Early Solid Diet Supplementation Influences the Proteomics of Rumen Epithelium in Goat Kids
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Animals and Diets
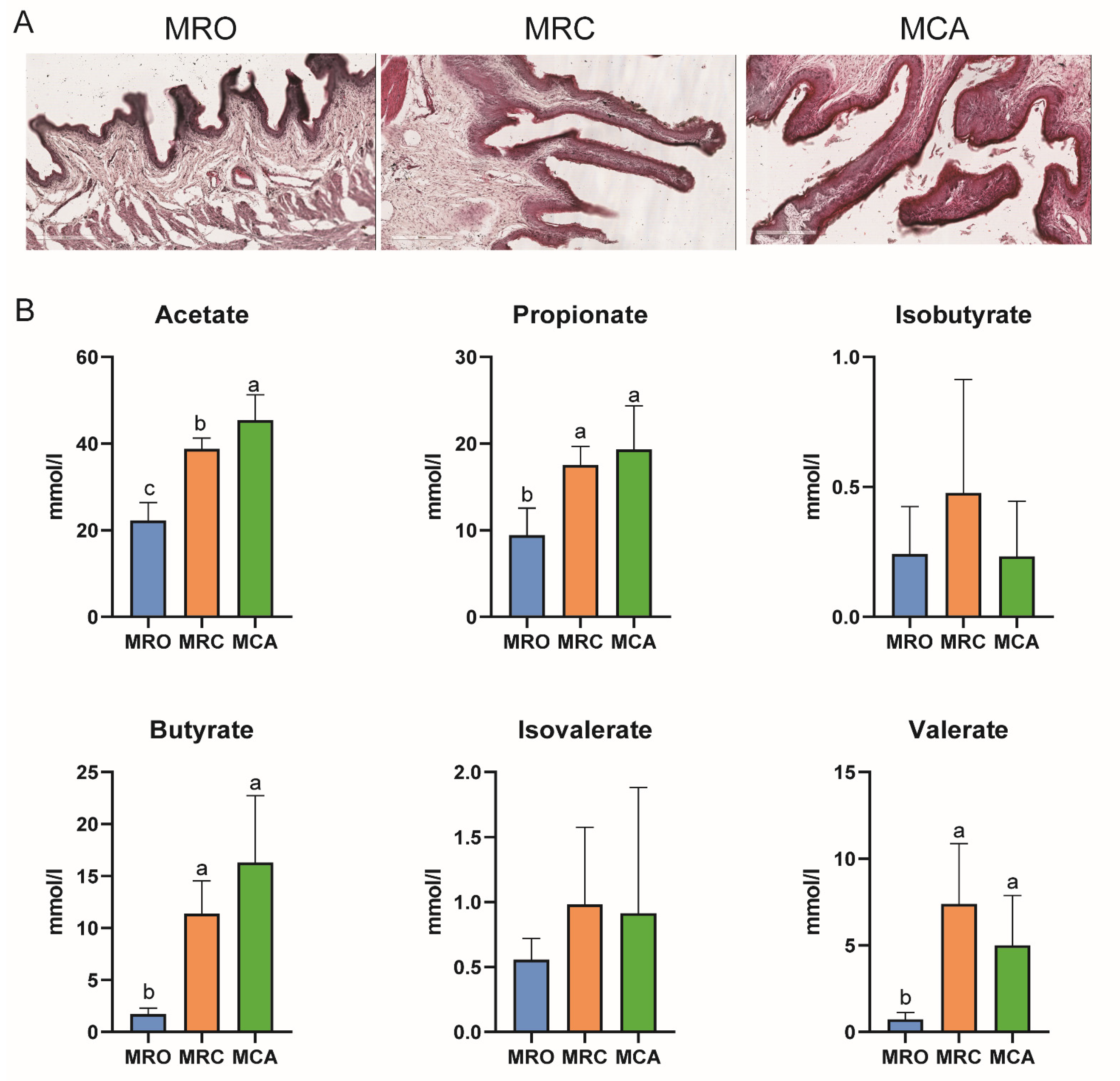
2.3. Determination of Rumen Fermentation Parameters and Morphology
2.4. Liquid Chromatography–Tandem Mass Spectrometry (LC/MS) Analysis
2.5. Protein Quantification and Data Analysis
2.6. Statistics
3. Results
3.1. Growth Performance and Rumen Development
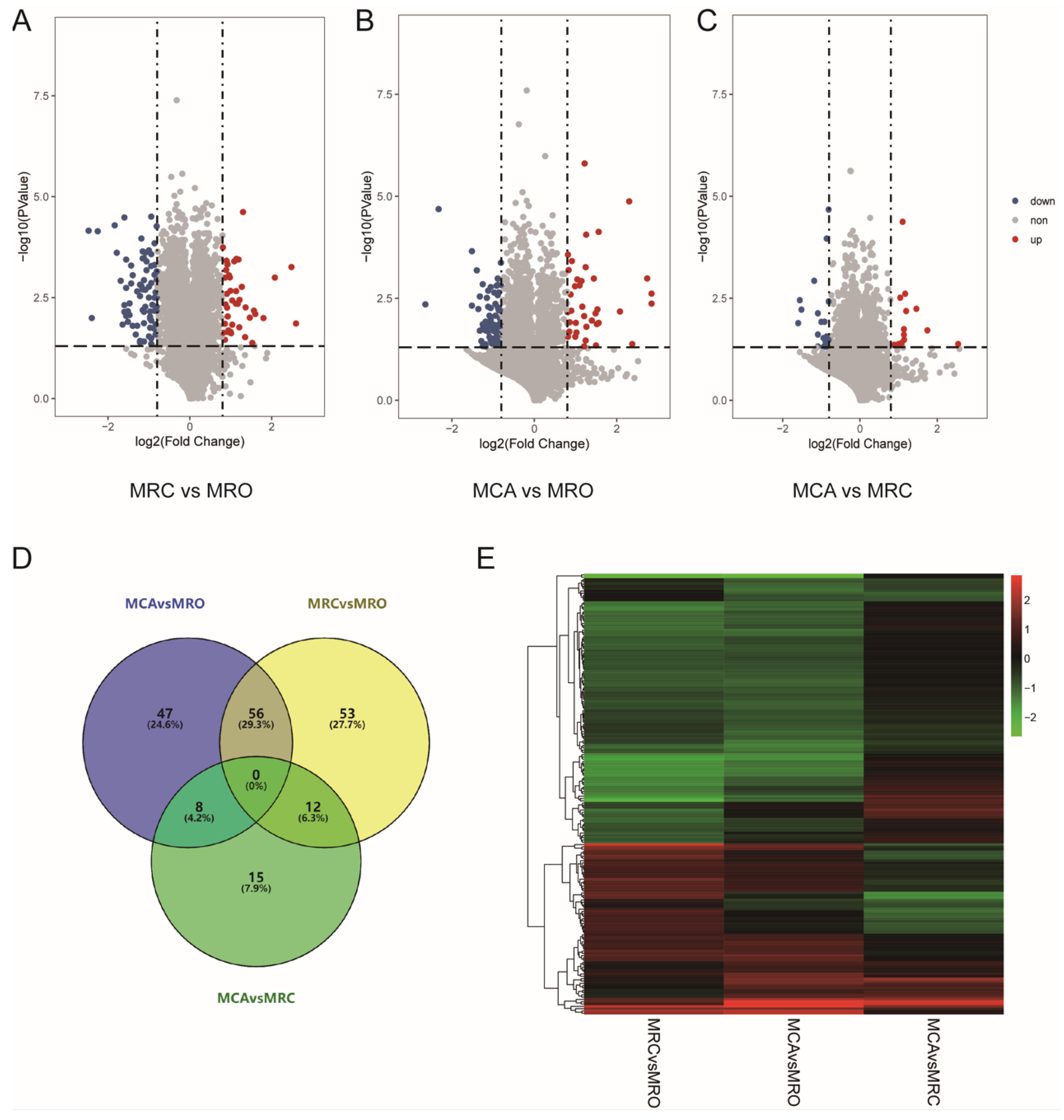
3.2. The Identification of Differently Expressed Proteins (DEPs)
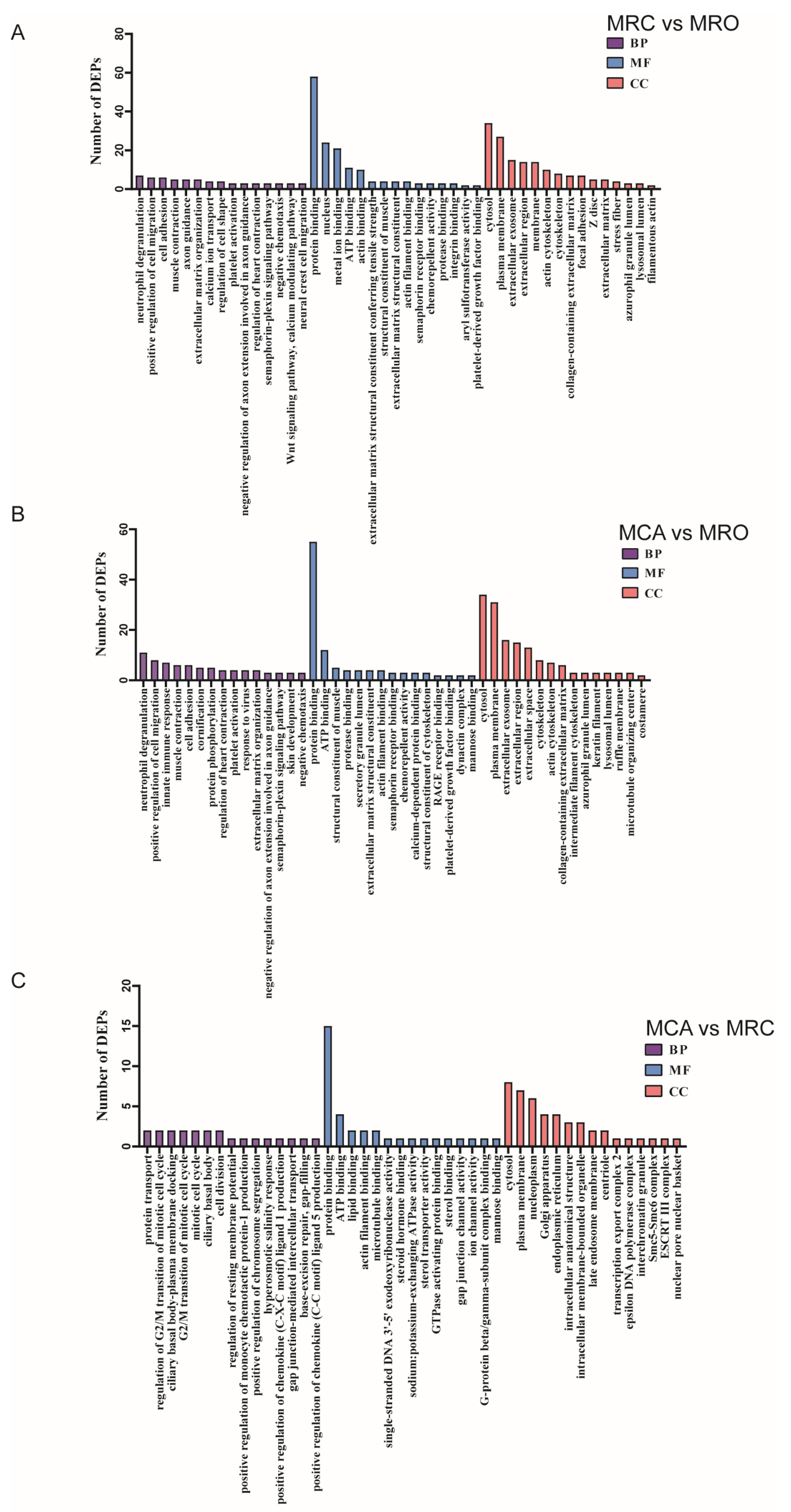
3.3. The Enrichment Analysis of DEPs According to Gene Ontology (GO)
3.4. The KEGG Enrichment Analysis of DEPs
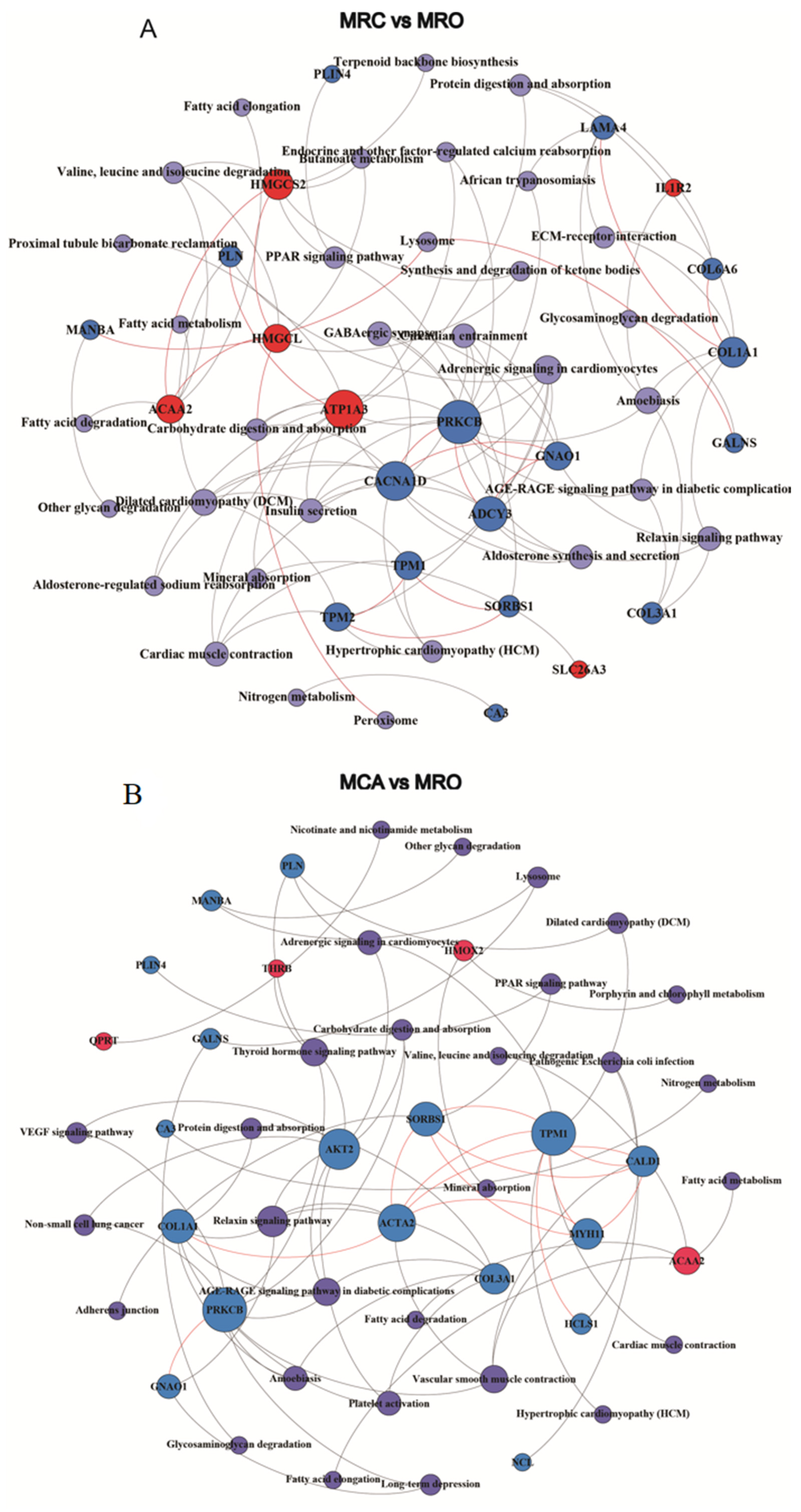
3.5. The PPI Analysis of DEPs
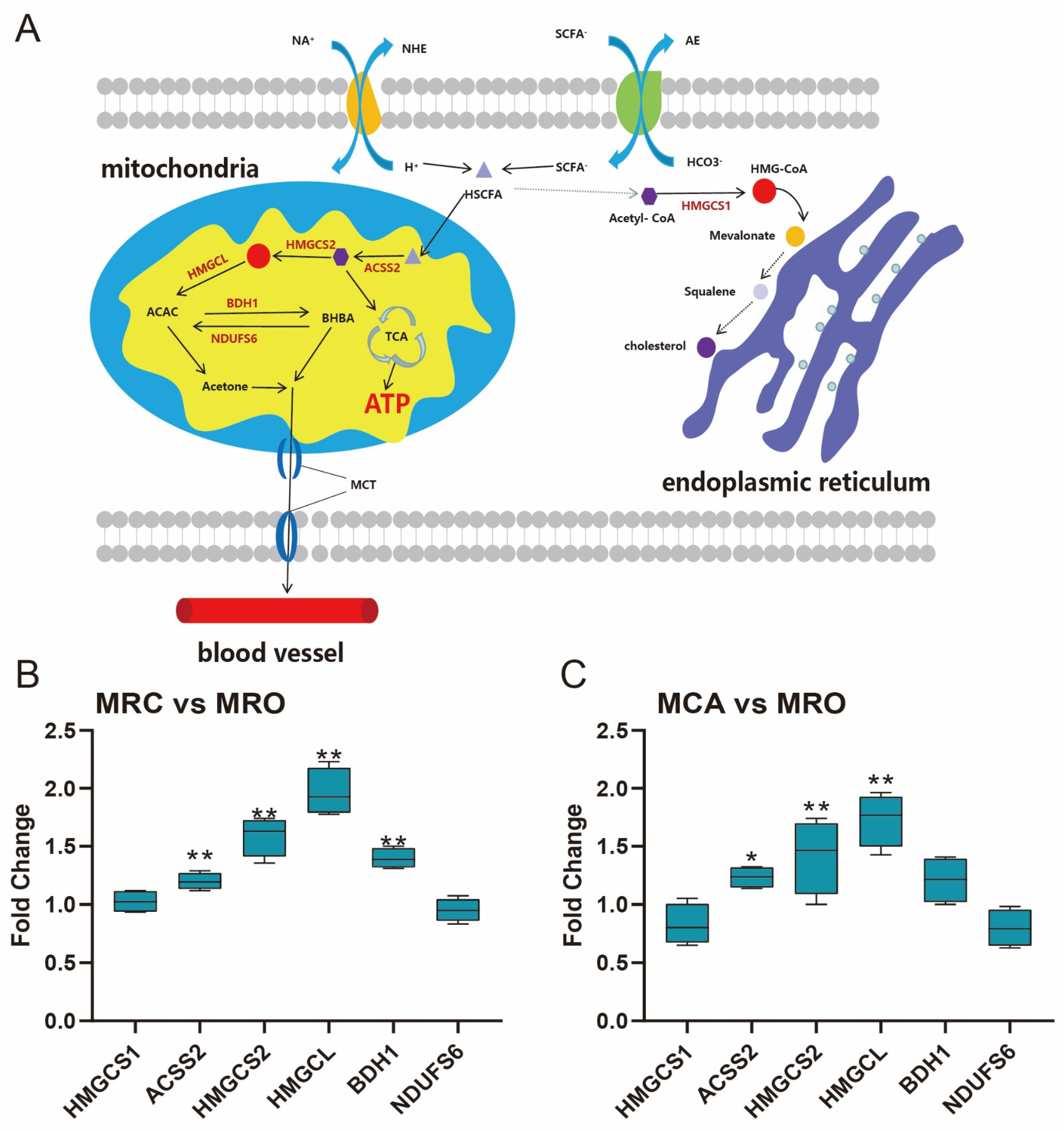
3.6. Rumen Ketogenesis in Response to Solid Feed Supplementation
4. Discussion
4.1. Rumen Fermentation Parameters and Papillae Development
4.2. Cell Development of Rumen Epithelium
4.3. Metabolism of Rumen Epithelium
4.4. Signal Transduction of Rumen Epithelium
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Chai, J.; Lv, X.; Diao, Q.; Usdrowski, H.; Zhuang, Y.; Huang, W.; Cui, K.; Zhang, N. Solid diet manipulates rumen epithelial microbiota and its interactions with host transcriptomic in young ruminants. Environ. Microbiol. 2021, 23, 6557–6568. [Google Scholar] [CrossRef] [PubMed]
- Malmuthuge, N.; Liang, G.; Guan, L.L. Regulation of rumen development in neonatal ruminants through microbial metagenomes and host transcriptomes. Genome Biol. 2019, 20, 172. [Google Scholar] [CrossRef] [PubMed]
- Abdelsattar, M.M.; Zhuang, Y.; Cui, K.; Bi, Y.; Haridy, M.; Zhang, N. Longitudinal investigations of anatomical and morphological development of the gastrointestinal tract in goats from colostrum to postweaning. J. Dairy Sci. 2022, 105, 2597–2611. [Google Scholar] [CrossRef]
- Lin, L.; Xie, F.; Sun, D.; Liu, J.; Zhu, W.; Mao, S. Ruminal microbiome-host crosstalk stimulates the development of the ruminal epithelium in a lamb model. Microbiome 2019, 7, 83. [Google Scholar] [CrossRef]
- Berends, H.; Reenen, C.G.V.; Stockhofe-Zurwieden, N.; Gerrits, W.J.J. Effects of early rumen development and solid feed composition on growth performance and abomasal health in veal calves. J. Dairy Sci. 2012, 95, 3190–3199. [Google Scholar] [CrossRef]
- Pazoki, A.; Ghorbani, G.R.; Kargar, S.; Sadeghi-Sefidmazgi, A.; Ghaffari, M.H.; Drackley, J.K. Technology. Growth performance, nutrient digestibility, ruminal fermentation, and rumen development of calves during transition from liquid to solid feed: Effects of physical form of starter feed and forage provision. Anim. Feed Sci. 2017, 234, 173–185. [Google Scholar] [CrossRef]
- Sun, D.M.; Mao, S.Y.; Zhu, W.Y.; Liu, J.H. Effect of starter diet supplementation on rumen epithelial morphology and expression of genes involved in cell proliferation and metabolism in pre-weaned lambs. Animal 2018, 12, 2274–2283. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Bian, G.; Sun, D.; Zhu, W.; Mao, S. Starter feeding altered ruminal epithelial bacterial communities and some key immune-related genes’ expression before weaning in lambs. J. Anim. Sci. 2017, 95, 910. [Google Scholar] [CrossRef]
- Jing, X.P.; Peng, Q.H.; Hu, R.; Zou, H.W.; Wang, H.Z.; Yu, X.Q.; Zhou, J.W.; Degen, A.; Wang, Z.S. Dietary supplements during the cold season increase rumen microbial abundance and improve rumen epithelium development in Tibetan sheep. J. Anim. Sci. 2018, 96, 293–305. [Google Scholar] [CrossRef]
- Lv, X.; Chai, J.; Diao, Q.; Huang, W.; Zhuang, Y.; Zhang, N. The Signature Microbiota Drive Rumen Function Shifts in Goat Kids Introduced to Solid Diet Regimes. Microorganisms 2019, 7, 516. [Google Scholar] [CrossRef]
- Yin, X.; Ji, S.; Duan, C.; Tian, P.; Ju, S.; Yan, H.; Zhang, Y.; Liu, Y. Age-Related Changes in the Ruminal Microbiota and Their Relationship With Rumen Fermentation in Lambs. Front. Microbiol. 2021, 12, 679135. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Zhang, K.; Li, C.; Wang, X.; Yang, Y. Characterization and Comparison of Microbiota in the Gastrointestinal Tracts of the Goat (Capra hircus) During Preweaning Development. Front. Microbiol. 2019, 10, 2125. [Google Scholar] [CrossRef] [PubMed]
- Reddy, K.E.; Jeong, J.Y.; Lee, S.D.; Baek, Y.C.; Oh, Y.K.; Kim, M.; So, K.M.; Kim, D.W.; Kim, J.H.; Park, S. Effect of different early weaning regimens for calves on adipogenic gene expression in Hanwoo loin at the fattening stage. Livest. Sci. 2016, 195, 87–98. [Google Scholar] [CrossRef]
- Dieho, K.; Van Baal, J.; Kruijt, L.; Bannink, A.D.; Schonewille, J.T.H.; Carreo, D.; Hendriks, W.H.; Dijkstra, J. Effect of supplemental concentrate during the dry period or early lactation on rumen epithelium gene and protein expression in dairy cattle during the transition period. J. Dairy Sci. 2017, 100, 7227–7245. [Google Scholar] [CrossRef]
- Duanmu, Y.; Cong, R.; Tao, S.; Tian, J.; Dong, H.; Zhang, Y.; Ni, Y.; Zhao, R. Biotechnology. Comparative proteomic analysis of the effects of high-concentrate diet on the hepatic metabolism and inflammatory response in lactating dairy goats. J. Anim. Sci. 2016, 7, 5. [Google Scholar] [CrossRef]
- Trinh, H.V.; Grossmann, J.; Gehrig, P.; Roschitzki, B.; Schlapbach, R.; Greber, U.F.; Hemmi, S. iTRAQ-Based and Label-Free Proteomics Approaches for Studies of Human Adenovirus Infections. Int. J. Proteom. 2013, 2013, 581862. [Google Scholar] [CrossRef] [PubMed]
- Wen, B.; Zhou, R.; Qiang, F.; Wang, Q.; Liu, S. IQuant: An automated pipeline for quantitative proteomics based upon isobaric tags. Proteomics 2015, 14, 2280–2285. [Google Scholar] [CrossRef]
- Huang, D.; Sherman, B.; Lempicki, R. Systematic and integrative analysis of large gene lists using DAVID Bioinformatics Resources. Nature Protoc. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef]
- Puchalska, P.; Crawford, P.A. Multi-dimensional Roles of Ketone Bodies in Fuel Metabolism, Signaling, and Therapeutics. Cell Metab. 2017, 25, 262–284. [Google Scholar] [CrossRef]
- Cui, Z.; Wu, S.; Li, J.; Yang, Q.E.; Chai, S.; Wang, L.; Wang, X.; Zhang, X.; Liu, S.; Yao, J. Effect of Alfalfa Hay and Starter Feeding Intervention on Gastrointestinal Microbial Community, Growth and Immune Performance of Yak Calves. Front. Microbiol. 2020, 11, 994. [Google Scholar] [CrossRef]
- Mizrahi, I.; Wallace, R.J.; Morais, S. The rumen microbiome: Balancing food security and environmental impacts. Nat. Rev. Microbiol. 2021, 19, 553–566. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.S.; Wang, R.; Ma, Z.Y.; Zhang, X.M.; Jiao, J.Z.; Zhang, Z.G.; Ungerfeld, E.M.; Yi, K.L.; Zhang, B.Z.; Long, L.; et al. Dietary selection of metabolically distinct microorganisms drives hydrogen metabolism in ruminants. ISME J. 2022, 16, 2535–2546. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Sun, D.; Mao, S.; Zhu, W.; Liu, J. Infusion of sodium butyrate promotes rumen papillae growth and enhances expression of genes related to rumen epithelial VFA uptake and metabolism in neonatal twin lambs. J. Anim. Sci. 2019, 97, 909–921. [Google Scholar] [CrossRef] [PubMed]
- Sun, D.; Mao, S.; Zhu, W.; Liu, J. Proteomic identification of ruminal epithelial protein expression profiles in response to starter feed supplementation in pre-weaned lambs. Anim. Nutr. 2021, 7, 1271–1282. [Google Scholar] [CrossRef]
- Baldwin, V.R.L. Use of isolated ruminal epithelial cells in the study of rumen metabolism. J. Nutr. 1998, 128, 293S–296S. [Google Scholar] [CrossRef]
- Jaluria, P.; Chu, C.; Betenbaugh, M.; Shiloach, J. Cells by design: A mini-review of targeting cell engineering using DNA microarrays. Mol. Biotechnol. 2008, 39, 105–111. [Google Scholar] [CrossRef]
- Kühn, S.; Geyer, M. Formins as effector proteins of Rho GTPases. Small Gtpases 2014, 5, e983876. [Google Scholar] [CrossRef]
- Hubbard, S.R.; Till, J.H. Protein tyrosine kinase structure and function. Annu. Rev. Biochem. 2000, 69, 373–398. [Google Scholar] [CrossRef]
- Wang, S.; Ma, T.; Zhao, G.; Zhang, N.; Tu, Y.; Li, F.; Diao, Q. Effect of Age and Weaning on Growth Performance, Rumen Fermentation, and Serum Parameters in Lambs Fed Starter with Limited Ewe-Lamb Interaction. Animals 2019, 9, 825. [Google Scholar] [CrossRef]
- Zhuang, Y.; Chai, J.; Cui, K.; Bi, Y.; Diao, Q.; Huang, W.; Usdrowski, H.; Zhang, N. Longitudinal Investigation of the Gut Microbiota in Goat Kids from Birth to Postweaning. Microorganisms 2020, 8, 1111. [Google Scholar] [CrossRef]
- Moraïs, S.; Mizrahi, I. The Road Not Taken: The Rumen Microbiome, Functional Groups, and Community States. Trends Microbiol. 2019, 27, 538–549. [Google Scholar] [CrossRef] [PubMed]
- Xue, Y.; Lin, L.; Hu, F.; Zhu, W.; Mao, S. Disruption of ruminal homeostasis by malnutrition involved in systemic ruminal microbiota-host interactions in a pregnant sheep model. Microbiome 2020, 8, 138. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Li, C.; Li, F.; Wang, X.; Zhang, X.; Liu, T.; Fang, N.; Yue, X.; Fei, L.; Pan, X. Effects of early feeding on the host rumen transcriptome and bacterial diversity in lambs. Sci. Rep. 2016, 6, 32479. [Google Scholar] [CrossRef]
- Wei, X.; Schultz, K.; Bazilevsky, G.A.; Vogt, A.; Marmorstein, R. Molecular basis for acetyl-CoA production by ATP-citrate lyase (vol 27, pg 33, 2020). Nat. Struct. Mol. Biol. Cell. 2020, 27, 33–41. [Google Scholar] [CrossRef]
- Ma, L.; Zhao, M.; Zhao, L.S.; Xu, J.C.; Loor, J.J.; Bu, D.P. Effects of dietary neutral detergent fiber and starch ratio on rumen epithelial cell morphological structure and gene expression in dairy cows. J. Dairy Sci. 2017, 100, 3705. [Google Scholar] [CrossRef] [PubMed]
- Cheng, C.W.; Biton, M.; Haber, A.L.; Gunduz, N.; Eng, G.; Gaynor, L.T.; Tripathi, S.; Calibasi-Kocal, G.; Rickelt, S.; Butty, V.L.; et al. Ketone Body Signaling Mediates Intestinal Stem Cell Homeostasis and Adaptation to Diet. Cell 2019, 178, 1115–1131.e15. [Google Scholar] [CrossRef]
- Wang, Q.; Zhou, Y.; Rychahou, P.; Fan, T.W.; Lane, A.N.; Weiss, H.L.; Evers, B.M. Ketogenesis contributes to intestinal cell differentiation. Cell. Death Differ. 2017, 24, 458–468. [Google Scholar] [CrossRef]
- Otsuka, H.; Kimura, T.; Ago, Y.; Nakama, M.; Fukao, T. Deficiency of 3-hydroxybutyrate dehydrogenase (BDH1) in mice causes low ketone body levels and fatty liver during fasting. J. Inherit. Metab. Dis. 2020, 43, 960–968. [Google Scholar] [CrossRef]
- Miltiadou, D.; Hager-Theodorides, A.L.; Symeou, S.; Constantinou, C.; Psifidi, A.; Banos, G.; Tzamaloukas, O. Variants in the 3′ untranslated region of the ovine acetyl-coenzyme A acyltransferase 2 gene are associated with dairy traits and exhibit differential allelic expression. J. Dairy Sci. 2017, 100, 6285–6297. [Google Scholar] [CrossRef]
- Yu, Q. Slc26a3 (DRA) in the Gut: Expression, Function, Regulation, Role in Infectious Diarrhea and Inflammatory Bowel Disease. Inflamm. Bowel Dis. 2021, 27, 575–584. [Google Scholar] [CrossRef]
- Hu, R.; Zou, H.; Wang, Z.; Cao, B.; Peng, Q.; Jing, X.; Wang, Y.; Shao, Y.; Pei, Z.; Zhang, X. Nutritional Interventions Improved Rumen Functions and Promoted Compensatory Growth of Growth-Retarded Yaks as Revealed by Integrated Transcripts and Microbiome Analyses. Front. Microbiol. 2019, 10, 318. [Google Scholar] [CrossRef] [PubMed]
- Aschenbach, J.R.; Bilk, S.; Tadesse, G.; Stumpff, F.; Gäbel, G. Bicarbonate-dependent and bicarbonate-independent mechanisms contribute to nondiffusive uptake of acetate in the ruminal epithelium of sheep. Am. J. Physiol. Gastrointest. Liver Physiol. 2009, 296, G1098–G1107. [Google Scholar] [CrossRef] [PubMed]
- Graham, C.; Gatherar, I.; Haslam, I.; Glanville, M.; Simmons, N.L. Expression and localization of monocarboxylate transporters and sodium/proton exchangers in bovine rumen epithelium. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2007, 292, R997–R1007. [Google Scholar] [CrossRef]
- Anhê, F.; Lellis-Santos, C.; Leite, A.R.; Hirabara, S.M.; Bordin, S. Smad5 regulates Akt2 expression and insulin-induced glucose uptake in L6 myotubes. Mol. Cell. Endocrinol. 2010, 319, 30–38. [Google Scholar] [CrossRef]
- Wang, S.; Tao, J.; Chen, H.; Kandadi, M.R.; Ren, J. Ablation of Akt2 and AMPKα2 rescues high fat diet-induced obesity and hepatic steatosis through Parkin-mediated mitophagy. Acta Pharm. Sin. B 2021, 11, 3508–3526. [Google Scholar] [CrossRef]
- Arshad, M.A.; Hassan, F.U.; Rehman, M.S.; Huws, S.A.; Cheng, Y.; Din, A.U. Gut microbiome colonization and development in neonatal ruminants: Strategies, prospects, and opportunities. Anim. Nutr. 2021, 7, 883–895. [Google Scholar] [CrossRef] [PubMed]
- Zhan, C.; Luo, X.; Chen, J.; Zhou, B.; Yang, G. Osteoprotegerin Promotes Liver Steatosis by Targeting the ERK-PPARγ-CD36 Pathway. Diabetes 2019, 68, 1902–1914. [Google Scholar] [CrossRef]
- Den Besten, G.; Bleeker, A.; Gerding, A.; Van Eunen, K.; Havinga, R.; Van Dijk, T.H.; Oosterveer, M.H.; Jonker, J.W.; Groen, A.K.; Reijngoud, D.J. Short-Chain Fatty Acids Protect against High-Fat Diet-Induced Obesity via a PPARγ-Dependent Switch From Lipogenesis to Fat Oxidation. Diabetes 2015, 64, 2398–2408. [Google Scholar] [CrossRef]
- Chen, W.; Chang, B.; Wu, X.; Li, L.; Sleeman, M.; Chan, L. Metabolism. Inactivation of Plin4 downregulates Plin5 and reduces cardiac lipid accumulation in mice. Am. J. Physiol. Endocrinol. 2013, 304, E770–E779. [Google Scholar] [CrossRef]

| Protein_ID | Protein Name | Gene Symbol | Mass | Protein_Coverage | Uniq_Pep | Uniq_Spec | log2Fold Change |
|---|---|---|---|---|---|---|---|
| W5PDY2 | Uncharacterized protein | - | 261,849.67 | 0 | 1 | 1 | 2.59 |
| A0A452EGR5 | Shisa family member 4 | SHISA4 | 26,983.28 | 0.03 | 1 | 2 | 2.48 |
| W5Q115 | semaphorin 3A | SEMA3A | 95,951.2 | 0.02 | 1 | 1 | 2.08 |
| W5NZJ6 | Tyrosine-protein kinase | FGR | 52,840.21 | 0.03 | 1 | 1 | 1.8 |
| W5P9E2 | SERPIN domain-containing protein | LOC101103612 | 47,177.77 | 0.18 | 1 | 1 | 1.6 |
| A0A452FDU1 | Phosphodiesterase | PDE6B | 97,921.12 | 0.01 | 1 | 1 | 1.57 |
| W5PMM4 | Aldo_ket_red domain-containing protein | LOC106990122 | 37,134.33 | 0.36 | 1 | 1 | 1.53 |
| A0A452F710 | RNA 3′-terminal phosphate cyclase | RTCA | 39,748.46 | 0.04 | 1 | 1 | 1.46 |
| W5P6R6 | Ubiquitin carboxyl-terminal hydrolase | USP27X | 59,865.01 | 0.04 | 1 | 1 | 1.36 |
| W5P8B8 | Zinc finger protein 48 | ZNF48 | 51,013.33 | 0.02 | 1 | 1 | 1.35 |
| A0A452FGG7 | Sulfotransferase | SULT1C2 | 34,285.13 | 0.08 | 1 | 1 | 1.3 |
| A0A452DK46 | Carbonic anhydrase | LOC102184245 | 29,121.58 | 0.41 | 1 | 12 | 1.27 |
| Q0PD85 | Monocarboxylate transporter 1 | SLC16A1 | 35,188.38 | 0.05 | 1 | 2 | 1.21 |
| W5QJ02 | Dehydrogenase/reductase 7 | DHRS7 | 37,262.55 | 0.31 | 1 | 2 | 1.21 |
| A0A452DKF9 | 4HBT domain-containing protein | - | 13,848.09 | 0.26 | 1 | 1 | 1.19 |
| W5PJY0 | ER membrane protein complex subunit 1 | EMC1 | 112,940.55 | 0.12 | 1 | 1 | 1.17 |
| A0A452ELD7 | BPI fold containing family A member 2 | BPIFA2 | 28,888.13 | 0.12 | 2 | 2 | 1.14 |
| W5QEK2 | Tubulin tyrosine ligase like 4 | TTLL4 | 133,873.12 | 0 | 1 | 1 | 1.13 |
| W5P9M9 | SERPIN domain-containing protein | LOC101103862 | 44,569.58 | 0.23 | 1 | 1 | 1.12 |
| A0A452EBL1 | Dihydrodiol dehydrogenase 3 | LOC102177638 | 37,351.57 | 0.33 | 1 | 1 | 1.08 |
| W5QAX2 | HDAC_interact domain-containing protein | SIN3B | 117,126.51 | 0.01 | 1 | 1 | 1.04 |
| W5P1M4 | Acetyl-CoA acyltransferase 2 | ACAA2 | 42,086.45 | 0.41 | 1 | 3 | 1.04 |
| A0A452G3K1 | Actin related protein 10 | ACTR10 | 44,622.44 | 0.02 | 1 | 1 | 1.01 |
| A0A452F7V6 | Centrin 2 | CETN2 | 19,100.59 | 0.08 | 1 | 1 | 0.99 |
| A0A452E3H3 | UDP-glucuronosyltransferase | LOC102172432 | 61,207.33 | 0.22 | 2 | 2 | 0.99 |
| A0A452GAC9 | 3-hydroxy-3-methylglutarate-CoA lyase | HMGCL | 34,621.11 | 0.3 | 8 | 24 | 0.98 |
| W5Q6A5 | Charged multivesicular body protein 2B | CHMP2B | 23,990.35 | 0.03 | 1 | 1 | 0.96 |
| A0A452FP65 | UDP-glucuronosyltransferase | LOC108633190 | 61,515.7 | 0.08 | 1 | 6 | 0.94 |
| W5QHL0 | Phospholipid-transporting ATPase | ATP11B | 125,527.01 | 0.01 | 1 | 1 | 0.93 |
| A0A452FF33 | Sodium/potassium-transporting ATPase subunit alpha | ATP1A3 | 108,265.03 | 0.16 | 1 | 1 | 0.93 |
| W5PUH3 | Interleukin 1 receptor type 2 | IL1R2 | 45,313.01 | 0.04 | 1 | 1 | 0.92 |
| W5PAD4 | Chloride anion exchanger | SLC26A3 | 84,603.08 | 0.05 | 4 | 7 | 0.92 |
| A0A452EX18 | Serine and arginine rich splicing factor 5 | SRSF5 | 31,584.45 | 0.03 | 1 | 1 | 0.91 |
| W5P699 | M-phase phosphoprotein 9 | MPHOSPH9 | 133,521.74 | 0.02 | 1 | 1 | 0.9 |
| W5PLM4 | Aldo_ket_red domain-containing protein | - | 32,650.38 | 0.33 | 1 | 1 | 0.9 |
| D4P8J3 | 3-hydroxy-3-methylglutaryl coenzyme A synthase | HMGCS2 | 57,306.89 | 0.31 | 13 | 105 | 0.89 |
| A0A452EWC8 | Monocarboxylate transporter 1 | SLC16A1 | 54,633.26 | 0.07 | 2 | 3 | 0.87 |
| A0A452FCI0 | Zinc finger protein 416 | LOC102179804 | 67,775.27 | 0.06 | 1 | 1 | 0.86 |
| A0A452DQX5 | Sulfotransferase | SULT1B1 | 34,876.74 | 0.11 | 3 | 3 | 0.86 |
| W5QAL6 | Formin like 3 | FMNL3 | 117,941.04 | 0.01 | 1 | 1 | 0.86 |
| W5PGI1 | Protein kinase X-linked | PRKX | 35,458.36 | 0.06 | 1 | 1 | 0.82 |
| A0A452DRA0 | Trichohyalin | TCHH | 188,388.47 | 0.05 | 2 | 3 | 0.81 |
| W5PZZ4 | Homeobox domain-containing protein | - | 11,162.59 | 0.12 | 1 | 1 | −0.81 |
| W5QHV0 | SET and MYND domain containing 1 | SMYD1 | 57,333.11 | 0.03 | 1 | 2 | −0.81 |
| W5PFB6 | CD177 molecule | CD177 | 47,924.93 | 0.03 | 1 | 1 | −0.81 |
| A0A452F2B2 | G protein subunit alpha o1 | GNAO1 | 40,653.28 | 0.07 | 1 | 1 | −0.81 |
| A0A452DVI8 | Anti-Muellerian hormone type-2 receptor | AMHR2 | 61,515.15 | 0.01 | 1 | 2 | −0.81 |
| A0A452FAG1 | Laminin subunit alpha 4 | LAMA4 | 205,009.31 | 0.15 | 21 | 32 | −0.81 |
| K4PF82 | RNA-binding protein with serine-rich domain 1 | RNPS1 | 34,157.69 | 0.13 | 2 | 2 | −0.81 |
| A0A452FD11 | Tropomyosin 1 | TPM1 | 32,731.66 | 0.23 | 3 | 10 | −0.84 |
| A0A452FTD0 | Ig-like domain-containing protein | - | 19,018.29 | 0.09 | 2 | 2 | −0.84 |
| A0A452E1D7 | Ankyrin repeat and SOCS box containing 7 | ASB7 | 36,426.64 | 0.02 | 1 | 1 | −0.84 |
| W5PX13 | Gap junction alpha-3 protein | GJA3 | 35,165.12 | 0.03 | 1 | 2 | −0.84 |
| A0A452FV55 | Collagen type XVI alpha 1 chain | COL16A1 | 147,706.13 | 0.01 | 1 | 1 | −0.84 |
| A0A452FBG8 | Smoothelin | SMTN | 98,772.31 | 0.09 | 6 | 7 | −0.86 |
| A0A452EMX2 | Tropomyosin 2 | TPM2 | 32,994.61 | 0.24 | 2 | 3 | −0.86 |
| A0A452G0P9 | Parvin alpha | PARVA | 46,577.9 | 0.11 | 3 | 5 | −0.86 |
| A0A452EAY9 | A0A452EAY9 | - | 28,201.62 | 0.29 | 1 | 1 | −0.86 |
| W5NYX4 | NACHT domain-containing protein | - | 104,305.02 | 0.01 | 1 | 1 | −0.86 |
| Q4LBD9 | Q4LBD9 | ovar-MHCI-H10 | 41,373.53 | 0.12 | 2 | 2 | −0.86 |
| A0A452EYA6 | Protein kinase C | PRKCB | 78,062.58 | 0.03 | 1 | 1 | −0.92 |
| W5PIF9 | Transforming growth factor beta 1 induced transcript 1 | TGFB1I1 | 51,299.58 | 0.04 | 2 | 3 | −0.94 |
| A0A452EUR3 | Guanylate cyclase | - | 46,878.37 | 0.02 | 1 | 1 | −0.94 |
| A0A452E7G7 | Family with sequence similarity 71 member A | FAM71A | 63,850.57 | 0.02 | 1 | 1 | −0.94 |
| W5Q4S0 | Collagen type III alpha 1 chain | COL3A1 | 139,701.25 | 0.08 | 8 | 35 | −0.94 |
| A0A452G5Y9 | Netrin 3 | NTN3 | 63,599.98 | 0.01 | 1 | 1 | −0.94 |
| W5PW78 | Sema domain-containing protein | SEMA4C | 81,534.11 | 0.02 | 1 | 1 | −0.94 |
| W5Q2A6 | EF-hand domain family member B | EFHB | 97,299.48 | 0.01 | 1 | 1 | −0.94 |
| A0A452G7W8 | Family with sequence similarity 110 member C | FAM110C | 30,168.37 | 0.03 | 1 | 1 | −0.97 |
| A1YZ35 | Mimecan | OGN | 34,450.92 | 0.29 | 8 | 26 | −0.97 |
| A0A452EWQ2 | SPEM family member 2 | SPEM2 | 56,745.55 | 0.02 | 1 | 1 | −0.97 |
| A0A452EV55 | A0A452EV55 | - | 213,096.55 | 0 | 1 | 1 | −0.97 |
| A0A452FFP6 | Beta-mannosidase | MANBA | 102,118.69 | 0.03 | 1 | 1 | −0.97 |
| A0A452G7S1 | HLA class II histocompatibility antigen, DM beta chain | LOC102187998 | 29,197.63 | 0.06 | 1 | 1 | −1 |
| A0A452F535 | General transcription factor IIF subunit 2 | - | 28,635.95 | 0.06 | 1 | 1 | −1 |
| A0A452ECP7 | Carbonic anhydrase | CA3 | 29,707.9 | 0.25 | 5 | 8 | −1 |
| W5NYU5 | USP domain-containing protein | USP43 | 112,010.29 | 0.01 | 1 | 2 | −1 |
| A0A452F0J4 | SPHK1 interactor, AKAP domain containing | SPHKAP | 182,094.96 | 0.01 | 1 | 2 | −1.06 |
| A0A452G4N4 | Adenylate cyclase type 3 | ADCY3 | 130,124.24 | 0.02 | 1 | 1 | −1.06 |
| A0A452F637 | Collagen type VI alpha 6 chain | COL6A6 | 246,602.86 | 0 | 1 | 1 | −1.09 |
| A0A452G885 | A0A452G885 | KRT4 | 56,270.57 | 0.39 | 13 | 30 | −1.09 |
| A0A452E7J1 | Olfactory receptor | OR1G1 | 35,499.99 | 0.13 | 1 | 1 | −1.09 |
| A0A452FWB4 | Corepressor interacting with RBPJ, 1 | CIR1 | 47,287.89 | 0.02 | 1 | 2 | −1.12 |
| A0A452FHU9 | Collagen type I alpha 1 chain | TPM1A1 | 139,952 | 0.18 | 15 | 179 | −1.12 |
| W5QHQ7 | Nucleolin | NCL | 73,334.54 | 0.15 | 1 | 1 | −1.12 |
| A0A452FMJ8 | Synaptopodin 2 | SYNPO2 | 115,845.7 | 0.24 | 1 | 1 | −1.12 |
| W5PY97 | HP domain-containing protein | SVIL | 246,128.41 | 0.02 | 1 | 1 | −1.12 |
| A0A452F3X3 | Sorbin and SH3 domain containing 2 | SORBS2 | 138,370.92 | 0.17 | 12 | 20 | −1.15 |
| Q6S5L3 | Major histocompatibility class II DQA1 | Cahi-DQA1 | 28,131.18 | 0.09 | 2 | 2 | −1.15 |
| A0A0H5FSL3 | MHC class I antigen | Ovar-I | 17,440.29 | 0.17 | 1 | 1 | −1.15 |
| A0A452G1W6 | Solute carrier family 9 member C2 (putative) | SLC9C2 | 129,059.02 | 0 | 1 | 2 | −1.15 |
| A0A452FBH2 | 2′-5′ oligoadenylate synthase | OAS1 | 41,810.87 | 0.13 | 1 | 2 | −1.18 |
| A0A452E3D3 | Perilipin 4 | PLIN4 | 126,234.86 | 0.13 | 12 | 16 | −1.18 |
| A0A224ATJ6 | Olfactory receptor | OR4X2 | 29,701.85 | 0.1 | 1 | 1 | −1.22 |
| A0A452EL11 | Chondroitinsulfatase | GALNS | 55,667.92 | 0.02 | 1 | 1 | −1.22 |
| A0A1S6YF29 | ATP synthase subunit a | ATP6 | 24,695.89 | 0.06 | 1 | 1 | −1.22 |
| W5PA36 | Zinc finger CCCH-type containing 7A | ZC3H7A | 112,014.91 | 0.01 | 1 | 1 | −1.22 |
| A0A452E3Q2 | Argonaute RISC catalytic component 2 | AGO2 | 97,447.94 | 0.09 | 1 | 1 | −1.29 |
| A0A452FXD3 | GMP reductase | GMPR | 34,625.42 | 0.07 | 1 | 1 | −1.29 |
| A0A452EA30 | PDZ and LIM domain 7 | PDLIM7 | 52,633.91 | 0.24 | 1 | 1 | −1.32 |
| W5Q756 | WH2 domain-containing protein | JMY | 108,165.74 | 0.02 | 1 | 1 | −1.4 |
| W5PI04 | Rab-GAP TBC domain-containing protein | TBC1D13 | 37,975.02 | 0.03 | 1 | 1 | −1.4 |
| W5PD82 | Caldesmon 1 | CALD1 | 89,883.93 | 0.14 | 9 | 20 | −1.43 |
| A0A452E4C4 | ADAM metallopeptidase with thrombospondin type 1 motif 16 | ADAMTS16 | 143,816.25 | 0 | 1 | 1 | −1.43 |
| A0A452FL85 | Profilin | PFN2 | 15,346.37 | 0.23 | 2 | 5 | −1.47 |
| A0A452DQP4 | Polycystin 1 like 2 | PKD1L2 | 272,917.19 | 0 | 1 | 1 | −1.47 |
| A0A452FL64 | Resistin | RETN | 12,192.84 | 0.24 | 1 | 1 | −1.51 |
| W5Q6L1 | IF rod domain-containing protein | KRT85 | 40,944.41 | 0.05 | 1 | 1 | −1.51 |
| W5NS84 | Ret finger protein-like 4A | LOC101116932 | 32,111.24 | 0.03 | 1 | 1 | −1.56 |
| Q9XSQ8 | MAP28 protein | map28 | 18,000.42 | 0.08 | 1 | 1 | −1.56 |
| W5NV41 | RING-type domain-containing protein | RNF175 | 38,416.48 | 0.07 | 1 | 1 | −1.56 |
| A0A452E9K4 | Cardiac phospholamban | PLN | 6229.38 | 0.21 | 1 | 1 | −1.6 |
| R9WH56 | MHC class I antigen | OLA-I | 41,077.25 | 0.1 | 1 | 1 | −1.6 |
| A0A452EH97 | Sorbin and SH3 domain containing 1 | SORBS1 | 142,840.96 | 0.15 | 1 | 3 | −1.6 |
| A0A452EKJ0 | Glycerophosphodiester phosphodiesterase domain containing 3 | GDPD3 | 36,818.99 | 0.04 | 1 | 1 | −1.64 |
| W5QB39 | Glutamate receptor interacting protein 2 | GRIP2 | 110,290.75 | 0.01 | 1 | 1 | −1.69 |
| A0A452FA76 | Voltage-dependent L-type calcium channel subunit alpha | CACNA1D | 239,629.01 | 0.01 | 1 | 1 | −1.79 |
| A0A452F4F7 | Sorbin and SH3 domain containing 2 | SORBS2 | 17,636.7 | 0.26 | 1 | 1 | −1.84 |
| A0A452EWT8 | NLR family pyrin domain containing 12 | NLRP12 | 119,902.48 | 0.01 | 1 | 1 | −2.25 |
| A0A452F5B1 | Semaphorin 3D | SEMA3D | 90,605.53 | 0.04 | 1 | 1 | −2.4 |
| A0A068B4V9 | Glutathione S-transferase | GSTA3 | 25,485.39 | 0.29 | 1 | 1 | −2.47 |
| Protein_ID | Protein Name | Gene Symbol | Mass | Protein_Coverage | Uniq_Pep | Uniq_Spec | log2Fold Change |
|---|---|---|---|---|---|---|---|
| Q28571 | Thyroid hormone receptor beta | THRB | 47,694.59 | 0.02 | 1 | 1 | 2.84 |
| A0A452EGR5 | Shisa family member 4 | SHISA4 | 26,983.28 | 0.03 | 1 | 2 | 2.84 |
| A0A452DTY3 | Olfactory receptor | LOC102188949 | 35,991.46 | 0.11 | 1 | 1 | 2.74 |
| A0A452EZP2 | Anoctamin | ANO9 | 89,634.08 | 0.01 | 1 | 2 | 2.38 |
| W5Q115 | Semaphorin 3A | SEMA3A | 95,951.2 | 0.02 | 1 | 1 | 2.3 |
| A0A452EMA9 | Oocyte-secreted protein 1 | LOC102171358 | 15,888.39 | 0.07 | 1 | 1 | 2.08 |
| A0A452FJ40 | Membrane cofactor protein | LOC102169209 | 37,010.88 | 0.05 | 1 | 1 | 1.56 |
| W5P5J3 | Adhesion G protein-coupled receptor A3 | ADGRA3 | 139,118.94 | 0 | 1 | 1 | 1.56 |
| W5P9E2 | Serpin B3-like | LOC101103612 | 47,177.77 | 0.18 | 1 | 1 | 1.53 |
| W5QJ02 | Dehydrogenase/reductase 7 | DHRS7 | 37,262.55 | 0.31 | 1 | 2 | 1.52 |
| A0A452FNE3 | PDZ domain containing 3 | PDZD3 | 52,094.46 | 0.04 | 1 | 2 | 1.5 |
| A0A452DVC9 | Nanos C2HC-type zinc finger 3 | NANOS3 | 19,372.36 | 0.05 | 1 | 1 | 1.49 |
| A0A452F7Y9 | Fer-1 like family member 6 | FER1L6 | 206,943 | 0.01 | 1 | 1 | 1.44 |
| W5NZJ6 | Tyrosine-protein kinase | FGR | 52,840.21 | 0.03 | 1 | 1 | 1.4 |
| W5PDY2 | Uncharacterized protein | - | 261,849.67 | 0 | 1 | 1 | 1.26 |
| A0A452EBL1 | Dihydrodiol dehydrogenase 3 | LOC102177638 | 37,351.57 | 0.33 | 1 | 1 | 1.26 |
| A0A452F4Q5 | Dynein axonemal heavy chain 8 | DNAH8 | 544,382.22 | 0 | 1 | 1 | 1.24 |
| W5P1M4 | Acetyl-CoA acyltransferase 2 | ACAA2 | 42,086.45 | 0.41 | 1 | 3 | 1.24 |
| W5NZV0 | Heme oxygenase (biliverdin-producing) | HMOX2 | 43,155.73 | 0.3 | 1 | 1 | 1.22 |
| W5PFZ8 | Chromosome 20 C6orf141 homolog | C20H6orf141 | 20,592.16 | 0.05 | 1 | 1 | 1.21 |
| A0A452G516 | Oxysterol-binding protein | OSBPL11 | 85,188.23 | 0.02 | 1 | 1 | 1.21 |
| W5NWX7 | C-type lectin domain family 4 member A | CLEC4A | 27,849.45 | 0.04 | 1 | 1 | 1.16 |
| A0A452G154 | NEDD1 gamma-tubulin ring complex targeting factor | NEDD1 | 71,754.15 | 0.01 | 1 | 1 | 1.14 |
| W5PLM4 | Aldo_ket_red domain-containing protein | - | 32,650.38 | 0.33 | 1 | 1 | 1.1 |
| W5PGI1 | Protein kinase X-linked | PRKX | 35,458.36 | 0.06 | 1 | 1 | 1.04 |
| W5PTZ0 | Membrane spanning 4-domains A5 | MS4A5 | 22,614.45 | 0.07 | 1 | 1 | 1.02 |
| A0A452DK46 | Carbonic anhydrase | LOC102184245 | 29,121.58 | 0.41 | 1 | 12 | 1.01 |
| A0A452G3K1 | Actin related protein 10 | ACTR10 | 44,622.44 | 0.02 | 1 | 1 | 0.99 |
| A0A452EBG2 | Tyrosine-protein kinase | ZAP70 | 69,121.32 | 0.03 | 1 | 1 | 0.99 |
| A0A452EGG2 | Rho GTPase activating protein 9 | ARHGAP9 | 79,394.55 | 0.02 | 1 | 1 | 0.91 |
| A0A452FP65 | UDP-glucuronosyltransferase | LOC108633190 | 61,515.7 | 0.08 | 1 | 6 | 0.9 |
| A0A452FEA5 | Nicotinate-nucleotide pyrophosphorylase [carboxylating] | QPRT | 30,980.17 | 0.02 | 1 | 1 | 0.88 |
| A0A452FGG7 | Sulfotransferase | SULT1C2 | 34,285.13 | 0.08 | 1 | 1 | 0.86 |
| A0A452G0M2 | Fraser extracellular matrix complex subunit 1 | FRAS1 | 452,350.77 | 0 | 1 | 1 | 0.85 |
| A0A452EWC8 | Monocarboxylate transporter 1 | SLC16A1 | 54,633.26 | 0.07 | 2 | 3 | 0.83 |
| W5PLW4 | Aldo_ket_red domain-containing protein | - | 37,979.63 | 0.26 | 1 | 2 | 0.82 |
| A0A452FG87 | Microcephalin | MCPH1 | 84,188.78 | 0.01 | 1 | 1 | 0.82 |
| W5QEK2 | Tubulin tyrosine ligase like 4 | TTLL4 | 133,873.12 | 0 | 1 | 1 | 0.81 |
| A0A452ELR2 | EvC ciliary complex subunit 2 | EVC2 | 138,071.86 | 0.01 | 1 | 1 | −0.81 |
| W5PCE0 | Phospholipase B-like | PLBD2 | 63,556.04 | 0.01 | 1 | 1 | −0.81 |
| P02080 | Hemoglobin subunit beta-C(NA) | - | 15,662.26 | 0.22 | 1 | 1 | −0.81 |
| W5PY97 | Supervillin | SVIL | 246,128.41 | 0.02 | 1 | 1 | −0.81 |
| A0A452G7Z1 | Hematopoietic cell-specific Lyn substrate 1 | HCLS1 | 52,381.17 | 0.02 | 1 | 1 | −0.81 |
| A0A452FBH2 | 2′-5′ oligoadenylate synthase | OAS1 | 41,810.87 | 0.13 | 1 | 2 | −0.81 |
| A0A452FBG8 | Smoothelin | SMTN | 98,772.31 | 0.09 | 6 | 7 | −0.84 |
| A0A452G7W8 | Family with sequence similarity 110 member C | FAM110C | 30,168.37 | 0.03 | 1 | 1 | −0.84 |
| W5P981 | Non-specific serine/threonine protein kinase | AKT2 | 53,026.59 | 0.02 | 1 | 1 | −0.84 |
| A0A452E8I8 | Uncharacterized protein | - | 20,701.89 | 0.21 | 3 | 3 | −0.84 |
| W5Q1F0 | ER lumen protein-retaining receptor | KDELR3 | 21,740.39 | 0.04 | 1 | 1 | −0.84 |
| A0A452F2B2 | G protein subunit alpha o1 | GNAO1 | 40,653.28 | 0.07 | 1 | 1 | −0.84 |
| A0A452DW39 | 60S ribosomal protein L29 | - | 16,612.21 | 0.08 | 1 | 4 | −0.86 |
| Q3LRQ1 | Vitronectin | - | 51,056.51 | 0.03 | 1 | 1 | −0.86 |
| W5PW78 | Semaphorin 4C | SEMA4C | 81,534.11 | 0.02 | 1 | 1 | −0.86 |
| A0A452ETB0 | Collagen type XXVIII alpha 1 chain | COL28A1 | 115,775.7 | 0.01 | 1 | 2 | −0.86 |
| A0A452EHK0 | Melanoma cell adhesion molecule | MCAM | 71,399.1 | 0.1 | 6 | 9 | −0.86 |
| A0A452EFB7 | Serine peptidase inhibitor, Kazal type 9 | SPINK9 | 9851.55 | 0.29 | 2 | 3 | −0.86 |
| G1DFY5 | Transforming growth factor beta-1-induced transcript 1 protein | TGFB1I1 | 50,629.24 | 0.02 | 1 | 1 | −0.86 |
| W5PMH6 | Lipocalin 2 | LCN2 | 23,169.06 | 0.09 | 1 | 1 | −0.86 |
| A0A452F2Y9 | Actin alpha 2, smooth muscle | ACTA2 | 42,073.87 | 0.63 | 6 | 94 | −0.89 |
| A0A452E3D3 | Perilipin 4 | PLIN4 | 126,234.86 | 0.13 | 12 | 16 | −0.89 |
| A0A452E6P5 | C-type lectin domain family 1 member B | CLEC1B | 26,077.02 | 0.02 | 1 | 1 | −0.89 |
| A0A452ECP7 | Carbonic anhydrase | CA3 | 29,707.9 | 0.25 | 5 | 8 | −0.89 |
| W5QHQ7 | Nucleolin | NCL | 73,334.54 | 0.15 | 1 | 1 | −0.89 |
| A0A452F4A0 | Sorbin and SH3 domain containing 2 | SORBS2 | 69,256.66 | 0.08 | 4 | 8 | −0.92 |
| W5P3N6 | Beta-hexosaminidase | LOC101112162 | 62,702.01 | 0.08 | 1 | 1 | −0.92 |
| W5Q4S0 | Collagen type III alpha 1 chain | COL3A1 | 139,701.25 | 0.08 | 8 | 35 | −0.92 |
| W5PFB6 | CD177 molecule | CD177 | 47,924.93 | 0.03 | 1 | 1 | −0.92 |
| A0A452DUI8 | Carcinoembryonic antigen-related cell adhesion molecule 7 | LOC102172184 | 41,661.16 | 0.02 | 1 | 1 | −0.94 |
| A0A452FL86 | TBC1 domain family member 32 | TBC1D32 | 146,388.94 | 0.01 | 1 | 1 | −0.94 |
| W5P0I2 | Complex I-B14.5a | NDUFA7 | 12,670.69 | 0.18 | 2 | 3 | −0.94 |
| A0A452FHU9 | Collagen type I alpha 1 chain | COL1A1 | 139,952 | 0.18 | 15 | 179 | −0.94 |
| W5Q2A6 | EF-hand domain family member B | EFHB | 97,299.48 | 0.01 | 1 | 1 | −0.94 |
| W5PI04 | TBC1 domain family member 13 | TBC1D13 | 37,975.02 | 0.03 | 1 | 1 | −0.94 |
| A0A452FFP6 | Beta-mannosidase | MANBA | 102,118.69 | 0.03 | 1 | 1 | −0.97 |
| A0A452E5B5 | Myosin heavy chain 11 | MYH11 | 228,250.52 | 0.27 | 40 | 123 | −0.97 |
| W5NS84 | Ret finger protein-like 4A | LOC101116932 | 32,111.24 | 0.03 | 1 | 1 | −0.97 |
| A0A452E7J1 | Olfactory receptor | OR1G1 | 35,499.99 | 0.13 | 1 | 1 | −1 |
| A0A452EWP1 | Synemin | SYNM | 171,645.21 | 0.14 | 18 | 34 | −1 |
| A0A452EYA6 | Protein kinase C | PRKCB | 78,062.58 | 0.03 | 1 | 1 | −1 |
| W5PD82 | Caldesmon 1 | CALD1 | 89,883.93 | 0.14 | 9 | 20 | −1.03 |
| E7EC28 | Thymosin beta | LOC102182562 | 5031.51 | 0.59 | 2 | 2 | −1.03 |
| A0A452DVI8 | Anti-Muellerian hormone type-2 receptor | AMHR2 | 61,515.15 | 0.01 | 1 | 2 | −1.06 |
| A0A452EL11 | Chondroitinsulfatase | GALNS | 55,667.92 | 0.02 | 1 | 1 | −1.06 |
| A0A452EUR3 | Guanylate cyclase | - | 46,878.37 | 0.02 | 1 | 1 | −1.06 |
| A0A452E0C7 | Olfactory receptor | - | 35,416.77 | 0.05 | 1 | 1 | −1.06 |
| F8T866 | MHC class II antigen | DQA | 19,937.83 | 0.07 | 1 | 1 | −1.09 |
| A0A452FD11 | Tropomyosin 1 | TPM1 | 32,731.66 | 0.23 | 3 | 10 | −1.09 |
| A0A452F535 | General transcription factor IIF subunit 2 | - | 28,635.95 | 0.06 | 1 | 1 | −1.09 |
| Q30DP7 | Type II small proline-rich protein | SPRR2A | 6489.94 | 0.3 | 1 | 4 | −1.12 |
| A0A452G1W6 | Solute carrier family 9 member C2 (putative) | SLC9C2 | 129,059.02 | 0 | 1 | 2 | −1.12 |
| A0A224ATJ6 | Olfactory receptor | OR4X2 | 29,701.85 | 0.1 | 1 | 1 | −1.12 |
| A0A452E4C4 | ADAM metallopeptidase with thrombospondin type 1 motif 16 | ADAMTS16 | 143,816.25 | 0 | 1 | 1 | −1.15 |
| A0A452G885 | Keratin 4 | KRT4 | 56,270.57 | 0.39 | 13 | 30 | −1.15 |
| A0A452EH97 | Sorbin and SH3 domain containing 1 | SORBS1 | 142,840.96 | 0.15 | 1 | 3 | −1.18 |
| W5PBX5 | Mitogen-activated protein kinase kinase kinase 1 | MAP3K1 | 157,095.44 | 0.01 | 1 | 1 | −1.18 |
| A1E458 | Adipocyte-type fatty acid-binding protein | LOC100861279 | 14,805.48 | 0.32 | 3 | 9 | −1.18 |
| A0A452G893 | S100 calcium binding protein A8 | S100A8 | 10,399.34 | 0.16 | 2 | 3 | −1.18 |
| W5Q6L1 | Keratin, type II microfibrillar, component 5-like | KRT85 | 40,944.41 | 0.05 | 1 | 1 | −1.22 |
| B5TQZ6 | 3beta-hydroxysteroid dehydrogenase/isomerase | 3BHSD | 43,108.07 | 0.07 | 1 | 1 | −1.25 |
| A0A452E9K4 | Cardiac phospholamban | PLN | 6229.38 | 0.21 | 1 | 1 | −1.25 |
| W5NV41 | Ring finger protein 175 | RNF175 | 38,416.48 | 0.07 | 1 | 1 | −1.25 |
| Q6S5L3 | Major histocompatibility class II DQA1 | Cahi-DQA1 | 28,131.18 | 0.09 | 2 | 2 | −1.25 |
| W5Q624 | Keratin, type II cytoskeletal 73 | KRT73 | 57,075.43 | 0.05 | 1 | 1 | −1.29 |
| A0A452EKJ0 | Glycerophosphodiester phosphodiesterase domain containing 3 | GDPD3 | 36,818.99 | 0.04 | 1 | 1 | −1.32 |
| A0A452EDZ4 | Protein S100 | S100A12 | 10,922.68 | 0.24 | 2 | 6 | −1.32 |
| R9WH56 | MHC class I antigen | OLA-I | 41,077.25 | 0.1 | 1 | 1 | −1.36 |
| Q9XSQ8 | MAP28 protein | map28 | 18,000.42 | 0.08 | 1 | 1 | −1.4 |
| A0A452FL64 | Resistin | RETN | 12,192.84 | 0.24 | 1 | 1 | −1.51 |
| W5QB39 | Glutamate receptor interacting protein 2 | GRIP2 | 11,0290.8 | 0.01 | 1 | 1 | −1.51 |
| A0A068B4V9 | Glutathione S-transferase | GSTA3 | 25,485.39 | 0.29 | 1 | 1 | −2.32 |
| A0A452F5B1 | Semaphorin 3D | SEMA3D | 90,605.53 | 0.04 | 1 | 1 | −2.64 |
| Protein_ID | UniProt Protein Name | Gene Symbol | Mass | Protein_Coverage | Uniq_Pep | Uniq_Spec | log2Fold Change |
|---|---|---|---|---|---|---|---|
| A0A452DTY3 | Olfactory receptor | LOC102188949 | 35,991.4566 | 0.109 | 1 | 1 | 2.54 |
| A0A452DVC9 | Nanos C2HC-type zinc finger 3 | NANOS3 | 19,372.3607 | 0.051 | 1 | 1 | 1.74 |
| A0A452G516 | Oxysterol-binding protein | OSBPL11 | 85,188.2306 | 0.017 | 1 | 1 | 1.46 |
| A0A0H5FSL3 | MHC class I antigen | Ovar-I | 17,440.2913 | 0.17 | 1 | 1 | 1.20 |
| W5QJ03 | Dehydrogenase/reductase 7 | DHRS7 | 38,509.2022 | 0.301 | 1 | 1 | 1.17 |
| A0A452E7G7 | Family with sequence similarity 71 member A | FAM71A | 63,850.5707 | 0.024 | 1 | 1 | 1.14 |
| A0A452FJ40 | Membrane cofactor protein | LOC102169209 | 37,010.882 | 0.051 | 1 | 1 | 1.14 |
| W5PFZ8 | Chromosome 20 C6orf141 homolog | C20H6orf141 | 20,592.1564 | 0.049 | 1 | 1 | 1.14 |
| A0A452G154 | NEDD1 gamma-tubulin ring complex targeting factor | NEDD1 | 71,754.1493 | 0.012 | 1 | 1 | 1.10 |
| A0A452FB93 | CD276 molecule | CD276 | 57,663.7688 | 0.038 | 1 | 1 | 1.08 |
| A0A452DUD2 | DNA polymerase epsilon catalytic subunit | POLE | 263,670.995 | 0.006 | 1 | 1 | 1.06 |
| A0A452EWQ2 | SPEM family member 2 | SPEM2 | 56,745.5547 | 0.018 | 1 | 1 | 1.04 |
| W5PRU6 | Gap junction protein | GJA8 | 49,509.247 | 0.014 | 1 | 1 | 1.00 |
| W5NWX7 | C-type lectin domain-containing protein | CLEC4A | 27,849.4487 | 0.042 | 1 | 1 | 0.97 |
| A0A452EIF3 | Transient receptor potential cation channel subfamily V member 4 | TRPV4 | 98,757.5519 | 0.018 | 1 | 1 | 0.89 |
| A0A452EB62 | Uncharacterized protein | - | 12,026.4189 | 0.09 | 2 | 2 | −0.81 |
| A0A452E2N5 | Structural maintenance of chromosomes 6 | SMC6 | 125,408.243 | 0.006 | 1 | 1 | −0.81 |
| A0A452E8Z1 | Golgi associated kinase 1A | GASK1A | 61,628.5216 | 0.011 | 1 | 1 | −0.81 |
| A0A452EZS9 | RAB, member of RAS oncogene family like 3 | RABL3 | 25,857.2573 | 0.026 | 1 | 1 | −0.86 |
| A0A452DKF9 | 4HBT domain-containing protein | - | 13,848.0892 | 0.264 | 1 | 1 | −0.86 |
| A0A452FF33 | Sodium/potassium-transporting ATPase subunit alpha | ATP1A3 | 108,265.03 | 0.1575 | 1 | 1 | −0.86 |
| A0A452EP10 | Keratin, type II cytoskeletal 71 | KRT71 | 55,027.6975 | 0.279 | 1 | 2 | −0.86 |
| W5PAY2 | WW domain-containing protein | WWC2 | 129,564.039 | 0.005 | 1 | 1 | −0.89 |
| W5PMM4 | Aldo_ket_red domain-containing protein | LOC106990122 | 37,134.3273 | 0.356 | 1 | 1 | −0.89 |
| W5Q6A5 | Charged multivesicular body protein 2B | CHMP2B | 23,990.3458 | 0.028 | 1 | 1 | −0.92 |
| W5Q9M6 | WD_REPEATS_REGION domain-containing protein | - | 47,001.9663 | 0.08 | 1 | 1 | −0.94 |
| G1DFX7 | BolA-like protein 2 | BOLA2 | 10,138.2385 | 0.081 | 1 | 1 | −0.94 |
| A0A452F7V6 | Centrin 2 | CETN2 | 19,100.5855 | 0.078 | 1 | 1 | −1.00 |
| E7EC28 | Thymosin beta | LOC102182562 | 5031.50567 | 0.591 | 2 | 2 | −1.09 |
| G8FRI8 | Copper chaperone of superoxide dismutase 1 | - | 23,909.9303 | 0.031 | 1 | 1 | −1.09 |
| W5QAL6 | Formin like 3 | FMNL3 | 117,941.038 | 0.012 | 1 | 1 | −1.09 |
| W5P9M9 | SERPIN domain-containing protein | LOC101103862 | 44,569.5781 | 0.225 | 1 | 1 | −1.18 |
| A0A452F710 | RNA 3′-terminal phosphate cyclase | RTCA | 39,748.4576 | 0.044 | 1 | 1 | −1.51 |
| A0A452EYK5 | Interferon induced protein with tetratricopeptide repeats 2 | IFIT2 | 54,161.2524 | 0.013 | 1 | 1 | −1.56 |
| W5P8B8 | Zinc finger protein 48 | ZNF48 | 51,013.327 | 0.018 | 1 | 1 | −1.60 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhuang, Y.; Lv, X.; Cui, K.; Chai, J.; Zhang, N. Early Solid Diet Supplementation Influences the Proteomics of Rumen Epithelium in Goat Kids. Biology 2023, 12, 684. https://doi.org/10.3390/biology12050684
Zhuang Y, Lv X, Cui K, Chai J, Zhang N. Early Solid Diet Supplementation Influences the Proteomics of Rumen Epithelium in Goat Kids. Biology. 2023; 12(5):684. https://doi.org/10.3390/biology12050684
Chicago/Turabian StyleZhuang, Yimin, Xiaokang Lv, Kai Cui, Jianmin Chai, and Naifeng Zhang. 2023. "Early Solid Diet Supplementation Influences the Proteomics of Rumen Epithelium in Goat Kids" Biology 12, no. 5: 684. https://doi.org/10.3390/biology12050684
APA StyleZhuang, Y., Lv, X., Cui, K., Chai, J., & Zhang, N. (2023). Early Solid Diet Supplementation Influences the Proteomics of Rumen Epithelium in Goat Kids. Biology, 12(5), 684. https://doi.org/10.3390/biology12050684







