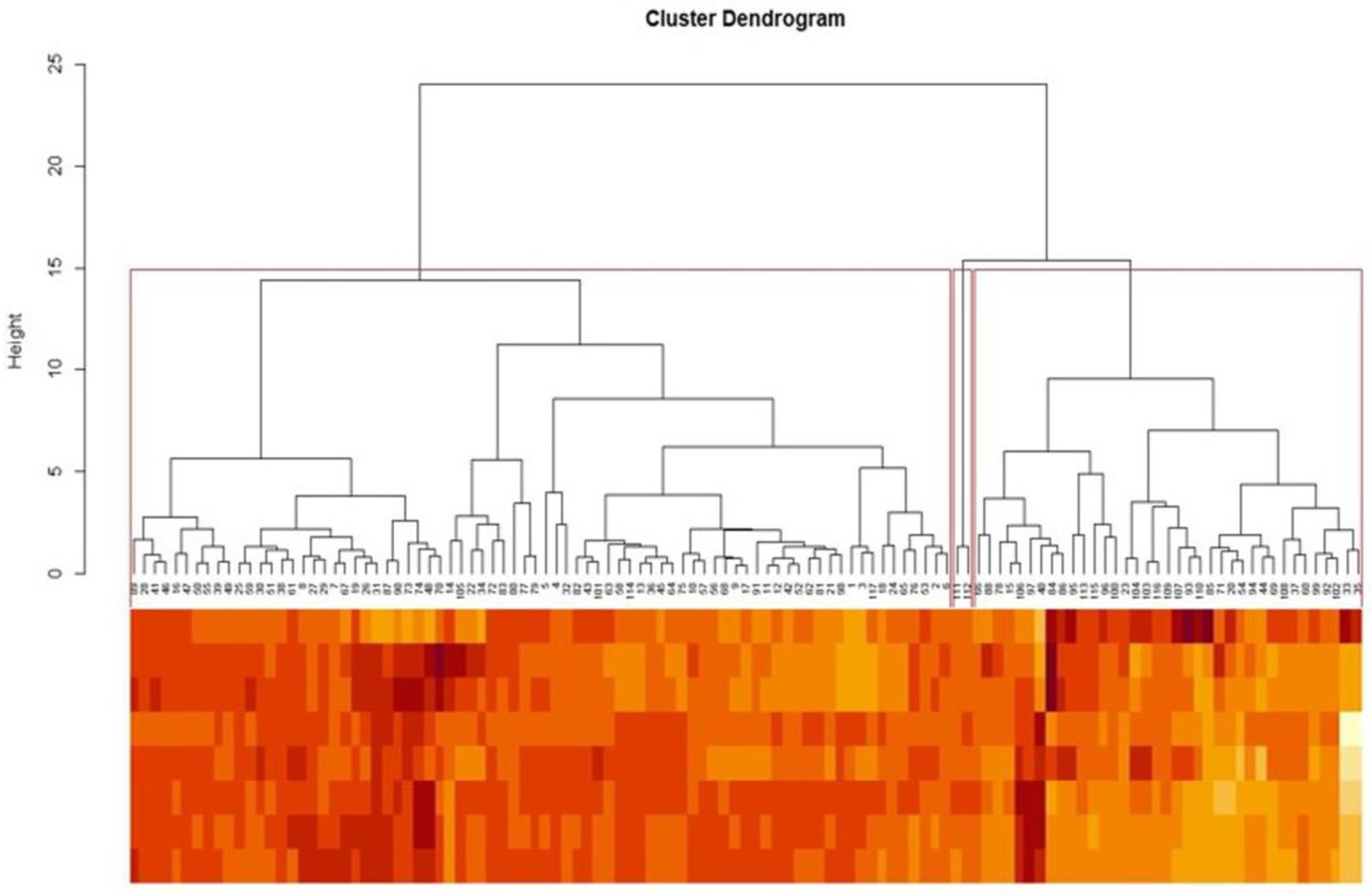
3.3. Principal Component (PC) and Cluster Analyses
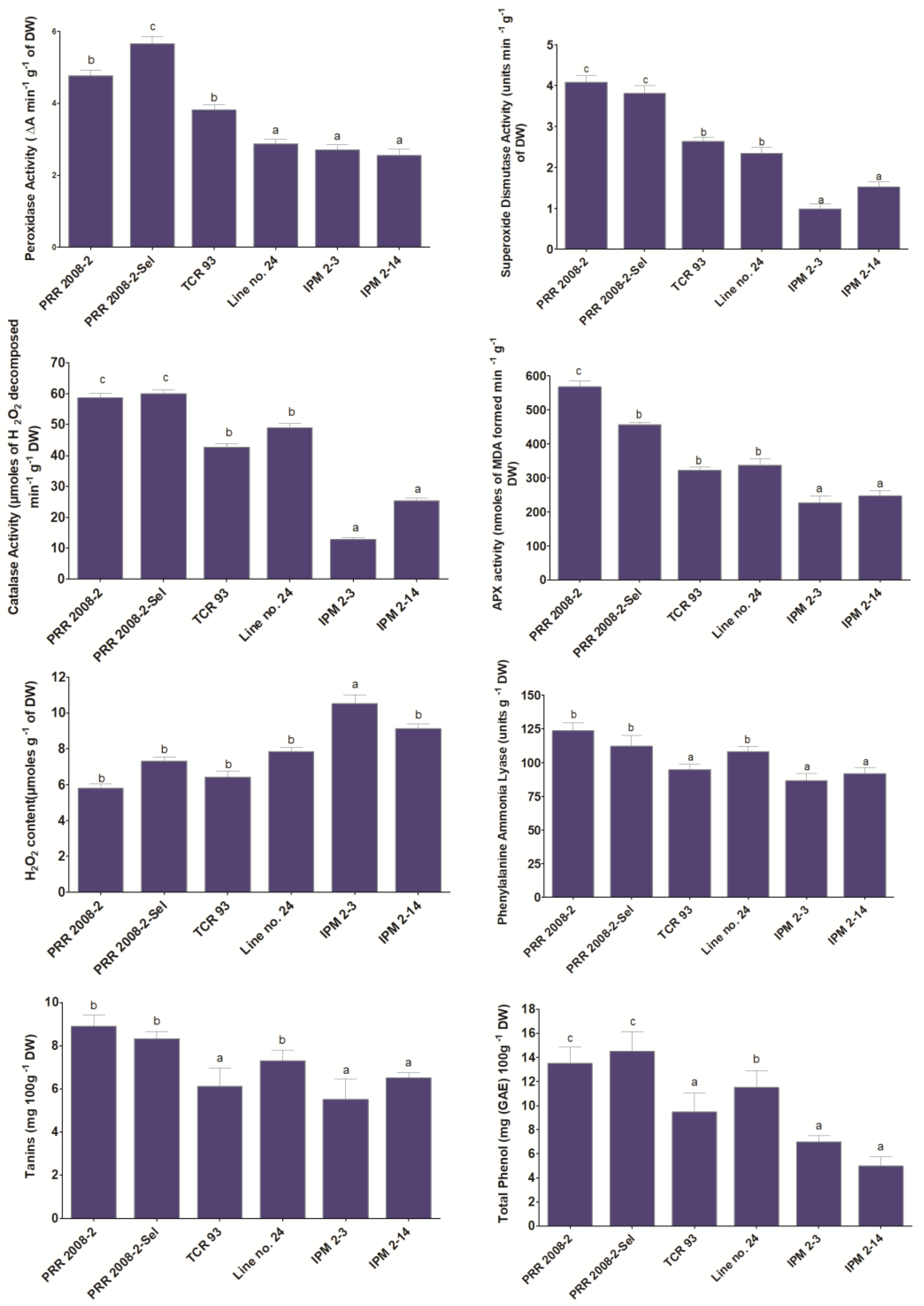
3.4. Antioxidant Enzymatic Analysis
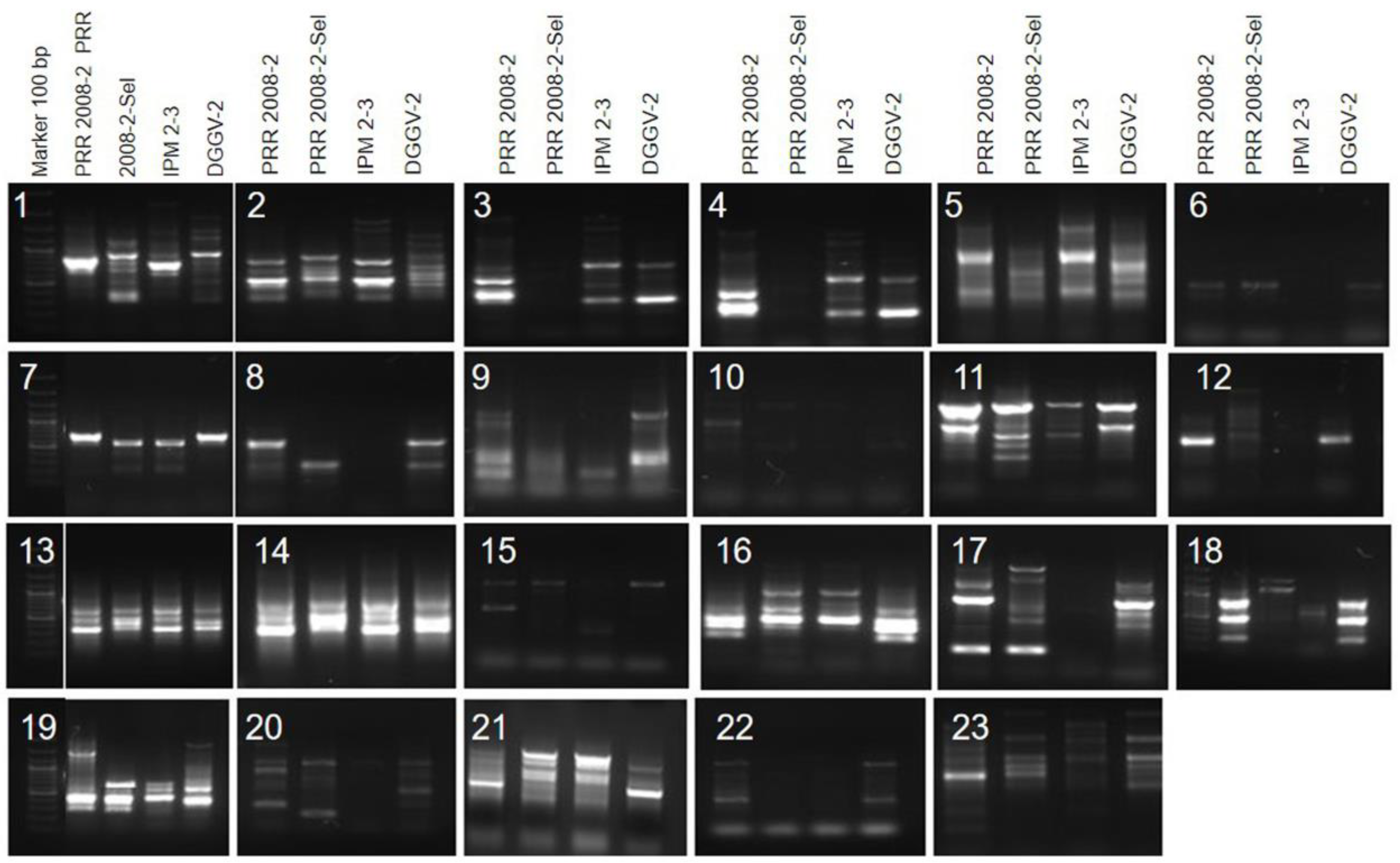
3.5. SCoT-Based Polymorphism in Selected Genotypes
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Singh, C.M.; Pratap, A.; Kumar, H.; Singh, S.; Singh, B.K.; Prasad, D.; Dhaliwal, I.; Kumar, M. Recent advances in omics approaches for mungbean improvement. In Technologies in Plant Biotechnology and Breeding of Field Crops; Springer: Berlin/Heidelberg, Germany, 2022; pp. 181–200. [Google Scholar] [CrossRef]
- Foyer, C.H.; Lam, H.-M.; Nguyen, H.T.; Siddique, K.H.; Varshney, R.K.; Colmer, T.D.; Cowling, W.; Bramley, H.; Mori, T.A.; Hodgson, J.M. Neglecting legumes has compromised human health and sustainable food production. Nat. Plants 2016, 2, 16112. [Google Scholar] [CrossRef] [PubMed]
- Pratap, A.; Dhaliwal, I.; Singh, C.M.; Mahalingam, A.; Manivannan, N.; Basavaraja, T.; Rathore, M.; Singh, Y.; Tiwari, P.; Yadav, S.; et al. Biofortification of Mungbean. In Biofortification of Staple Crops; Kumar, S., Dikshit, H.K., Mishra, G.P., Singh, A., Eds.; Springer: Singapore, 2022; pp. 295–333. [Google Scholar] [CrossRef]
- Allito, B.B.; Ewusi-Mensah, N.; Alemneh, A.A. Rhizobia strain and host-legume interaction effects on nitrogen fixation and yield of grain legume: A review. Mol. Soil Biol. 2014, 2, 1–12. [Google Scholar] [CrossRef]
- Sehrawat, N.; Yadav, M.; Bhat, K.V.; Sairam, R.K.; Jaiwal, P.K. Effect of salinity stress on mungbean [Vigna radiata (L.) Wilczek] during consecutive summer and spring seasons. J. Agric. Sci. Belgrade 2015, 60, 23–32. [Google Scholar] [CrossRef]
- Kumar, C.; Mishra, S.; Singh, C.M. Evaluation of selection indices for improving terminal heat tolerance in greengram (Vignaradiata L. Wilczek). J. Agrometeorol. 2016, 18, 216–221. [Google Scholar] [CrossRef]
- Kumar, C.; Mishra, S.; Singh, C.M. Investigating morpho-physiological traits and agro-meteorological indices in green gram [Vigna radiata (L.) Wilczek] for terminal heat tolerance. Legume Res. 2017, 40, 271–276. [Google Scholar] [CrossRef]
- Kumar, R.; Singh, C.M.; Arya, M.; Kumar, R.; Mishra, S.; Singh, U.; Paswan, S. Investigating stress indices to discriminate the physiologically efficient heat tolerant genotypes of mungbean [Vigna radiata (L.) Wilczek]. Legume Res. 2020, 43, 43–49. [Google Scholar]
- Singh, C.M.; Singh, P.; Tiwari, C.; Purwar, S.; Kumar, M.; Pratap, A.; Singh, S.; Chugh, V.; Mishra, A.K. Improving drought tolerance in mungbean (Vigna radiata L. Wilczek): Morpho-physiological, biochemical and molecular perspectives. Agronomy 2021, 11, 1534. [Google Scholar] [CrossRef]
- Singh, C.M.; Mishra, S.B.; Pandey, A.; Arya, M. Eberhart-REussell’ and AMMI approches of genotype by environment interaction (GEI) for yield and yield component traits in Vigna radiata L. Wilczek. Int. J. Agric. Environ. Biotechnol. 2014, 7, 277–292. [Google Scholar] [CrossRef]
- Singh, C.M.; Kumar, R.; Mishra, S.; Pandey, A.; Arya, M. Characterization of mungbean genotypes against Mungbean Yellow Mosaic Virus and Cercospora leaf spot diseases under north east plain zone. Int. J. Agric. Environ. Biotechnol. 2015, 8, 119–125. [Google Scholar] [CrossRef]
- Singh, C.M.; Singh, P.; Pratap, A.; Pandey, R.; Purwar, S.; Douglas, C.A.; Baek, K.-H.; Mishra, A.K. Breeding for enhancing Legumovirus resistance in mungbean: Current understanding and future directions. Agronomy 2019, 9, 622. [Google Scholar] [CrossRef]
- Singh, C.M.; Pratap, A.; Gupta, S.; Biradar, R.S.; Singh, N.P. Association mapping for mungbean yellow mosaic India virus resistance in mungbean (Vigna radiata L. Wilczek). 3 Biotech 2020, 10, 33. [Google Scholar] [CrossRef] [PubMed]
- Nair, R.; Schreinemachers, P. Global status and economic importance of mungbean. In The Mungbean Genome; Springer: Berlin/Heidelberg, Germany, 2020; pp. 1–8. [Google Scholar]
- Mishra, G.P.; Dikshit, H.K.; Sv, R.; Tripathi, K.; Kumar, R.R.; Aski, M.; Singh, A.; Roy, A.; Kumari, N.; Dasgupta, U. Yellow mosaic disease (YMD) of mungbean (Vigna radiata (L.) Wilczek): Current status and management opportunities. Front. Plant Sci. 2020, 11, 918. [Google Scholar] [CrossRef] [PubMed]
- Duraimurugan, P.; Raja, K.; Regupathy, A. An eco-friendly approach for management of pulse beetle, Callosobruchus maculatus through neem formulations assisted with pitfall trap. J. Food Legumes 2011, 24, 23–27. [Google Scholar]
- Southgate, B. Biology of the Bruchidae. Annu. Rev. Entomol. 1979, 24, 449–473. [Google Scholar] [CrossRef]
- Talekar, N.S. Biology, damage and control of bruchid pests of mungbean. In Mungbean, Proceeding of the Second International Symposium, Bangkok, Thailand, 16–20 November 1987; Shanmugasundaram, S., McLean, B.T., Eds.; Asian Vegetable Research and Development Center: Taiwan, 1988; pp. 329–342. [Google Scholar]
- Akinkurolere, R.O.; Adedire, C.O.; Odeyemi, O.O. Laboratory evaluation of the toxic properties of forest anchomanes, Anchomanes difformis against pulse beetle Callosobruchus maculatus (Coleoptera: Bruchidae). Insect Sci. 2006, 13, 25–29. [Google Scholar] [CrossRef]
- Somta, C.; Somta, P.; Tomooka, N.; Ooi, P.-C.; Vaughan, D.; Srinives, P. Characterization of new sources of mungbean (Vigna radiata (L.) Wilczek) resistance to bruchids, Callosobruchus spp. (Coleoptera: Bruchidae). J. Stored Prod. Res. 2008, 44, 316–321. [Google Scholar] [CrossRef]
- Swella, G.; Mushobozy, D. Comparative susceptibility of different legume seeds to infestation by cowpea bruchid Callosobruchus maculatus (F.) (Coleoptera: Chrysomelidae). Plant Prot. Sci. 2009, 45, 19–24. [Google Scholar] [CrossRef]
- Lattanzio, V.; Terzano, R.; Cicco, N.; Cardinali, A.; Venere, D.D.; Linsalata, V. Seed coat tannins and bruchid resistance in stored cowpea seeds. J. Sci. Food Agric. 2005, 85, 839–846. [Google Scholar] [CrossRef]
- Mohamed, H.I.; Abd-El Hameed, A.G. Molecular and biochemical markers of some Vicia faba L. genotypes in response to storage insect pests infestation. J. Plant Interact. 2014, 9, 618–626. [Google Scholar] [CrossRef]
- Lattanzio, V.; Cardinali, A.; Linsalata, V. Plant phenolics: A biochemical and physiological perspective. Recent Adv. Polyphen. Res. 2012, 3, 1–39. [Google Scholar]
- Adenekan, M.; Okpeze, V.; Ogundipe, W.; Oguntade, M. Evaluation of Moringa oleifera powders for the control of bruchid beetles during storage. Int. J. Agric. Policy Res. 2013, 10, 305–310. [Google Scholar]
- Ahad, M.; Nahar, M.; Amin, M.; Suh, S.; Kwon, Y. Effect of weed extracts against pulse beetle, Callosobruchus chinensis L. (Coleoptera: Bruchidae) of mung bean. Bangladesh J. Agric. Res. 2016, 41, 75–84. [Google Scholar] [CrossRef]
- Soumia, P.; Srivastava, C.; Dikshit, H.; Guru Pirasanna Pandi, G. Screening for resistance against pulse beetle, Callosobruchus analis (F.) in greengram (Vigna radiata (L.) Wilczek) accessions. Proc. Natl. Acad. Sci. India Sect. B Biol. Sci. 2017, 87, 551–558. [Google Scholar] [CrossRef]
- Pratap, A.; Basu, P.S.; Gupta, S.; Malviya, N.; Rajan, N.; Tomar, R.; Madhavan, L.; Nadarajan, N.; Singh, N.P. Identification and characterization of sources for photo-and thermo-insensitivity in Vigna species. Plant Breed. 2014, 133, 756–764. [Google Scholar] [CrossRef]
- Eker, T.; Erler, F.; Adak, A.; Imrek, B.; Guven, H.; Tosun, H.S.; Sari, D.; Sari, H.; Upadhyaya, H.D.; Toker, C. Screening of chickpea accessions for resistance against the pulse beetle, Callosobruchus chinensis L. (Coleoptera: Bruchidae). J. Stored Prod. Res. 2018, 76, 51–57. [Google Scholar] [CrossRef]
- Esen, A.; Sari, H.; Erler, F.; Adak, A.; Sari, D.; Eker, T.; Canci, H.; Ikten, C.; Kahraman, A.; Toker, C. Screening and selection of accessions in the genus Pisum L. for resistance to pulse beetle (Callosobruchus chinensis L.). Euphytica 2019, 215, 82. [Google Scholar] [CrossRef]
- Pandiyan, M.; Krishnaveni, A.; Sivakumar, C.; Vaityalingan, M.; Sivakumar, P.; Jamuna, E.; Radhakrishnan, V.; Senthilkumar, P. Development of bruchid resistant genotypes in mungbean through introgrossion of wild genotypes. Int. J Chem. Stud. 2020, 8, 62–66. [Google Scholar] [CrossRef]
- Sari, H.; Sari, D.; Eker, T.; Aydinoglu, B.; Canci, H.; Ikten, C.; Gokturk, R.S.; Zeybek, A.; Bakir, M.; Smykal, P. Inheritance and Expressivity of Neoplasm Trait in Crosses between the Domestic Pea (Pisum sativum subsp. sativum) and Tall Wild Pea (Pisum sativum subsp. elatius). Agronomy 2020, 10, 1869. [Google Scholar] [CrossRef]
- Pratap, A.; Das, A.; Kumar, S.; Gupta, S. Current perspectives on introgression breeding in food legumes. Front. Plant Sci. 2021, 11, 589189. [Google Scholar] [CrossRef]
- Toker, C.; Berger, J.; Eker, T.; Sari, D.; Sari, H.; Gokturk, R.S.; Kahraman, A.; Aydin, B.; von Wettberg, E.J. Cicer turcicum: A new Cicer species and its potential to improve chickpea. Front. Plant Sci. 2021, 12, 662891. [Google Scholar] [CrossRef]
- Eker, T.; Erler, F.; Sari, H.; Sari, D.; Berger, J.; Toker, C. Deployment of Cicer echinospermum PH Davis for resistance to Callosobruchus chinensis L. J. Plant Dis. Prot. 2022, 129, 843–851. [Google Scholar] [CrossRef]
- Singh, C.M.; Kumar, M.; Pratap, A.; Tripathi, A.; Singh, S.; Mishra, A.; Kumar, H.; Nair, R.M.; Singh, N.P. Genome-wide analysis of late embryogenesis abundant protein gene family in vigna species and expression of VrLEA encoding genes in vigna glabrescens reveal its role in heat tolerance. Front. Plant Sci. 2022, 13, 843107. [Google Scholar] [CrossRef] [PubMed]
- Singh, P.; Pandey, B.; Pratap, A.; Gyaneshwari, U.; Nair, R.M.; Mishra, A.K.; Singh, C.M. Genetic and Genomics Resources of Cross-Species Vigna Gene Pools for Improving Biotic Stress Resistance in Mungbean (Vigna radiata L. Wilczek). Agronomy 2022, 12, 3000. [Google Scholar] [CrossRef]
- Kang, Y.J.; Kim, S.K.; Kim, M.Y.; Lestari, P.; Kim, K.H.; Ha, B.-K.; Jun, T.H.; Hwang, W.J.; Lee, T.; Lee, J. Genome sequence of mungbean and insights into evolution within Vigna species. Nat. Commun. 2014, 5, 5443. [Google Scholar] [CrossRef]
- Singh, C.M.; Prajapati, U.; Gupta, S.; Pratap, A. Microsatellite-based association mapping for agronomic traits in mungbean (Vigna radiata L. Wilczek). J. Genet. 2021, 100, 87. [Google Scholar] [CrossRef]
- Kumari, G.; Roopa Lavanya, G.; Shanmugavadivel, P.; Singh, Y.; Singh, P.; Patidar, B.; Madhavan, L.; Gupta, S.; Singh, N.; Pratap, A. Genetic diversity and population genetic structure analysis of an extensive collection of wild and cultivated Vigna accessions. Mol. Genet. Genom. 2021, 296, 1337–1353. [Google Scholar] [CrossRef]
- Pratap, A.; Singh, C.M.; Gupta, S.; Gupta, A.K.; Birader, R.S.; Prajapati, U.; Tomar, R.; Singh, N.P. Genetic enhancement in mungbean (Vigna radiata) as revealed by genome-wide mapped microsatellite markers. Agric. Res. 2021, 10, 369–377. [Google Scholar] [CrossRef]
- Butani, P.; Motka, M.; Kapadia, M. Storage pests and their management. In Bulletin; Department of Agricultural Entomology, College of Agriculture, Gujarat Agricultural University: Junagadh, India, 2001; pp. 25–27. [Google Scholar]
- Ghosh, S.; Roy, A.; Kundagrami, S. Screening of Mungbean [Vigna radiata (L.) Wilczek] Genotypes against Bruchid (Callosobruchus maculatus) Attack to Reduce Postharvest Losses. Legume Res. Int. J. 2022, 45, 1019–1027. [Google Scholar] [CrossRef]
- Howe, R. A parameter for experssing the suitability of an environment for insect development. J. Stored Prod. Res. 1971, 7, 63–65. [Google Scholar] [CrossRef]
- Sulehrie, M.; Golob, P.; Tran, B.; Farrell, G. The effect of attributes of Vigna spp. on the bionomics of Callosobruchus maculatus. Entomol. Exp. Et Appl. 2003, 106, 159–168. [Google Scholar] [CrossRef]
- Shannon, L.M.; Kay, E.; Lew, J.Y. Peroxidase isozymes from horseradish roots: I. Isolation and physical properties. J. Biol. Chem. 1966, 241, 2166–2172. [Google Scholar] [CrossRef]
- Marklund, S.; Marklund, G. Involvement of the superoxide anion radical in the autoxidation of pyrogallol and a convenient assay for superoxide dismutase. Eur. J. Biochem. 1974, 47, 469–474. [Google Scholar] [CrossRef] [PubMed]
- Chance, B.; Maehly, A. Assay of catalases and peroxidases. Methods Enzimol. 1955, 2, 764–775. [Google Scholar]
- Nakano, Y.; Asada, K. Purification of ascorbate peroxidase in spinach chloroplasts; its inactivation in ascorbate-depleted medium and reactivation by monodehydroascorbate radical. Plant Cell Physiol. 1987, 28, 131–140. [Google Scholar]
- Biehn, W.L.; Kuc, J.; Williams, E.B. Accumulation of phenols in resistant plant-fungi interaction. Phytopathology 1968, 58, 1255–1260. [Google Scholar]
- Shafique, M.; Ahmad, M. Screening of pulse grains for resistance to Callosobruchus analis (F.) (Coleoptera: Bruchidae). Pak. J. Zool. 2002, 34, 293–296. [Google Scholar]
- Shivanna, B.; Ramamurthy, B.; Gangadhara, N.; Gayathri, D.; Mallikarjunaiah, H.; Krishna, N. Varietal screening of cowpea against pulse beetles, Callosobruchus maculatus (Fab.) and C. analis (Fab.). Int. J. Sci. Nat. 2011, 2, 245–247. [Google Scholar]
- Badii, K.; Asante, S.; Sowley, E. Varietal susceptibility of cowpea (Vigna unguiculata L.) to the storage beetle, Callosobruchus maculatus F. (Coleoptera: Bruchidae). Int. J. Sci. Technol. Res. 2013, 2, 82–89. [Google Scholar]
- Soumia, P.; Srivastava, C.; Pandi, G.; Subramanian, S. Varietal preference of pulse beetle, Callosobruchus maculatus (F.) in greengram. Indian J. Entomol. 2017, 79, 86–91. [Google Scholar] [CrossRef]
- Jackai, L.; Asante, S. A case for the standardization of protocols used in screening cowpea, Vigna unguiculata for resistance to Callosobruchus maculatus (Fabricius) (Coleoptera: Bruchidae). J. Stored Prod. Res. 2003, 39, 251–263. [Google Scholar] [CrossRef]
- Pawara, N.; Bantewad, S.; Patil, D. Assessment of different interspecific progenies of mungbean against pulse beetle, Callosobruchus chinensis Linn. and it’s influence of seed physical characteristics on infestation. J. Entomol. Zool. Stud. 2019, 7, 1335–1344. [Google Scholar]
- Aidbhavi, R.; Pratap, A.; Verma, P.; Lamichaney, A.; Bandi, S.M.; Nitesh, S.; Akram, M.; Rathore, M.; Singh, B.; Singh, N.P. Screening of endemic wild Vigna accessions for resistance to three bruchid species. J. Stored Prod. Res. 2021, 93, 101864. [Google Scholar] [CrossRef]
- Chavan, P.; Yeshbir, S.; Singh, S. Ovipositional preference of Callosobruchus chinensis for cowpea lines. Indian J. Entomol. 1997, 59, 295–303. [Google Scholar]
- Wang, M.H.; Horng, S.B. Egg dumping and life history strategy of Callosobruchus maculatus. Physiol. Entomol. 2004, 29, 26–31. [Google Scholar] [CrossRef]
- Shafique, M.; Ahmad, M. Chickpea grains resistance to pulse beetle, Callosobruchus analis (F.) (Coleoptera: Bruchidae). Pak. J. Zool. 2005, 37, 123. [Google Scholar]
- Chakraborty, S.; Chaudhuri, N.; Senapati, S. Correlation Between Seed Parameters and Relative Susceptibility of Mung Bean Genotypes (Vigna radiata L.) to Callosobruchus chinensis L. During Storage. Ann. Plant Prot. Sci. 2004, 12, 48–50. [Google Scholar]
- Jha, A.; Chitra, S.; Naresh, C. Screening of greengram (Vigna radiata) cultivars to three species of pulse beetle (Callosobruchus spp.). Indian J. Agric. Sci. 2011, 81, 283–286. [Google Scholar]
- Rawat, S.; Srivastava, M. Evaluation of qualitative and quantitative losses caused by Callosobruchus chinensis to some pulses. J. Entomol. Res. 2011, 35, 117–120. [Google Scholar]
- Usha, R.; Singh, P.; Singh, S.; Saxena, R. Screening of green gram genotypes against Callosobruchus maculatus (F.) under laboratory conditions. Ann. Plant Prot. Sci. 2018, 26, 227–228. [Google Scholar]
- Lambrides, C.; Imrie, B.C. Susceptibility of mungbean varieties to the bruchid species Callosobruchus maculatus (F.), C. phaseoli (Gyll.), C. chinensis (L.), and Acanthoscelides obtectus (Say.) (Coleoptera: Chrysomelidae). Aust. J. Agric. Res. 2000, 51, 85–90. [Google Scholar] [CrossRef]
- Kashiwaba, K.; Tomooka, N.; Kaga, A.; Han, O.-K.; Vaughan, D. Characterization of resistance to three bruchid species (Callosobruchus spp., Coleoptera, Bruchidae) in cultivated rice bean (Vigna umbellata). J. Econ. Entomol. 2003, 96, 207–213. [Google Scholar] [CrossRef] [PubMed]
- Tomooka, N.; Kashiwaba, K.; Vaughan, D.A.; Ishimoto, M.; Egawa, Y. The effectiveness of evaluating wild species: Searching for sources of resistance to bruchid beetles in the genus Vigna subgenus Ceratotropis. Euphytica 2000, 115, 27–41. [Google Scholar] [CrossRef]
- Sharma, H.; Singh, D.; Kumar, A.; Shrotria, P. Evaluation of F4 progenies emanated from an interspecific hybridization of mungbean and blackgram. Legume Res. 2013, 36, 191–199. [Google Scholar]
- Chen, T.; Hu, L.; Wang, S.; Wang, L.; Cheng, X.; Chen, H. Construction of High-Density Genetic Map and Identification of a Bruchid Resistance Locus in Mung Bean (Vigna radiata L.). Front. Genet. 2022, 13, 903267. [Google Scholar] [CrossRef]
- Chaisan, T.; Somta, P.; Srinives, P.; Chanprame, S.; Kaveeta, R.; Dumrongkittikule, S. Development of tetraploid plants from an interspecific hybrid between mungbean (Vigna radiata) and rice bean (Vigna umbellata). J. Crop Sci. Biotechnol. 2013, 16, 45–51. [Google Scholar] [CrossRef]
- Basavaraja, T.; Murthy, N.; Kumar, L.V.; Mallikarjun, K. Studies on cross compatibility in interspecific crosses of Vigna radiata × Vigna umbellata species. Legume Res. 2019, 42, 699–704. [Google Scholar] [CrossRef]
- Bhanu, A.N.; Singh, M.; Srivastava, K. Crossability studies of interspecific hybridization among Vigna species. Biomed. J. 2018. [Google Scholar] [CrossRef]
- Mariyammal, I.; Seram, D.; Samyuktha, S.M.; Karthikeyan, A.; Dhasarathan, M.; Murukarthick, J.; Kennedy, J.S.; Malarvizhi, D.; Yang, T.-J.; Pandiyan, M. QTL mapping in Vigna radiata × Vigna umbellata population uncovers major genomic regions associated with bruchid resistance. Mol. Breed. 2019, 39, 110. [Google Scholar] [CrossRef]
- Sharma, V.; Singh, C.M.; Chugh, V.; Prajapati, P.K.; Mishra, A.; Kaushik, P.; Dhanda, P.S.; Yadav, A. Morpho-Physiological and Biochemical Responses of Field Pea Genotypes under Terminal Heat Stress. Plants 2023, 12, 256. [Google Scholar] [CrossRef]
- Mallikarjun, A.B.; Sudhirkumar, S.; Shivaraju, C. Influence of biochemical constituents’ total phenol and tannin on incidence of bruchid (Callosobruchus theobromae L.) on Selected Fielbean Accessions. Int. J. Agric. Sci. 2011, 1, 377–380. [Google Scholar]
- Sofo, A.; Scopa, A.; Nuzzaci, M.; Vitti, A. Ascorbate peroxidase and catalase activities and their genetic regulation in plants subjected to drought and salinity stresses. Int. J. Mol. Sci. 2015, 16, 13561–13578. [Google Scholar] [CrossRef] [PubMed]
- Purwar, S.; Singh, C.M.; Kumar, M.; Singh, A.K.; Pratap, A.; Singh, P.; Gore, P.G.; Singh, N.P. Genome-wide identification and analysis of NBS-LRR-encoding genes in mungbean (Vigna radiata L. Wilczek) and their expression in two wild non-progenitors reveal their role in MYMIV resistance. J. Plant Growth Regul. 2023. [Google Scholar] [CrossRef]
- Pal, S.; Sandhu, J.; Singh, I. Exploitation of genetic variability in interspecific cross between Vigna Mungo X V. umbellata. Indian J. Pulses Res. 2005, 18, 9. [Google Scholar]
- Sehrawat, N.; Yadav, M.; Bhat, K.V.; Sairam, R.K.; Jaiwal, P.K. Introgression of mungbean yellow mosaic virus resistance in Vigna mungo (L.) Hepper and purity testing of F1 hybrids using SSRs. Turk. J. Agric. For. 2016, 40, 95–100. [Google Scholar] [CrossRef]
- Feng, S.; Zhu, Y.; Yu, C.; Jiao, K.; Jiang, M.; Lu, J.; Shen, C.; Ying, Q.; Wang, H. Development of species-specific SCAR markers, based on a SCoT analysis, to authenticate Physalis (Solanaceae) species. Front. Genet. 2018, 9, 192. [Google Scholar] [CrossRef]
- Liu, Z.; Sun, Q.; Ni, Z.; Yang, T.; McIntosh, R. Development of SCAR markers linked to the Pm21 gene conferring resistance to powdery mildew in common wheat. Plant Breed. 1999, 118, 215–219. [Google Scholar] [CrossRef]
- Dhole, V.J.; Reddy, K.S. Development of a SCAR marker linked with a MYMV resistance gene in mungbean (Vigna radiata L. W ilczek). Plant Breed. 2013, 132, 127–132. [Google Scholar] [CrossRef]


| GN | Genotypes | Species | Number of Eggs Laid | No. Adults Emerged | Adult Emergence (%) | Mean Development Period (days) | Growth Index | Seed Weight Loss (%) (30 DAIR) | Seed Weight Loss (%) (60 DAIR) | Seed Weight Loss (%) (90 DAIR) |
|---|---|---|---|---|---|---|---|---|---|---|
| G1 | PDM 139 | V. radiata | 19.33 ± 0.67 | 8.67 ± 0.33 | 45.00 ± 2.89 | 22.00 ± 0.58 | 0.075 ± 0.002 | 24.75 ± 1.26 | 29.91 ± 0.94 | 56.20 ± 1.86 |
| G2 | PDM 04-123 | V. radiata | 12.00 ± 1.16 | 7.00 ± 0.58 | 58.49 ± 0.83 | 25.33 ± 0.33 | 0.070 ± 0.001 | 37.77 ± 1.07 | 39.79 ± 2.22 | 56.79 ± 1.75 |
| G3 | PDM 281 | V. radiata | 18.67 ± 0.67 | 9.67 ± 0.33 | 51.85 ± 1.85 | 23.93 ± 1.10 | 0.072 ± 0.003 | 25.23 ± 0.62 | 30.21 ± 1.70 | 58.92 ± 1.42 |
| G4 | PDM 54 | V. radiata | 25.33 ± 0.88 | 18.67 ± 0.67 | 73.69 ± 0.89 | 25.67 ± 0.33 | 0.073 ± 0.001 | 28.39 ± 0.61 | 31.40 ± 1.73 | 52.39 ± 1.19 |
| G5 | Pusa Vishal | V. radiata | 26.67 ± 0.88 | 22.33 ± 1.20 | 83.63 ± 1.82 | 31.00 ± 0.76 | 0.062 ± 0.001 | 12.49 ± 0.36 | 19.02 ± 0.49 | 24.50 ± 0.71 |
| G6 | PDM 262 | V. radiata | 14.00 ± 1.16 | 9.33 ± 0.88 | 66.57 ± 1.29 | 24.67 ± 0.67 | 0.074 ± 0.002 | 36.92 ± 1.36 | 39.42 ± 0.44 | 54.77 ± 1.27 |
| G7 | PDM 288 | V. radiata | 9.67 ± 0.88 | 4.67 ± 0.33 | 48.48 ± 1.52 | 23.33 ± 0.88 | 0.072 ± 0.003 | 27.04 ± 0.82 | 30.26 ± 0.65 | 57.44 ± 1.72 |
| G8 | PDM 178 | V. radiata | 12.67 ± 0.33 | 8.00 ± 0.01 | 63.25 ± 1.71 | 24.67 ± 0.33 | 0.073 ± 0.001 | 27.38 ± 2.06 | 32.52 ± 1.55 | 52.99 ± 1.35 |
| G9 | PDM 191 | V. radiata | 14.33 ± 0.88 | 12.00 ± 0.58 | 83.86 ± 1.34 | 26.93 ± 0.93 | 0.071 ± 0.003 | 32.09 ± 1.73 | 34.02 ± 1.44 | 58.09 ± 2.08 |
| G10 | IPM 2-14 | V. radiata | 16.33 ± 0.88 | 11.33 ± 0.67 | 69.45 ± 2.78 | 24.93 ± 0.52 | 0.074 ± 0.002 | 28.84 ± 1.29 | 34.61 ± 2.02 | 56.37 ± 1.39 |
| G11 | IPM 06-5 | V. radiata | 13.67 ± 0.88 | 11.33 ± 0.88 | 82.86 ± 2.35 | 26.83 ± 0.83 | 0.072 ± 0.002 | 25.08 ± 1.56 | 31.73 ± 1.07 | 54.10 ± 0.96 |
| G12 | IPM 409-4 | V. radiata | 13.33 ± 0.88 | 11.00 ± 0.58 | 82.65 ± 1.38 | 26.33 ± 0.88 | 0.073 ± 0.003 | 27.35 ± 1.18 | 31.49 ± 0.88 | 55.52 ± 2.32 |
| G13 | IPM 312-43K | V. radiata | 11.33 ± 0.67 | 8.67 ± 0.33 | 76.67 ± 1.67 | 26.93 ± 1.10 | 0.070 ± 0.003 | 26.70 ± 1.70 | 30.98 ± 0.65 | 56.76 ± 1.30 |
| G14 | Selection 18-5 | V. radiata | 19.67 ± 0.33 | 16.33 ± 0.67 | 82.98 ± 2.02 | 23.00 ± 0.58 | 0.083 ± 0.002 | 38.91 ± 1.46 | 51.15 ± 1.18 | 81.51 ± 0.91 |
| G15 | IPM 2K-14-5 | V. radiata | 5.33 ± 0.67 | 3.67 ± 0.33 | 69.45 ± 2.78 | 27.07 ± 1.07 | 0.068 ± 0.003 | 8.94 ± 1.86 | 15.47 ± 1.39 | 40.95 ± 1.04 |
| G16 | Selection 18-2 | V. radiata | 9.00 ± 0.58 | 8.67 ± 0.67 | 96.30 ± 3.70 | 27.13 ± 1.13 | 0.073 ± 0.003 | 22.41 ± 0.61 | 26.47 ± 1.70 | 58.77 ± 2.81 |
| G17 | IPM 2-23 | V. radiata | 15.33 ± 0.33 | 12.67 ± 0.33 | 82.64 ± 2.05 | 26.59 ± 0.50 | 0.072 ± 0.001 | 33.37 ± 1.73 | 36.05 ± 2.25 | 55.88 ± 0.81 |
| G18 | IPM 03-1 | V. radiata | 19.00 ± 1.16 | 13.67 ± 0.33 | 72.27 ± 2.92 | 24.33 ± 0.67 | 0.077 ± 0.003 | 39.25 ± 0.65 | 45.44 ± 0.75 | 59.66 ± 1.15 |
| G19 | IPM 03-3 | V. radiata | 9.33 ± 0.88 | 5.33 ± 0.33 | 57.54 ± 2.50 | 24.33 ± 0.33 | 0.072 ± 0.001 | 25.75 ± 1.61 | 32.65 ± 1.52 | 55.06 ± 2.23 |
| G20 | IPM 2-17 | V. radiata | 11.33 ± 0.67 | 8.33 ± 0.67 | 73.33 ± 1.67 | 26.57 ± 1.11 | 0.070 ± 0.003 | 21.29 ± 1.29 | 24.58 ± 1.07 | 55.05 ± 0.40 |
| G21 | IPM 02-3 | V. radiata | 15.33 ± 0.67 | 12.67 ± 0.33 | 82.74 ± 1.49 | 26.00 ± 0.01 | 0.073 ± 0.001 | 22.30 ± 0.84 | 29.62 ± 0.90 | 54.04 ± 1.50 |
| G22 | IPM 02-3-2 | V. radiata | 17.00 ± 0.58 | 15.33 ± 0.33 | 90.40 ± 3.54 | 21.67 ± 0.88 | 0.091 ± 0.004 | 38.55 ± 0.86 | 46.00 ± 1.47 | 71.47 ± 0.50 |
| G23 | IPM 2-19 | V. radiata | 14.67 ± 0.67 | 12.67 ± 0.33 | 86.61 ± 3.38 | 28.67 ± 0.33 | 0.068 ± 0.001 | 12.62 ± 0.50 | 19.63 ± 0.79 | 37.31 ± 1.51 |
| G24 | IPM 5-2-8 | V. radiata | 18.33 ± 0.88 | 14.33 ± 0.33 | 78.38 ± 2.14 | 26.47 ± 0.87 | 0.072 ± 0.003 | 32.65 ± 1.50 | 42.39 ± 2.19 | 60.85 ± 1.59 |
| G25 | CO-4 | V. radiata | 7.33 ± 0.67 | 4.67 ± 0.33 | 63.89 ± 1.39 | 24.57 ± 0.30 | 0.074 ± 0.001 | 32.52 ± 0.94 | 37.38 ± 1.91 | 59.28 ± 3.62 |
| G26 | IPM 05-3-22 | V. radiata | 7.67 ± 0.88 | 4.00 ± 0.58 | 51.85 ± 1.85 | 25.22 ± 0.78 | 0.068 ± 0.002 | 27.16 ± 0.19 | 31.62 ± 0.30 | 59.32 ± 3.62 |
| G27 | IPM 306-6 | V. radiata | 9.33 ± 0.33 | 6.00 ± 0.01 | 64.45 ± 2.22 | 25.00 ± 0.58 | 0.072 ± 0.002 | 30.66 ± 3.13 | 35.10 ± 1.82 | 56.66 ± 2.53 |
| G28 | CO-5 | V. radiata | 4.67 ± 0.67 | 3.67 ± 0.67 | 77.78 ± 2.78 | 24.67 ± 0.33 | 0.077 ± 0.001 | 30.88 ± 1.60 | 34.57 ± 1.58 | 58.02 ± 1.21 |
| G29 | CO-6 | V. radiata | 11.67 ± 0.33 | 6.67 ± 0.33 | 57.07 ± 1.26 | 24.67 ± 0.88 | 0.071 ± 0.003 | 30.20 ± 0.50 | 34.23 ± 1.54 | 57.58 ± 0.37 |
| G30 | IPM 2K-14-9 | V. radiata | 6.67 ± 0.88 | 3.67 ± 0.33 | 55.71 ± 2.97 | 23.93 ± 0.52 | 0.073 ± 0.001 | 26.04 ± 2.17 | 29.70 ± 1.07 | 57.07 ± 1.16 |
| G31 | COGG-912 | V. radiata | 8.67 ± 0.67 | 4.67 ± 0.67 | 53.33 ± 3.33 | 24.50 ± 0.29 | 0.071 ± 0.001 | 28.44 ± 2.56 | 31.03 ± 0.20 | 60.03 ± 1.06 |
| G32 | JBT 46/23 | V. radiata | 23.33 ± 1.20 | 14.33 ± 0.67 | 61.47 ± 0.75 | 26.92 ± 0.92 | 0.066 ± 0.002 | 14.67 ± 0.53 | 21.67 ± 1.12 | 40.54 ± 1.76 |
| G33 | CO-7 | V. radiata | 13.67 ± 1.20 | 9.67 ± 0.33 | 71.47 ± 4.52 | 27.00 ± 0.58 | 0.069 ± 0.002 | 15.18 ± 0.88 | 17.14 ± 1.05 | 47.71 ± 2.00 |
| G34 | Selection 18-4 | V. radiata | 14.67 ± 0.67 | 12.33 ± 0.33 | 84.22 ± 1.49 | 22.33 ± 0.33 | 0.086 ± 0.001 | 41.22 ± 1.26 | 44.23 ± 0.42 | 66.24 ± 0.86 |
| G35 | Yellow Selection | V. radiata | 16.33 ± 0.88 | 10.67 ± 0.88 | 65.14 ± 2.64 | 27.13 ± 1.13 | 0.067 ± 0.002 | 10.82 ± 0.64 | 11.75 ± 1.71 | 37.95 ± 1.12 |
| G36 | SML 832 | V. radiata | 13.00 ± 1.00 | 9.33 ± 0.67 | 71.86 ± 0.43 | 26.60 ± 0.60 | 0.070 ± 0.002 | 27.84 ± 2.41 | 30.45 ± 0.64 | 55.26 ± 1.54 |
| G37 | Pusa 9531 | V. radiata | 10.00 ± 0.58 | 5.67 ± 0.33 | 57.07 ± 4.98 | 25.00 ± 0.01 | 0.070 ± 0.002 | 12.22 ± 1.45 | 15.60 ± 1.63 | 43.54 ± 2.01 |
| G38 | Pusa 9972 | V. radiata | 6.33 ± 0.67 | 4.33 ± 0.67 | 67.62 ± 3.81 | 24.67 ± 0.33 | 0.074 ± 0.001 | 23.65 ± 1.09 | 32.02 ± 1.71 | 58.21 ± 1.09 |
| G39 | Pusa Bold 2 | V. radiata | 8.67 ± 0.33 | 6.33 ± 0.33 | 73.61 ± 6.94 | 24.33 ± 0.88 | 0.077 ± 0.004 | 23.71 ± 1.24 | 25.45 ± 2.32 | 58.91 ± 2.39 |
| G40 | Pusa 672 | V. radiata | 6.33 ± 1.20 | 2.67 ± 0.67 | 44.05 ± 9.74 | 26.33 ± 0.88 | 0.062 ± 0.004 | 10.56 ± 1.11 | 12.82 ± 0.96 | 34.21 ± 1.95 |
| G41 | Sona Green | V. radiata | 6.33 ± 0.33 | 5.00 ± 0.01 | 79.36 ± 3.97 | 25.67 ± 0.33 | 0.074 ± 0.002 | 31.11 ± 2.92 | 33.88 ± 2.31 | 58.25 ± 1.50 |
| G42 | ML 818 | V. radiata | 14.67 ± 0.88 | 11.33 ± 0.67 | 77.31 ± 1.46 | 25.81 ± 0.43 | 0.073 ± 0.001 | 28.32 ± 1.79 | 30.97 ± 2.25 | 57.38 ± 1.24 |
| G43 | ML 5 | V. radiata | 10.00 ± 1.16 | 7.67 ± 0.88 | 76.67 ± 1.67 | 26.07 ± 1.21 | 0.072 ± 0.003 | 23.64 ± 1.68 | 25.89 ± 0.74 | 59.60 ± 1.17 |
| G44 | ML 512 | V. radiata | 11.67 ± 0.88 | 7.67 ± 0.33 | 66.07 ± 2.46 | 24.67 ± 1.33 | 0.074 ± 0.005 | 23.16 ± 1.95 | 25.95 ± 1.22 | 55.89 ± 2.63 |
| G45 | ML 515 | V. radiata | 12.33 ± 0.33 | 9.33 ± 0.33 | 75.64 ± 0.64 | 26.33 ± 0.88 | 0.071 ± 0.002 | 32.87 ± 1.61 | 35.22 ± 1.37 | 55.96 ± 1.62 |
| G46 | ML 682 | V. radiata | 6.33 ± 0.33 | 5.00 ± 0.01 | 79.36 ± 3.97 | 25.73 ± 0.73 | 0.074 ± 0.002 | 26.03 ± 1.43 | 32.75 ± 0.93 | 55.69 ± 0.72 |
| G47 | ML 729 | V. radiata | 7.33 ± 0.88 | 6.33 ± 0.88 | 85.98 ± 1.61 | 26.80 ± 1.33 | 0.072 ± 0.004 | 22.48 ± 0.49 | 24.76 ± 1.57 | 54.40 ± 1.20 |
| G48 | ML 1059 | V. radiata | 5.33 ± 0.33 | 3.33 ± 0.33 | 62.22 ± 2.22 | 24.67 ± 0.33 | 0.073 ± 0.001 | 21.26 ± 2.01 | 24.54 ± 1.42 | 55.59 ± 1.72 |
| G49 | ML 1256 | V. radiata | 8.67 ± 0.88 | 7.00 ± 0.58 | 81.16 ± 2.36 | 24.67 ± 0.33 | 0.077 ± 0.001 | 25.01 ± 1.48 | 28.59 ± 1.74 | 59.14 ± 0.46 |
| G50 | ML 1257 | V. radiata | 5.00 ± 0.58 | 4.00 ± 0.58 | 79.44 ± 2.42 | 25.40 ± 0.95 | 0.075 ± 0.003 | 20.17 ± 1.20 | 25.10 ± 1.83 | 56.47 ± 0.93 |
| G51 | AKM 96-4 | V. radiata | 6.00 ± 1.00 | 3.67 ± 0.67 | 60.83 ± 0.83 | 23.67 ± 0.33 | 0.075 ± 0.001 | 24.53 ± 1.60 | 32.49 ± 0.74 | 58.96 ± 1.49 |
| G52 | AKM 96-1 | V. radiata | 13.33 ± 1.20 | 10.33 ± 0.88 | 77.75 ± 3.20 | 26.05 ± 0.62 | 0.072 ± 0.001 | 27.29 ± 0.67 | 32.58 ± 1.40 | 58.88 ± 1.74 |
| G53 | AKM 96-2 | V. radiata | 13.67 ± 0.88 | 8.67 ± 0.67 | 63.49 ± 3.18 | 23.67 ± 0.67 | 0.076 ± 0.002 | 39.58 ± 2.54 | 48.61 ± 2.63 | 57.27 ± 1.93 |
| G54 | AKM/NP/8/9 | V. radiata | 12.00 ± 1.00 | 9.00 ± 1.00 | 74.61 ± 2.31 | 25.67 ± 0.33 | 0.073 ± 0.001 | 21.48 ± 1.76 | 24.59 ± 1.16 | 54.59 ± 2.27 |
| G55 | Pratiksha | V. radiata | 6.67 ± 0.33 | 5.33 ± 0.33 | 80.16 ± 4.42 | 25.33 ± 0.67 | 0.075 ± 0.001 | 21.52 ± 0.73 | 26.32 ± 1.43 | 59.10 ± 1.41 |
| G56 | LGG 460 | V. radiata | 13.67 ± 0.88 | 11.33 ± 0.33 | 83.41 ± 4.15 | 26.33 ± 0.88 | 0.073 ± 0.003 | 30.83 ± 1.69 | 39.21 ± 0.28 | 58.87 ± 1.21 |
| G57 | TARAM 18 | V. radiata | 16.33 ± 0.33 | 11.00 ± 0.58 | 67.28 ± 2.45 | 25.33 ± 0.33 | 0.072 ± 0.002 | 24.19 ± 0.74 | 30.82 ± 1.63 | 59.03 ± 1.76 |
| G58 | TARAM 1 | V. radiata | 11.33 ± 0.67 | 9.67 ± 0.88 | 85.00 ± 3.47 | 27.27 ± 1.37 | 0.071 ± 0.004 | 30.13 ± 1.10 | 34.97 ± 0.25 | 58.09 ± 0.82 |
| G59 | TMB 37 | V. radiata | 6.67 ± 0.33 | 4.00 ± 0.01 | 60.32 ± 3.18 | 24.33 ± 0.33 | 0.073 ± 0.002 | 28.88 ± 1.43 | 37.76 ± 1.70 | 56.57 ± 1.32 |
| G60 | TMB 96-2 | V. radiata | 13.67 ± 0.67 | 6.67 ± 0.67 | 48.54 ± 2.39 | 24.67 ± 1.20 | 0.068 ± 0.002 | 11.76 ± 0.72 | 16.44 ± 1.05 | 42.82 ± 2.35 |
| G61 | PS 16 | V. radiata | 8.00 ± 0.58 | 5.00 ± 0.58 | 62.10 ± 2.76 | 25.00 ± 0.58 | 0.072 ± 0.002 | 22.30 ± 1.31 | 36.62 ± 0.78 | 58.83 ± 1.47 |
| G62 | K851 | V. radiata | 11.67 ± 1.45 | 10.00 ± 1.16 | 85.98 ± 1.61 | 25.67 ± 0.33 | 0.075 ± 0.001 | 24.82 ± 1.21 | 34.84 ± 1.23 | 55.43 ± 1.46 |
| G63 | MG 331 | V. radiata | 8.67 ± 0.67 | 6.67 ± 0.67 | 76.67 ± 1.67 | 27.33 ± 0.67 | 0.069 ± 0.002 | 30.46 ± 0.37 | 39.16 ± 0.66 | 57.86 ± 0.18 |
| G64 | Saptari | V. radiata | 12.33 ± 0.88 | 8.67 ± 0.67 | 70.28 ± 1.84 | 26.33 ± 0.88 | 0.070 ± 0.003 | 29.57 ± 1.65 | 34.65 ± 0.67 | 57.85 ± 2.95 |
| G65 | Asha | V. radiata | 10.67 ± 0.67 | 8.00 ± 0.01 | 75.56 ± 4.44 | 25.33 ± 0.67 | 0.074 ± 0.001 | 38.82 ± 1.18 | 46.48 ± 0.78 | 57.17 ± 2.08 |
| G66 | SPS-5 | V. radiata | 8.33 ± 0.88 | 4.00 ± 0.58 | 47.62 ± 2.38 | 23.33 ± 0.88 | 0.072 ± 0.003 | 12.12 ± 1.01 | 14.11 ± 0.69 | 24.62 ± 1.43 |
| G67 | Bhutan LM-1 | V. radiata | 10.67 ± 0.67 | 5.33 ± 0.33 | 50.00 ± 0.01 | 23.67 ± 0.33 | 0.072 ± 0.001 | 29.98 ± 1.02 | 33.39 ± 2.61 | 57.99 ± 2.38 |
| G68 | Bhutan LM-2 | V. radiata | 16.00 ± 1.00 | 12.33 ± 0.67 | 77.41 ± 4.64 | 26.33 ± 0.88 | 0.072 ± 0.003 | 29.14 ± 2.30 | 35.36 ± 1.57 | 57.85 ± 0.68 |
| G69 | SM 47 | V. radiata | 9.67 ± 1.20 | 5.67 ± 0.33 | 59.72 ± 5.01 | 24.33 ± 0.88 | 0.073 ± 0.002 | 19.75 ± 0.65 | 23.66 ± 1.29 | 56.97 ± 1.90 |
| G70 | SM 48 | V. radiata | 4.00 ± 0.58 | 2.33 ± 0.33 | 58.89 ± 4.84 | 25.00 ± 0.58 | 0.071 ± 0.003 | 24.44 ± 1.63 | 29.62 ± 0.97 | 55.37 ± 1.23 |
| G71 | PM-4 | V. radiata | 13.33 ± 0.88 | 8.67 ± 0.67 | 64.96 ± 1.71 | 25.17 ± 0.73 | 0.072 ± 0.002 | 18.34 ± 0.88 | 22.88 ± 1.82 | 48.65 ± 7.50 |
| G72 | BMS 18-1 | V. radiata | 17.33 ± 0.33 | 15.67 ± 0.33 | 90.52 ± 3.60 | 22.33 ± 0.33 | 0.088 ± 0.001 | 26.00 ± 0.40 | 36.83 ± 1.09 | 64.82 ± 5.53 |
| G73 | UPM 98-1 | V. radiata | 2.00 ± 0.01 | 1.00 ± 0.01 | 50.00 ± 0.01 | 25.00 ± 0.58 | 0.068 ± 0.002 | 17.55 ± 1.24 | 26.23 ± 0.84 | 59.26 ± 0.89 |
| G74 | UPM02-17 | V. radiata | 6.67 ± 0.88 | 4.00 ± 0.58 | 59.88 ± 1.55 | 23.67 ± 0.33 | 0.075 ± 0.001 | 16.11 ± 1.26 | 27.67 ± 1.50 | 56.49 ± 0.65 |
| G75 | UPM02-18 | V. radiata | 15.67 ± 1.45 | 11.67 ± 0.88 | 74.71 ± 1.36 | 24.67 ± 0.88 | 0.076 ± 0.003 | 25.25 ± 0.96 | 36.50 ± 0.81 | 61.16 ± 1.78 |
| G76 | HUM 12 | V. radiata | 12.67 ± 1.20 | 11.00 ± 1.00 | 86.97 ± 2.19 | 25.00 ± 0.00 | 0.078 ± 0.001 | 36.94 ± 2.63 | 45.39 ± 0.46 | 56.55 ± 1.64 |
| G77 | Selection 18-1 | V. radiata | 11.33 ± 0.88 | 10.67 ± 0.67 | 94.41 ± 2.83 | 24.00 ± 0.58 | 0.082 ± 0.001 | 38.11 ± 0.56 | 49.22 ± 0.74 | 83.28 ± 2.15 |
| G78 | EC 398885 | V. radiata | 3.33 ± 0.33 | 2.00 ± 0.01 | 61.11 ± 5.56 | 27.11 ± 0.59 | 0.066 ± 0.001 | 7.04 ± 0.25 | 10.16 ± 1.02 | 24.18 ± 1.70 |
| G79 | BMS 18-2 | V. radiata | 11.33 ± 0.67 | 9.67 ± 0.33 | 85.55 ± 2.22 | 23.67 ± 0.33 | 0.082 ± 0.001 | 42.48 ± 1.53 | 54.51 ± 1.81 | 82.86 ± 1.82 |
| G80 | BMS 18-3 | V. radiata | 3.67 ± 0.33 | 3.67 ± 0.33 | 100.00 ± 0.01 | 20.67 ± 0.33 | 0.097 ± 0.002 | 41.33 ± 0.66 | 54.69 ± 1.05 | 83.46 ± 1.01 |
| G81 | BMS 18-4 | V. radiata | 13.00 ± 1.16 | 10.67 ± 0.88 | 82.15 ± 1.34 | 24.67 ± 0.88 | 0.078 ± 0.003 | 25.42 ± 0.36 | 32.52 ± 0.53 | 57.98 ± 2.96 |
| G82 | LGG-544 | V. radiata | 8.67 ± 0.33 | 6.33 ± 0.33 | 73.15 ± 3.34 | 26.33 ± 0.33 | 0.071 ± 0.001 | 29.47 ± 2.18 | 38.70 ± 1.32 | 59.16 ± 0.98 |
| G83 | EC 398897 | V. radiata | 20.67 ± 1.20 | 16.00 ± 1.00 | 77.40 ± 1.22 | 23.43 ± 0.98 | 0.081 ± 0.004 | 30.20 ± 1.17 | 36.57 ± 0.54 | 78.44 ± 2.19 |
| G84 | EC 391178 (Y) | V. radiata | 5.33 ± 1.20 | 2.67 ± 0.33 | 53.18 ± 7.05 | 24.67 ± 0.88 | 0.07 ± 0.005 | 12.92 ± 0.97 | 21.05 ± 1.33 | 38.86 ± 1.14 |
| G85 | EC 496841 | V. radiata | 13.67 ± 0.88 | 10.33 ± 0.67 | 75.63 ± 1.55 | 25.60 ± 0.40 | 0.073 ± 0.001 | 14.46 ± 0.95 | 20.13 ± 1.66 | 60.94 ± 2.21 |
| G86 | EC 520041 | V. radiata | 4.67 ± 0.33 | 2.67 ± 0.33 | 56.67 ± 3.33 | 25.67 ± 0.33 | 0.068 ± 0.001 | 8.60 ± 0.33 | 13.64 ± 0.42 | 37.44 ± 1.29 |
| G87 | Banda Local-1 | V. mungo | 2.67 ± 0.33 | 1.67 ± 0.33 | 61.11 ± 5.56 | 22.77 ± 0.77 | 0.078 ± 0.002 | 28.28 ± 1.45 | 38.40 ± 1.55 | 58.53 ± 0.27 |
| G88 | EC 426841 | V. radiata | 4.67 ± 0.33 | 2.33 ± 0.33 | 50.00 ± 5.77 | 23.67 ± 0.67 | 0.072 ± 0.001 | 12.41 ± 0.37 | 18.74 ± 0.53 | 29.40 ± 1.00 |
| G89 | EC 496839 | V. radiata | 1.33 ± 0.33 | 1.00 ± 0.01 | 83.33 ± 16.67 | 24.50 ± 0.76 | 0.078 ± 0.004 | 27.19 ± 0.92 | 32.03 ± 0.38 | 57.40 ± 0.63 |
| G90 | IPU 11-2 | V. mungo | 2.33 ± 0.33 | 1.33 ± 0.33 | 55.56 ± 5.56 | 22.67 ± 0.33 | 0.077 ± 0.001 | 25.30 ± 0.95 | 33.82 ± 1.57 | 59.43 ± 0.95 |
| G91 | IPU 2-43 | V. mungo | 13.00 ± 1.00 | 11.33 ± 1.33 | 86.66 ± 3.33 | 26.33 ± 0.88 | 0.074 ± 0.003 | 18.39 ± 0.73 | 40.76 ± 3.04 | 56.60 ± 1.39 |
| G92 | EC 520014 | V. radiata | 10.67 ± 0.88 | 6.67 ± 0.88 | 61.96 ± 3.32 | 26.67 ± 1.20 | 0.067 ± 0.002 | 10.47 ± 0.62 | 16.01 ± 1.25 | 39.01 ± 0.83 |
| G93 | EC 520016 | V. radiata | 10.67 ± 0.67 | 7.67 ± 0.33 | 72.22 ± 4.01 | 27.83 ± 0.44 | 0.067 ± 0.002 | 12.31 ± 0.99 | 19.09 ± 0.30 | 38.98 ± 1.68 |
| G94 | EC 520024(DR) | V. radiata | 11.67 ± 0.88 | 7.00 ± 0.58 | 60.51 ± 5.80 | 25.33 ± 0.33 | 0.070 ± 0.002 | 12.13 ± 0.99 | 27.73 ± 2.31 | 57.44 ± 2.41 |
| G95 | EC 520024 | V. radiata | 2.00 ± 0.58 | 1.00 ± 0.01 | 61.11 ± 20.03 | 31.00 ± 0.58 | 0.056 ± 0.004 | 7.91 ± 0.44 | 10.54 ± 0.72 | 22.00 ± 1.19 |
| G96 | EC 520026 | V. radiata | 10.00 ± 0.58 | 4.33 ± 0.33 | 43.30 ± 1.67 | 28.13 ± 1.16 | 0.058 ± 0.002 | 9.19 ± 1.03 | 11.77 ± 1.38 | 21.92 ± 2.24 |
| G97 | EC 520029 | V. radiata | 6.67 ± 0.88 | 4.00 ± 0.58 | 59.88 ± 1.55 | 25.67 ± 0.33 | 0.069 ± 0.001 | 21.42 ± 1.09 | 23.07 ± 1.37 | 38.53 ± 1.73 |
| G98 | EC 520034 | V. radiata | 12.67 ± 0.88 | 11.00 ± 0.58 | 87.08 ± 1.94 | 25.50 ± 0.29 | 0.076 ± 0.001 | 21.87 ± 0.41 | 25.87 ± 0.47 | 59.49 ± 0.85 |
| G99 | EC 520034-1 | V. radiata | 9.33 ± 0.33 | 5.67 ± 0.33 | 60.74 ± 3.23 | 28.00 ± 1.16 | 0.064 ± 0.003 | 11.45 ± 1.56 | 14.34 ± 1.18 | 33.07 ± 0.61 |
| G100 | EC 550831 | V. radiata | 7.67 ± 0.33 | 2.00 ± 0.01 | 26.19 ± 1.19 | 25.67 ± 0.67 | 0.055 ± 0.002 | 7.43 ± 0.43 | 11.67 ± 1.19 | 24.84 ± 1.60 |
| G101 | TCR 80 | V. radiata | 10.33 ± 0.33 | 8.00 ± 0.01 | 77.58 ± 2.42 | 26.00 ± 0.58 | 0.073 ± 0.002 | 26.42 ± 0.60 | 31.16 ± 1.25 | 56.90 ± 0.35 |
| G102 | TCR 82 | V. radiata | 13.00 ± 0.58 | 7.33 ± 0.33 | 56.62 ± 3.44 | 27.27 ± 0.73 | 0.064 ± 0.002 | 9.47 ± 0.85 | 14.18 ± 0.50 | 35.57 ± 0.62 |
| G103 | TCR 7 | V. sublobata | 7.67 ± 0.33 | 5.67 ± 0.33 | 73.81 ± 1.19 | 31.40 ± 1.14 | 0.06 ± 0.002 | 8.21 ± 0.86 | 10.51 ± 0.77 | 29.35 ± 0.46 |
| G104 | TCR 64 | V. trilobata | 13.00 ± 0.01 | 11.33 ± 0.33 | 87.18 ± 2.56 | 29.67 ± 0.33 | 0.065 ± 0.001 | 14.14 ± 0.19 | 21.17 ± 0.80 | 34.74 ± 0.42 |
| G105 | DGGV 2 | V. radiata | 22.33 ± 0.88 | 13.67 ± 0.67 | 61.16 ± 1.05 | 21.87 ± 0.94 | 0.082 ± 0.004 | 40.41 ± 0.85 | 51.93 ± 0.35 | 81.50 ± 0.57 |
| G106 | TCR 64-1 | V. trilobata | 5.67 ± 0.33 | 3.67 ± 0.33 | 64.45 ± 2.22 | 27.67 ± 0.88 | 0.066 ± 0.002 | 9.24 ± 0.82 | 13.01 ± 0.10 | 39.76 ± 0.57 |
| G107 | TCR 254-1 | V. radiata | 8.00 ± 0.00 | 7.00 ± 0.01 | 87.50 ± 0.01 | 28.80 ± 0.76 | 0.068 ± 0.002 | 12.60 ± 0.58 | 14.99 ± 0.35 | 45.77 ± 0.43 |
| G108 | TCR 254-2 | V. radiata | 17.33 ± 0.33 | 8.00 ± 0.01 | 46.19 ± 0.87 | 23.67 ± 0.67 | 0.071 ± 0.002 | 13.69 ± 1.01 | 19.52 ± 0.64 | 49.59 ± 0.32 |
| G109 | TCR 262 | V. sublobata | 2.67 ± 0.33 | 2.33 ± 0.33 | 88.89 ± 11.11 | 28.00 ± 0.58 | 0.069 ± 0.001 | 16.00 ± 0.58 | 18.02 ± 0.22 | 47.29 ± 0.85 |
| G110 | TCR 20 | V. glabrescens | 8.33 ± 0.33 | 6.00 ± 0.01 | 72.22 ± 2.78 | 27.83 ± 0.73 | 0.067 ± 0.001 | 11.97 ± 0.91 | 13.77 ± 0.71 | 43.24 ± 0.47 |
| G111 | PRR 2008-2 | V. umbellata | 2.33 ± 0.33 | 0.00 ± 0.00 | 0.00 ± 0.00 | 0.000 ± 0.000 | 0.000 ± 0.000 | 0.00 ± 0.00 | 0.00 ± 0.00 | 0.00 ± 0.00 |
| G112 | PRR 2008-2 sel | V. umbellata | 2.67 ± 0.33 | 0.00 ± 0.00 | 0.00 ± 0.00 | 0.000 ± 0.000 | 0.000 ± 0.000 | 0.00 ± 0.00 | 0.00 ± 0.00 | 0.00 ± 0.00 |
| G113 | TCR 93 | V. umbellata | 4.00 ± 0.01 | 1.33 ± 0.33 | 33.33 ± 8.33 | 31.77 ± 0.50 | 0.047 ± 0.003 | 5.26 ± 0.76 | 9.62 ± 0.53 | 19.14 ± 0.15 |
| G114 | TL 2 | V. stipulacea | 10.67 ± 0.33 | 9.00 ± 0.01 | 84.55 ± 2.73 | 27.33 ± 1.20 | 0.071 ± 0.003 | 25.74 ± 1.09 | 29.79 ± 0.93 | 55.61 ± 0.94 |
| G115 | L-24 | V. umbellata | 17.00 ± 0.58 | 6.33 ± 0.33 | 37.23 ± 1.05 | 25.50 ± 1.04 | 0.062 ± 0.002 | 3.45 ± 0.25 | 4.70 ± 0.21 | 9.27 ± 0.64 |
| G116 | W 17 | V. stipulacea | 9.00 ± 0.58 | 6.67 ± 0.33 | 74.26 ± 2.28 | 31.00 ± 0.58 | 0.059 ± 0.001 | 8.29 ± 0.18 | 13.00 ± 0.15 | 33.71 ± 0.70 |
| G117 | LRM 13-26 | V. stipulacea | 23.00 ± 0.00 | 11.33 ± 0.33 | 49.28 ± 1.45 | 23.67 ± 0.67 | 0.072 ± 0.001 | 27.91 ± 0.76 | 33.14 ± 1.27 | 56.83 ± 0.39 |
| SN | Primer Code | Sequence | Tm | Amplified Loci | Polymorphic Loci | Polymorphism (%) |
|---|---|---|---|---|---|---|
| 1 | SCoT-2 | CAACAATGGCTACCACCC | 56 | 10 | 07 | 70.00 |
| 2 | SCoT-3 | CAACAATGGCTACCACCG | 53 | 11 | 09 | 81.82 |
| 3 | SCoT-7 | CAACAATGGCTACCACGG | 53 | 04 | 01 | 25.00 |
| 4 | SCoT-10 | CAACAATGGCTACCAGCC | 52 | 08 | 03 | 37.50 |
| 5 | SCoT-12 | ACGACATGGCGACCAACG | 53 | 02 | 02 | 100.00 |
| 6 | SCoT-13 | ACGACATGGCGACCATCG | 58 | 04 | 03 | 75.00 |
| 7 | SCoT-14 | ACGACATGGCGACCACGC | 60 | 04 | 03 | 75.00 |
| 8 | SCoT-15 | ACGACATGGCGACCGCGA | 60 | 05 | 03 | 60.00 |
| 9 | SCoT-16 | ACCATGGCTACCACCGAC | 58 | 05 | 02 | 40.00 |
| 10 | SCoT-17 | ACCATGGCTACCACCGAG | 60 | 08 | 04 | 50.00 |
| 11 | SCoT-19 | ACCATGGCTACCACCGGC | 60 | 06 | 04 | 66.67 |
| 12 | SCoT-21 | ACGACATGGCGACCCACA | 60 | 05 | 01 | 20.00 |
| 13 | SCoT-22 | AACCATGGCTACCACCAC | 58 | 05 | 04 | 80.00 |
| 14 | SCoT-24 | CACCATGGCTACCACCAT | 58 | 04 | 01 | 25.00 |
| 15 | SCoT-25 | ACCATGGCTACCACCGGG | 56 | 05 | 03 | 60.00 |
| 16 | SCoT-29 | CCATGGCTACCACCGGCC | 60 | 08 | 04 | 50.00 |
| 17 | SCoT-30 | CCATGGCTACCACCGGCG | 63 | 07 | 06 | 85.71 |
| 18 | SCoT-31 | CCATGGCTACCACCGCCT | 63 | 07 | 05 | 71.43 |
| 19 | SCoT-32 | CCATGGCTACCACCGCAC | 60 | 06 | 05 | 83.33 |
| 20 | SCoT-33 | CCATGGCTACCACCGCAG | 60 | 07 | 05 | 71.43 |
| 21 | SCoT-34 | ACCATGGCTACCACCGCA | 58 | 06 | 02 | 33.33 |
| 22 | SCoT-35 | CATGGCTACCACCGGCCC | 53 | 05 | 01 | 20.00 |
| 23 | SCoT-36 | GCAACAATGGCTACCACC | 54 | 09 | 05 | 55.56 |
| Average | 6.13 | 3.61 | 58.12 | |||
| Minimum | 2.00 | 1.00 | 20.00 | |||
| Maximum | 11.00 | 9.00 | 100.00 | |||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sahu, P.; Singh, M.; Pandey, R.; Mishra, M.K.; Singh, A.K.; Singh, B.K.; Singh, S.K.; Rai, A.; Chugh, V.; Shukla, G.; et al. Screening of Comprehensive Panel of Cultivated and Wild Vigna Species for Resistance to Pulse Beetle, Callosobruchus chinensis L. Biology 2023, 12, 781. https://doi.org/10.3390/biology12060781
Sahu P, Singh M, Pandey R, Mishra MK, Singh AK, Singh BK, Singh SK, Rai A, Chugh V, Shukla G, et al. Screening of Comprehensive Panel of Cultivated and Wild Vigna Species for Resistance to Pulse Beetle, Callosobruchus chinensis L. Biology. 2023; 12(6):781. https://doi.org/10.3390/biology12060781
Chicago/Turabian StyleSahu, Prince, Mahendra Singh, Rakesh Pandey, Mukesh Kumar Mishra, Akhilesh Kumar Singh, Bhupendra Kumar Singh, Surendra Kumar Singh, Ashutosh Rai, Vishal Chugh, Gaurav Shukla, and et al. 2023. "Screening of Comprehensive Panel of Cultivated and Wild Vigna Species for Resistance to Pulse Beetle, Callosobruchus chinensis L." Biology 12, no. 6: 781. https://doi.org/10.3390/biology12060781
APA StyleSahu, P., Singh, M., Pandey, R., Mishra, M. K., Singh, A. K., Singh, B. K., Singh, S. K., Rai, A., Chugh, V., Shukla, G., Singh, S., Singh, K., Kumar, M., & Singh, C. M. (2023). Screening of Comprehensive Panel of Cultivated and Wild Vigna Species for Resistance to Pulse Beetle, Callosobruchus chinensis L. Biology, 12(6), 781. https://doi.org/10.3390/biology12060781







