Simple Summary
TIR-NBS-LRR (TNL) is a disease resistance gene family that responds to biotic stress in many plants, but the systematic analysis of this gene family and the expression response to biotic stress have rarely been reported in roses. In the present study, 96 intact TNL gene family members were identified by bioinformatics in Rosa chinensis, and analyzed from the perspectives of evolutionary relationships, conserved structures, expression regulation, collinear relationships, and expression patterns. Some of the TNL genes responded to hormones and fungal disease: RcTNL23 demonstrated strong responses to three hormones and three pathogens. In addition, some TNL genes responded significantly to the black spot pathogen that we isolated, and different members may be involved in different stages of disease defense. In conclusion, we found that the TNL gene family is involved in the response to fungal disease and may function as a disease resistance gene in the rose. The present study lays a theoretical foundation for the functional study of TNL genes and the mining of disease resistance gene in roses, which will inform the selection and breeding of disease-resistant varieties.
Abstract
Roses, which are one of the world’s most important ornamental plants, are often damaged by pathogens, resulting in serious economic losses. As a subclass of the disease resistance gene family of plant nucleotide-binding oligomerization domain (NOD)-like receptors, TIR-NBS-LRR (TNL) genes play a vital role in identifying pathogen effectors and activating defense responses. However, a systematic analysis of the TNL gene family is rarely reported in roses. Herein, 96 intact TNL genes were identified in Rosa chinensis. Their phylogenies, physicochemical characteristics, gene structures, conserved domains and motifs, promoter cis-elements, microRNA binding sites, and intra- and interspecific collinearity relationships were analyzed. An expression analysis using transcriptome data revealed that RcTNL genes were dominantly expressed in leaves. Some RcTNL genes responded to gibberellin, jasmonic acid, salicylic acid, Botrytis cinerea, Podosphaera pannosa, and Marssonina rosae (M. rosae); the RcTNL23 gene responded significantly to three hormones and three pathogens, and exhibited an upregulated expression. Furthermore, the black spot pathogen was identified as M. rosae. After inoculating rose leaves, an expression pattern analysis of the RcTNL genes suggested that they act during different periods of pathogen infection. The present study lays the foundations for an in-depth investigation of the TNL gene function and the mining of disease resistance genes in roses.
1. Introduction
As sessile organisms, plants have evolved complex immune systems in response to the invasion of pathogens from the external environment. These systems comprise two lines of defense. The first, called pattern-triggered immunity (PTI), is the activation of downstream immune responses by pattern recognition receptors (PRRs). PRRs are localized on cell membranes and recognize pathogen-associated molecular patterns [1,2]. In response to PTI, pathogens secrete effectors that inhibit the activity of PRRs and their complexes, interfering with downstream signaling [3]. Plants have also evolved intracellular nucleotide-binding oligomerization domain (NOD)-like receptors (NLRs). NLRs, also known as nucleotide-binding site–leucine-rich repeats (NBS-LRRs), can directly or indirectly identify pathogen effectors and trigger the plant immune response. The second line of defense in plants is called effector-triggered immunity (ETI) [1,4,5,6]. ETI reactions are more intense than PTI reactions, and can cause hypersensitive responses [5].
Approximately 61% of the more than 310 cloned plant disease resistance genes belong to the NLR gene family [7]. Consequently, NLR proteins, which are also known as resistance gene analogs, have been extensively studied in numerous plants to elucidate their effects on pathogens [4,8]. According to the phylogenetic analysis of NLR genes in angiosperms, they can be divided into three subcategories: TIR-NBS-LRR (TNL), CC-NBS-LRR, and RPW8-NBS-LRR. The N-terminus of TNL protein contains a Toll/interleukin-1 receptor (TIR) domain [9], which is mainly distributed in dicots and is missing in monocots [9,10]. TIR domains typically contain different but highly conserved TIR-1, TIR-2, and TIR-3 motifs [10]. The NBS domain generally contains eight conserved motifs: P-loop (phosphate-binding loop), RNBS-A (resistance nucleotide binding site A), RNBS-B, RNBS-C, RNBS-D, kinase 2, GLPL (Gly-Leu-Pro-Leu, also called kinase 3), and MHDV (Met-His-Asp-Val) [10,11,12,13]. In addition, TNL genes may contain integrated domains (IDs) that serve as bait for pathogen effectors, such as the WRKY domain found in the RRS1 (resistance to Ralstonia solanacearum 1) protein in Arabidopsis thaliana (A. thaliana) [14,15].
Approximately 60 TNL genes are associated with disease resistance in 21 plant species (Table S1). In Gossypium hirsutum, GhDSC1 responds to verticillium wilt (Verticillium dahliae) infection by upregulating expression, and silencing the GhDSC1 gene makes plants more susceptible to verticillium wilt. A single nucleotide polymorphism (SNP) mutation of the GhDSC1 protein P-loop motif removes the defense response to verticillium wilt in susceptible cotton [16,17]. An RT4-4 gene involved in the cucumber mosaic virus (CMV) resistance response, which interacts with the elicitor CMV 2a to induce a necrotic response, has been identified in the common bean [18]. The overexpression of the GmKR3 gene may enhance the resistance of soybeans to a variety of viruses, but does not affect the yield and quality of the plants [19]. Genetic analysis and transgenic verification in potato have proven that the Gro1-4 gene in the Globodera rostochiensis (G. rostochiensis) resistance locus is resistant to pathotype Ro1 G. rostochiensis [20]. The Glomerella leaf spot (GLS, Colletotrichum fructicola) resistance locus was mapped in apple, and MdTNL1 was considered as a candidate gene [21]. Its overexpression can significantly increase apple resistance to GLS, and an SNP on this gene can be used to distinguish resistant and susceptible germplasms [22]. Table S1 summarizes the TNL genes involved in disease resistance.
As one of the most prominent ornamental plants worldwide, roses are known as the “Queen of flowers” and are widely sold as cut flowers (accounting for approximately 30% of the market), garden plants, and potted plants with important economic value and cultural connotations [23]. However, during cultivation, roses are often damaged by black spot and powdery mildew, which affect their normal growth and flowering [24]. Cut roses are often affected by gray mold during storage and transportation [25]. These three diseases are prevalent among roses worldwide and have a significant impact on their ornamental and economic value. Therefore, it is extremely important to identify disease resistance genes in roses [24]. Map-based cloning is a traditional method for identifying the target genes of a trait, but it is limited by the long study periods required and the complex genomes of roses. Consequently, a few resistance genes have been identified using this method [24]. In recent years, with the rapid development of genomics and sequencing technology, genome-wide gene family analyses have been successfully applied to the mining of disease resistance genes in roses. Examples of such genes include RcbZIP17 of the bZIP family [26], RcWRKY41 of the WRKY family [27], and RcERF099 of the AP2/ERF family [28]. These genes participate in resistance to gray mold and have been used in virus-induced gene silencing for the preliminary verification of gene function in roses.
The NLR gene family has been identified in many plants, including members of the Rosaceae family [9,29,30,31,32,33,34,35]. Although the TNL gene family has been identified in Rosa chinensis (R. chinensis) and an evolutionary analysis has been conducted [33], more in-depth research is required. Therefore, in the present study, to determine the function of the TNL gene family in roses, 96 intact TNL genes were identified in R. chinensis. The genes were analyzed from the perspectives of evolution, physicochemical properties, protein structure conservation, gene expression regulation, and collinear relationships. Transcriptome data were also used to analyze the expression pattern of RcTNL genes and their response to exogenous hormones and three different pathogens. The black spot pathogen in the roses was identified as Marssonina rosae (M. rosae). The expression pattern of TNL genes in response to M. rosae infection was detected using qRT-PCR. Therefore, we performed a comprehensive analysis of the TNL gene family in R. chinensis. Our findings will provide a foundation for the functional study of TNL genes and the identification of disease resistance genes in roses.
2. Materials and Methods
2.1. Identification of TNL Genes in R. chinensis
Possible TNL gene family members in A. thaliana were obtained from The Arabidopsis Information Resource (TAIR) (https://www.arabidopsis.org/, last accessed on 13 August 2022), NIBLRRS website (https://niblrrs.ucdavis.edu/data_links.php, last accessed on 17 August 2022), and from Zhang et al. (2016) [31]. The TNL protein sequences of A. thaliana were aligned to the ‘Old Blush’ genome [36] to obtain possible homologous TNL genes in R. chinensis. The Batch CD-Search tool provided by NCBI was used to select the Pfam database in order to determine the conserved domains of the proteins, and genes containing TIR, NBS, and LRR domains were obtained. In addition, the Pfam-A.hmm file was obtained from the Pfam website (http://pfam.xfam.org/, last accessed on 18 August 2022), and the Simple HMM Search tool in the TBtools v1.106 software was used to search for genes containing the TIR (Pfam ID: PF01582) and NB-ARC (Pfam ID: PF00931) domains, respectively [37]. The Batch CD-Search tool was used to obtain the conserved domains of the proteins, and to screen for genes that contained all three domains. The results of the two identification methods were combined to obtain the possible TNL genes in R. chinensis.
2.2. Gene Characteristics and Phylogenetic Analysis
Information about the coding sequence (CDS), number of exons, and distribution of the RcTNL genes on the chromosomes was obtained from the ‘Old Blush’ genome annotation file [36]. The distribution of RcTNL genes on the chromosomes was visualized using the MG2C tool [38]. The molecular weight, isoelectric point, instability index, aliphatic index, and grand average of hydropathicity of each protein were calculated using the ExPASy ProtParam tool (https://web.expasy.org/protparam/, last accessed on 19 August 2022). The subcellular localization of proteins was predicted using CELLO v2.5 (http://cello.life.nctu.edu.tw/, last accessed on 19 August 2022). The protein sequences of RcTNL were first aligned using MUSCLE [39], and a phylogenetic tree was constructed using MEGA X. The evolutionary history was inferred using the maximum likelihood method with 1000 bootstrap replicates and the w/freq. model [40].
2.3. Analysis of Gene Structures, Promoters, and Conserved Motifs
The general feature format (GFF) file containing the ‘Old Blush’ genome annotation information was used to obtain the structure of the RcTNL gene, and was subsequently combined with the genome file to obtain the sequence 2 kb upstream of the gene initiation codon. This sequence was identified as the promoter sequence. The cis- elements were predicted using the PlantCARE database [41]. The protein sequences of RcTNL genes were submitted to the MEME Suite v5.5.1 website to obtain their conserved motif information [42]. The results of this analysis were displayed using TBtools v1.106 software [37].
2.4. Prediction of microRNA (miRNA) Target Sites on the Genes
Published Malus domestica (M. domestica) miRNA sequences were selected from the psRNATarget website to predict the possible miRNA binding sites on RcTNL mRNA. Schema V2 (released in 2017) was selected as the scoring schema, but others use the default settings [43].
2.5. Analysis of Gene Duplication Events and Collinearity
After BLASTP was used for all rose protein sequences and combined with the GFF file of the genome, MCScanX and TBtools v1.106 software were used to analyze and plot the duplication type and collinearity relationships between the RcTNL gene family members [37,44]. GFF files and protein sequences of Rosa rugosa (R. rugosa) and Rosa wichuraiana (R. wichuraiana) were obtained from the Genome Database for Rosaceae [45], and the collinearity of the R. chinensis genome and those of the two species mentioned above were analyzed and displayed using MCScanX and TBtools v1.106 software [37,44]. Based on the CDS and protein sequences of the RcTNL gene pairs, their Ka and Ks values were calculated using TBtools v1.106 software [37].
2.6. Transcriptome Data Acquisition and Analysis
The transcriptome data pertaining to different rose tissues were obtained from the SRA database (https://www.ncbi.nlm.nih.gov/sra/?term=, last accessed on 13 December 2022). The study ID for R. chinensis cv. ’Old Blush’ root, stem, leaf, and prickle data was SRP200448 and that for R. chinensis cv. ‘Old Blush’ flower bud and open flower data was SRP115334. The study ID for six hormone treatments of ‘Samantha‘ was SRP186551 [46]. The study ID for the ‘Samantha’ response to Botrytis cinerea (B. cinerea) was SRP120271 [46]. The study ID for the transcriptome data related to the ‘Pariser Charme’ response to M. rosae and Podosphaera pannosa (P. pannosa) was SRP136240 [47]. The fastq files for the transcriptome data mentioned above were analyzed with the kallisto program [48] using the ‘Old Blush’ genome as a reference [36] to obtain the count and transcripts per million (TPM) values of gene expression. DESeq2 was used to perform differential expression analysis on count data for various comparisons [49]. Differentially expressed genes (DEGs) were identified using the absolute value of a log2 fold change ≥ 1 and a padj < 0.05. The gene expression heatmaps were generated using TBtools v1.106, and the log2TPM values were used [37].
2.7. Isolation, Identification, and Inoculation of the Black Spot Pathogen
The black spot pathogen was isolated from infected rose leaves by monosporic isolation. After DNA of the fungal strain was extracted by E.Z.N.A. ® Fungal DNA Kit (Omega Bio-tek Inc., Norcross, GA, USA), internal transcribed spacer (ITS) sequence was amplified with 2 × KeyPo Mas-ter Mix (Vazyme Biotech Co., Ltd., Nanjing, China ) and then sequenced. MEGA X software was used to construct a phylogenetic tree based on the ITS sequence to determine the pathogen species. The isolated and purified fungal strain (DBE24-1) was transferred to a complete medium (1% glucose, 0.1% yeast extract, 0.1% Ca(NO3)2·4H2O, 0.02% KH2PO4, 0.025% MgSO4·7H2O, 0.015% NaCl, 0.05% casein enzymatic hydrolysate, and 0.05% casein acid hydrolysate), incubated at 23 °C while agitating at 180 rpm for approximately 15 days, and filtered through sterilized lens paper to obtain the conidia fluid. A hemocytometer was used to calculate the conidia concentration, which was then adjusted to 2 × 105 colony-forming units (CFUs)/mL by adding 0.1% Tween-20 to the solution. The fourth and fifth leaves from the top of each ‘Old Blush’ plant were picked and sterilized with an aqueous solution of 0.06% to 0.09% NaClO for 2 min, then washed twice with sterile water. Each leaf retained the top three leaflets, which were moisturized and stored after inoculation, as described by Zurn et al. (2018) [50]. The conidia fluid was sprayed evenly on the leaf surface. Three biological replicates were set up at each time-point, and each biological replicate was composed of three leaves of a plant. Sterile water containing 0.1% Tween-20 was used as the control to inoculate the same number of leaves. Samples were collected at 0 h, 24 h, 48 h, 6 days, and 12 days after inoculation, respectively.
2.8. qRT-PCR Analysis of RcTNL Genes Response to M. rosae
RNA was extracted from the samples collected in Section 2.7 using a Quick RNA Isolation Kit (Beijing Huayueyang Biotechnology Co., Ltd., Beijing, China) and a PrimeScript™ RT reagent Kit with gDNA Eraser (Takara Bio Inc., Kusatsu, Japan) for reverse transcription synthesis of complementary DNA, all according to the manufacturer’s instructions. Nine pathogen-responsive RcTNL genes were randomly selected for qRT-PCR analysis. UBC and GAPDH were used as reference genes, and the primer sequences are presented in Table S14. The fluorescence signals were detected using a CFX384™ Real-Time System (Bio-Rad, Hercules, CA, USA) after the PCR reaction had been configured using ChamQ Universal SYBR qPCR Master Mix (Vazyme Biotech Co., Ltd., Nanjing, China). Three technical replicates were performed for each biological replicate using the method described by Song et al. (2021) [51]. The relative expression of each gene was calculated using the 2−ΔΔCT method [52]. Statistical analysis of the relative expression levels of the various treatments was performed using a t-test on GraphPad Prism 8 (GraphPad Software, Inc., San Diego, CA, USA).
3. Results
3.1. Identification and Phylogenetic Analysis of the TIR-NBS-LRR (TNL) Genes
A total of 73 TNL genes annotated on the TAIR website, 92 TNL genes from the NIBLRRS website, and 79 TNL genes identified by Zhang et al. (2016) using the Pfam database [31] were combined to obtain 107 TNL genes from A. thaliana (Table S2). The sequences of these genes were aligned to the R. chinensis reference genome [36] to obtain 1275 possible homologous genes. The conserved domains of these genes were analyzed by the Batch CD-Search tool (NCBI), and 96 of the possible homologous genes contained TIR, NBS, and LRR domains. In addition, using the hidden Markov model (HMM) search method, a total of 183 genes were found to contain both TIR and NBS domains, 95 of which contained TIR, NBS, and LRR domains and were included in the 96 genes. These 96 genes were considered to be the TNL genes of R. chinensis and were named RcTNL01 through RcTNL96 in order of chromosome number and physical location from low to high (Figure 1, Table S3). They were distributed unevenly across the seven chromosomes of R. chinensis with up to 24 on chromosome 1, 22 on chromosome 5, and only 4 on chromosome 2 (Figure 1, Table S3).
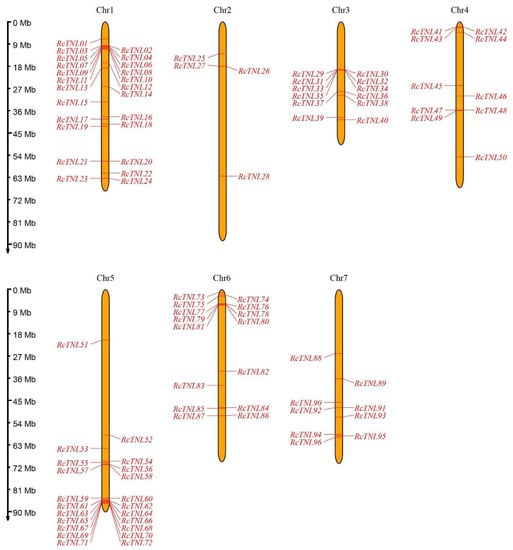
Figure 1.
Distribution of TNL genes on Rosa chinensis (R. chinensis) chromosomes. The rulers on the left indicate the physical size of each chromosome; the ruler unit is megabase (Mb) pair. The black text across the top represents the chromosome number.
A phylogenetic tree was constructed using the maximum likelihood method to study the evolutionary relationship between RcTNL family members in R. chinensis. The 96 RcTNL genes were divided into six clades, of which, clade I contained the most (up to 41 members) and clade II contained the least (only 4 members) (Figure 2).
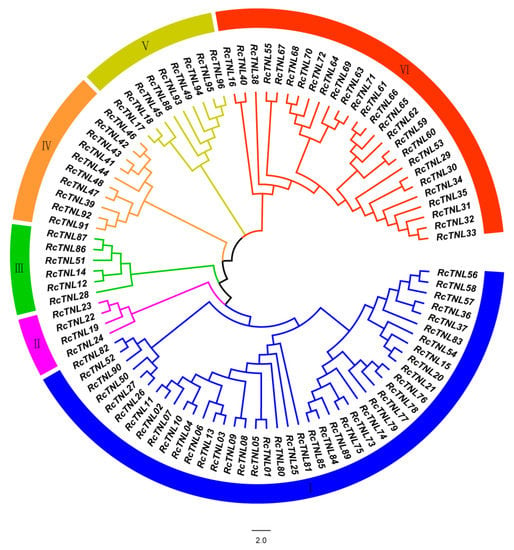
Figure 2.
Phylogenetic analysis of TNL proteins in R. chinensis. The innermost circle is a phylogenetic tree of RcTNL proteins constructed by MEGA X using the maximum likelihood method; the colors represent the six clades. The outermost circle uses blocks of the same six colors to represent the clades; the Roman numerals in the blocks are the clade numbers.
3.2. Analysis of the Physicochemical Properties of the RcTNL Proteins
The number of exons in the RcTNL genes varied greatly from 2 to 19. The RcTNL proteins contained 656 (RcTNL48) to 3431 (RcTNL64) amino acids. The molecular weights of their respective proteins ranged from 74.31 (RcTNL48) to 388.72 kDa (RcTNL64), and the isoelectric points ranged from 5.08 (RcTNL63) to 8.93 (RcTNL95). In total, 87.5% (84) of all of the RcTNL proteins had an instability index greater than 40, indicating that most RcTNL proteins may be unstable. The aliphatic indexes of the RcTNL proteins ranged from 87.38 (RcTNL46) to 106.95 (RcTNL48). The grand average of the hydropathicity of the RcTNL proteins ranged from −0.386 (RcTNL16) to −0.061 (RcTNL77); all of the values were negative, indicating that the RcTNL proteins were hydrophilic. Subcellular localization prediction revealed that 71 (73.96%) RcTNL proteins were in the nucleus, 15 (15.62%) were in the cytoplasm, and 10 (10.42%) were in the plasma membrane (Table S3). This suggests that most RcTNL proteins are primarily intracellular and may be involved in the ETI response.
3.3. Domain and Conserved Motif Analysis of the RcTNL Proteins
We analyzed the domains of the RcTNL proteins. Their domains were arranged in the order: TIR, NBS, and LRR. We found that 22.92% (22) of the genes contained two to five LRR domains, e.g., RcTNL26 contained five LRR domains. In addition, the WRKY, Rx_N, Pkinase, NAP, and zf-RVT domains were present in 8.33% (8) of the RcTNL proteins (Figure 3a). The additional domains are called IDs. They may act as bait to perceive pathogen effectors, particularly the WRKY, Pkinase, and zf-RVT domains [15,53].
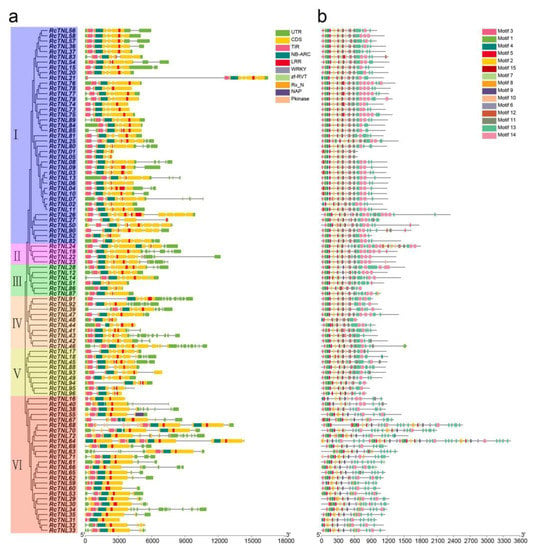
Figure 3.
Phylogeny, conserved domains, and motifs of RcTNL proteins and their gene structures. (a) Gene structures and conserved domains of the RcTNL genes. The phylogenetic tree of the RcTNL genes is on the left with the six clades indicated by different background colors. Roman numerals indicate the clade numbers; rectangles of assorted colors to the right represent untranslated regions (UTRs), coding sequences (CDSs), and domains. The horizontal lines connecting rectangles represent introns. The rulers indicate base pair (bp) values. (b) Conserved motifs of the RcTNL proteins. The 15 motifs are represented by different colored rectangles. The horizontal lines of tandem rectangles indicate the extent of the protein.
The MEME Suite was used to analyze the conserved motifs of the RcTNL proteins. The motif arrangement was more conserved at the 5′ ends than at the 3′ ends of the proteins. Fifteen motifs were predicted on the RcTNL proteins, and they existed simultaneously on 52.08% (50) of all RcTNL proteins. The probability of each motif appearing on an RcTNL protein was 69.79–100% (Figure 3b, Table S4). Among the identified motifs, TIR-1 (motif 3), TIR-2 (motif 1), TIR-3 (motif 4), TIR-4 (motif 5), RNBS-B (motif 7), RNBS-D (motif 10), GLPL (motif 9), P-loop (motif 2), Kinase 2 (motif 15), and MHDV (motif 6) were also conserved in other species [10,11,13]. TIR-4 (motif 5), MHDV (motif 6), RNBS-B (motif 7), and GLPL (motif 9) were present in all of the RcTNL proteins (Figure 3b), indicating that these four motifs might have crucial functions.
3.4. Cis-Element Analysis of the RcTNL Promoters
Transcription factors can regulate the expression of genes by binding cis-elements in the promoter region. The PlantCARE database was used to predict the promoter region cis-elements that were used to study the regulation of RcTNL gene expression. The light responsive elements in the promoter region of each RcTNL gene were most abundant and were present in all of the genes; each gene contained at least two. The RcTNL gene promoter regions contained cis-elements associated with jasmonic acid (JA), abscisic acid (ABA), gibberellin (GA), auxin, and salicylic acid (SA), indicating that the expression of RcTNL genes may be regulated by these five hormones. Cis-elements related to stress and defense were also present in the promoter regions of the RcTNL genes (Figure 4 and Figure S1, Table S5), indicating that RcTNL genes may be involved in the defense against biotic and abiotic stress, as reported in other species [12,14,54].
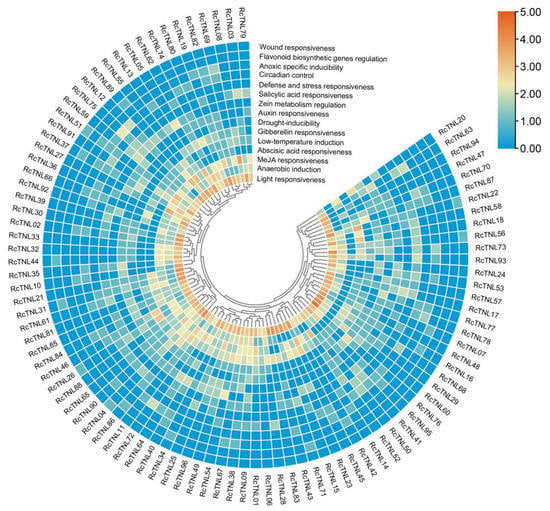
Figure 4.
Classification and number of cis-elements in the promoter regions of the RcTNL genes. The heatmap was plotted by taking log2 fold of the number of cis-elements. The color shift from blue to red indicates an increasing number of cis-elements.
3.5. Prediction of Target Binding Sites for miRNAs on the RcTNL Genes
In plants such as A. thaliana [54], soybean [55], and tomato [56], miRNAs can regulate plant immune responses to pathogens by targeting TNL genes. To determine whether RcTNL genes were targeted by miRNAs, 207 published M. domestica miRNA sequences were selected from the psRNATarget website to predict possible miRNA binding sites on the RcTNL genes [43]. A total of 164 miRNAs from 42 miRNA families were identified. These miRNAs regulate genes primarily at the transcription level in a cleavage manner (Table S6). Among them, the miR482 family targeted the most RcTNL genes at 67 (69.79%), followed by the miR171, miR169, and miR396 families, which targeted 26 (27.08%), 20 (20.83%), and 17 (17.70%) RcTNL genes, respectively. Moreover, we found that multiple miRNAs were capable of targeting a single RcTNL, e.g., RcTNL85 might be regulated by 13 miRNAs. One miRNA was also capable of targeting multiple RcTNL genes, e.g., miR482b could target 61 RcTNL genes (Figure 5, Table S6). This is similar to previous studies [13,57].

Figure 5.
Possible microRNA binding sites for five RcTNL genes. Double dots between bases indicate a successful pairing, single dot indicates an additional pairing between U and G, and a blank indicates no pairing.
3.6. Duplication and Collinearity Analysis of the RcTNL Genes
A collinearity analysis of the R. chinensis genome revealed four pairs of RcTNL genes with segmental duplication events (including eight RcTNL genes) and two pairs of RcTNL genes with tandem duplication events located on chromosomes 3 and 5 (Table 1, Figure S2). The RcTNL gene pairs were in the same evolutionary clade and were mainly located in clades I and VI. In addition, 12 RcTNL genes had a collinearity relationship with 14 non-TNL genes, of which, RcTNL04 and RcTNL42 had segmental duplication events with two non-TNL genes, respectively (Table S7).

Table 1.
Ka/Ks ratio of the duplicated RcTNL gene pairs in collinear blocks.
To study the selection pressure of the RcTNL genes at the protein level, the Ka to Ks ratio of the RcTNL gene pairs was calculated. The Ka/Ks values of the six RcTNL gene pairs ranged from 0.443 to 0.653. All values were less than 1, indicating that these RcTNL gene pairs underwent purification or negative selection (Table 1). This is similar to the observation that NLR genes in Rosaceae also undergo purification selection [33].
A collinearity analysis of R. chinensis with R. rugosa and R. wichuraiana revealed that, whether based only on TNL genes or on all genes, the collinear relationship between R. chinensis and R. wichuraiana was stronger than that between R. chinensis and R. rugosa. Of the 96 RcTNL genes, 41 were collinear with R. rugosa and 45 were collinear with R. wichuraiana (Figure 6, Table S8).
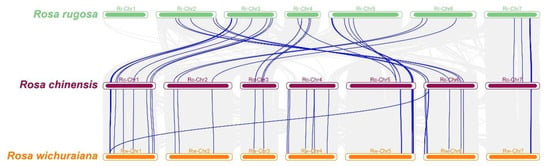
Figure 6.
Collinearity of R. chinensis with Rosa rugosa (R. rugosa) and Rosa wichuraiana (R. wichuraiana). Chromosomes of R. rugosa, R. chinensis, and R. wichuraiana are indicated by green, purple, and orange bars, respectively, and are arranged by chromosome number from low (left) to high (right). The blue lines between the chromosomes of the different species indicate collinearity blocks with RcTNL genes, and the gray lines indicate blocks of collinearity between genomes.
3.7. Expression Pattern Analysis of the RcTNL Genes
We analyzed the ‘Old Blush’ transcriptome data from the root, stem, leaf, prickle, flower bud, and open flower published on the SRA database. The expression of RcTNL genes in the leaf was higher than in any of the other tissues, followed by the stem and root. Among these genes, RcTNL12, RcTNL95, and RcTNL28 exhibited higher expression levels in the leaf, and RcTNL12, RcTNL33, and RcTNL74 exhibited higher expression levels in the stem. RcTNL33, RcTNL19, and RcTNL63 exhibited high expression levels in all tissues. Some RcTNL genes were only highly expressed in specific tissues, e.g., RcTNL95 in leaf (Figure 7, Table S9). These results indicate that, under non-pathogenic conditions, the overall expression of RcTNL genes is low, but a few genes exhibit higher expression levels in specific tissues.
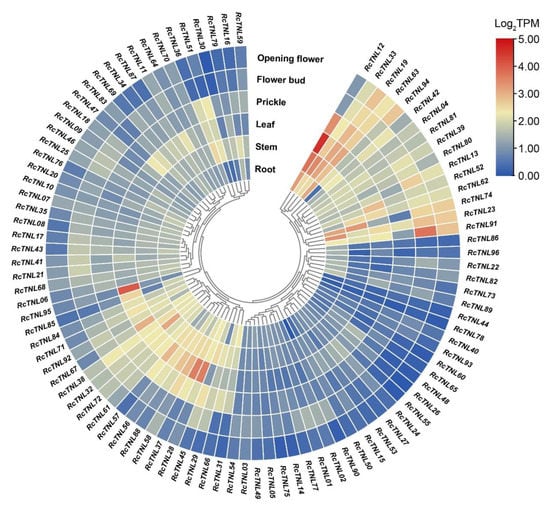
Figure 7.
Expression patterns of RcTNL genes in various R. chinensis tissues. The expression of RcTNL genes in the various tissues is represented on a heatmap using log2TPM (transcripts per million) values. The innermost circle shows clustering by gene expression patterns. The blocks change from blue to red, indicating a change in the log2TPM values from low to high.
3.8. Response of the RcTNL Genes to Exogenous Hormones
Because the promoter regions of some RcTNL genes contain hormone responsive elements (Figure 4), it is possible that they are regulated by exogenous hormones. The transcriptome sequencing was performed on petals after treatment with six hormones (GA, JA, SA, ABA, 1-naphthaleneacetic acid (NAA), and 2,4-dichlorophenoxy acetic acid (2,4-D)) and deionized water (CK) using ’Samantha’ cut flowers [58]. The differentially expressed RcTNL genes of six hormonal treatments compared to CK were analyzed separately. The results revealed that 15, 9, and 26 RcTNL genes were differentially expressed after GA, JA, and SA treatment, respectively, but the remaining hormone treatments did not result in the differential expression of the RcTNL genes. Integrating the differential expression of the RcTNL genes after the three effective hormone treatments (GA, JA, and SA), 35 RcTNL genes were found to be differentially expressed after at least one hormone treatment, and all of them exhibited different degrees of upregulated expression (Figure 8, Table S10). Combining the distribution of the three hormone response elements on the promoters of these RcTNL genes revealed that 60%, 44.44%, and 30.77% of the RcTNL genes that responded to GA, JA, and SA, respectively, contained corresponding cis-elements on the promoters (Figure 4, Table S5). This indicates that these RcTNL genes may be directly or indirectly regulated by these three hormones.

Figure 8.
Heatmap of RcTNL genes differentially expressed in ‘Samantha’ petals after hormone treatment. CK denotes treatment with deionized water (the control), and GA, JA, and SA denote treatment with gibberellin, jasmonic acid, and salicylic acid, respectively. The change in color of the circles from blue to red indicates the change in the log2TPM values from low to high.
3.9. Response of the RcTNL Genes to Fungal Pathogens
In order to investigate the response of RcTNL genes to fungal pathogens, transcriptome data were analyzed during infection by B. cinerea, M. rosae, and P. pannosa [46,47]. At 30 h and 48 h after the infection of ‘Samantha’ petals by B. cinerea, 1 and 11 RcTNL genes were differentially expressed, respectively, compared to the expression in the control petals. This indicates that the response of the RcTNL genes to B. cinerea mainly occurred at 48 h. After inoculation with B. cinerea, four RcTNL genes were differentially expressed between the two time-points. The differentially expressed RcTNL genes in the two comparisons described above were integrated; 12 genes were differentially expressed in at least one comparison and were considered to respond to B. cinerea. These genes will be referred to as RB-TNL genes. Compared to the uninfected control, 8 and 10 RB-TNL genes were upregulated at 30 h and 48 h, respectively. Eight RB-TNL genes were upregulated at 48 h after infection rather than at 30 h (Figure 9a, Table S11). It is likely that these differentially expressed members of the RcTNL family participate in the defense response against B. cinerea, mostly through an increased expression.
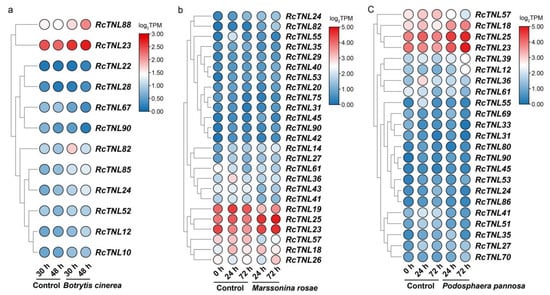
Figure 9.
Expression pattern of RcTNL genes in response to fungal disease. (a) Heatmap of RcTNL genes differentially expressed in ‘Samantha’ petals during inoculation with Botrytis cinerea. (b) Heatmap of RcTNL genes differentially expressed in ‘Pariser Charme’ leaves during inoculation with Marssonina rosae. (c) Heatmap of RcTNL genes differentially expressed in ‘Pariser Charme’ leaves during inoculation with Podosphaera pannosa. The change in color of the circles from blue to red indicates a change in the log2TPM values from low to high.
In ‘Pariser Charme’ leaves infected with M. rosae, 12 and 5 differentially expressed RcTNL genes were observed at 24 h and 72 h of infection, respectively, compared to in the control leaves. A total of 14 (24 h vs. 0 h), 6 (72 h vs. 24 h), and 12 (72 h vs. 0 h) differentially expressed RcTNL genes were observed between the time-points. After integrating these DEGs, 25 RcTNLs were found to be differentially expressed in at least one comparison and were considered to respond to M. rosae. These genes will be referred to as RM-TNL genes. Compared to the control leaves, 17 RM-TNL genes exhibited a downregulated expression at 24 h, and 17 RM-TNL genes exhibited an upregulated expression at 72 h of infection. The expression pattern of the RM-TNL gene during infection with M. rosae was characterized by both continuous upregulation (10 genes) and downregulation followed by upregulation (10 genes; Figure 9b, Table S12).
‘Pariser Charme’ leaves were inoculated with P. pannosa and followed through three time-points. Compared to the control leaves, 7 and 12 differentially expressed RcTNL genes were observed at 24 h and 72 h of infection, respectively, and 5 (24 h vs. 0 h), 8 (72 h vs. 24 h), and 13 (72 h vs. 0 h) differentially expressed RcTNL genes were observed between the time-points. After integrating these DEGs, 23 RcTNLs were found to be differentially expressed in at least one comparison in response to P. pannosa. These genes will be referred to as RP-TNL genes. Compared to the control leaves, 13 RP-TNL genes exhibited a downregulated expression at 24 h and 12 RP-TNL genes exhibited an upregulated expression at 72 h. The RP-TNL genes were mainly upregulated and then downregulated (8 genes), continuously upregulated (7 genes), and continuously downregulated (6 genes) during P. pannosa infection (Figure 9c, Table S13).
A total of 15 common genes were differentially expressed in ’Pariser Charme‘ leaves infected with M. rosae and P. pannosa, of which, seven exhibited the same expression patterns at all three time-points of infection with the two pathogens (Figure 9b,c). It is possible that these genes participate in defense against these two pathogens in similar ways. RcTNL23, RcTNL24, and RcTNL90 were differentially expressed in the roses inoculated with the three pathogens; only RcTNL23 had the same expression pattern during infection with the three pathogens, and all exhibited an upregulated expression. This indicates that RcTNL23 may participate in defense against the three pathogens in the same way. RcTNL24 was upregulated during infection with M. rosae and B. cinerea, and first upregulated and then downregulated during infection with P. pannosa. However, the expression pattern of RcTNL90 was different during the infection of three pathogens (Figure 9). The results indicate that, when dealing with different types of pathogens, members of the RcTNL family may have different functions.
3.10. Expression Patterns of the RcTNL Genes in Response to M. rosae
Rose leaves infected with black spot disease in the field were used for pathogen isolation. A phylogenetic tree was constructed after amplifying the ITS sequence of the isolated strain (DBE24-1), sequencing, and alignment. The pathogen was identified as M. rosae (Figure S3), as in a previous study [59,60,61]. The surfaces of leaves from the two groups of ’Old Blush‘ were inoculated with DBE24-1 (DR) and sterile water (CK), respectively, and samples were collected at 0 h, 24 h, 48 h, 6 days, and 12 days post-inoculation. Black spots began to appear on the leaves after 6 days, and at 12 days, the disease index was determined (Figure S4), as in a previous study [59]. The disease index of ‘Old Blush’ was 48.89. Of the RcTNL genes identified in response to pathogens (Figure 9), nine were randomly selected for qRT-PCR analysis. Compared to the control, all nine RcTNL genes were significantly upregulated at one or more time-point after M. rosae inoculation, while RcTNL88 showed up-regulation at all four time-points after inoculation. The response of these genes to M. rosae was the most at 12 days, followed by 6 days and 24 h, whereas only RcTNL88 responded at 48 h (Figure 10, Table S15).
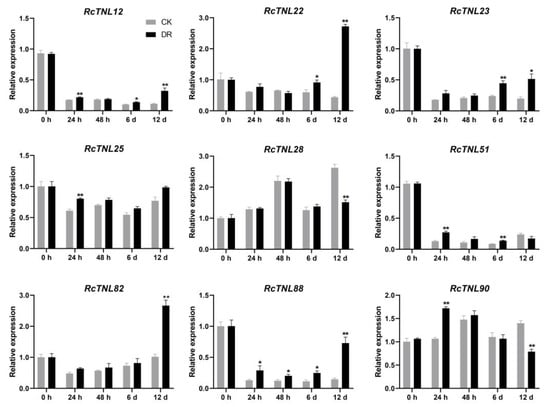
Figure 10.
Expression patterns of nine RcTNL genes during infection with Marssonina rosae. The gray and black columns indicate the relative expression levels (mean ± standard deviation) of the RcTNL genes after inoculation with sterile water (CK) and DBE24-1 conidia (DR), respectively. Statistical analysis of the relative expression levels of CK and DR was performed using a t-test; * and ** indicate adjusted p-values of < 0.05 and < 0.01, respectively.
4. Discussion
Roses are important ornamental plants, and are widely planted worldwide. Their growth and flower quality is often damaged by various diseases, including black spot, powdery mildew, and gray mold. However, only a small number of disease resistance genes have been cloned and studied [24,25,47,62]. In recent years, the publication of the reference genome and related omics data pertaining to Rosa plants has provided a shortcut for the identification of disease resistance genes [23,36,45]. As a subclass of the NLR gene family, TNL genes have similar functions in terms of the defense response to pathogens. In plants, the immune response to pathogens is mainly triggered by the direct or indirect identification of pathogen effectors [5,8]. TNL genes have been reported as disease resistance genes in a variety of plants (Table S1), such as cotton [17], soybean [19], apple [22], and potato [20]. In the present study, the TNL gene family was identified in R. chinensis and analyzed by bioinformatics. Transcriptome data were used to mine TNL genes that were possibly involved in the defense response, and the responses of nine TNL genes to M. rosae were detected by qRT-PCR.
4.1. Frequency and Duplication Type of the RcTNL Genes
The frequency of identified TNL genes varies greatly among plant species. For example, the frequency of intact TNL genes is as high as 0.319% in Vitis vinifera [29], 0.215% in Euryale ferox [34], 0.117% in potato [12], and only 0.003% in Actinidia chinensis [32]. In the present study, 96 intact TNL genes containing TIR, NBS, and LRR domains were identified in R. chinensis, and their frequency was 0.211%, similar to that in Euryale ferox. Guo et al. (2022) reported a frequency of intact TNL genes of only 0.01% in R. chinensis [33]. This difference may have been due to the different genomes and identification methods used for TNL genes.
A collinearity analysis of the genome revealed that a total of 18 segment and 13 tandem duplication events contributed to the expansion of the TNL gene family in R. chinensis, and occurred mainly between TNL and non-TNL genes (Table S7). However, there were four segment and two tandem duplication events between the TNL genes. This corroborates the results reported by Dubey et al. (2022) following their analysis of TNL gene duplication events in potato [12]. We believe that this was mainly due to the intact TNL genes identified in the present study. Genes that did not simultaneously comprise three domains were non-TNL genes, even if they contained two domains. This may explain why some TNL genes have a collinear relationship with non-TNL genes.
4.2. Possible Functions of RcTNL Protein Domains
NLR proteins usually contain TIR, CC, RPW8, NBS, and LRR domains. TNL proteins, which constitute a subclass of NLR proteins, mainly contain TIR, NBS, and LRR domains. The TIR domain can recognize and interact with pathogen effectors, and its heterodimization is required for the formation of functional immune receptor complexes [63]. NBS domains can bind and hydrolyze ATP, transform proteins from active to inactive states, and act as molecular switches in the signal transduction process [64,65]. The LRR domain affects the specificity of NLR proteins for effector recognition [66], and mediates the interaction of NLR proteins with pathogen effectors [67]. In the present study, 96 RcTNL proteins contained these three domains, of which, 22.92% contained two to five LRR domains. These conserved domains may be critical for the immune response in roses.
TNL proteins contain other IDs in addition to the three domains described above. Among these, the WRKY and kinase domains are the most common, and may be used as bait for pathogen effectors to participate in plant defense [15,53]. In the present study, the RcTNL proteins contained five IDs distributed on eight RcTNL proteins. Of these, three RcTNL proteins contained the WRKY domain (Figure 3a). In A. thaliana, the WRKY domain of RRS1 (a TNL protein) is capable of detecting pathogen effectors that target the WRKY protein, which is involved in pathogenic defense [14]. In the present study, the response of RcTNL genes to three pathogens was analyzed (Figure 9). Six out of eight RcTNL genes containing IDs were involved in the pathogen response. Among these genes, RcTNL23, which contains a WRKY domain, responded strongly to all of the pathogens tested. It may have played an important role in the defense against the three fungal diseases. RcTNL55, which contains an NAP domain, responded to both M. rosae and P. pannosa, and may be involved in the defense against these two pathogens. In A. thaliana, the domains of interaction between NLR proteins and pathogen effectors contain various IDs, including WRKY and NAP [53]. Therefore, it is speculated that RcTNL proteins may also bind to pathogen effectors by their IDs, thereby participating in defense against pathogens. Gene silencing or gene editing techniques can be used to further validate the function of RcTNL genes.
4.3. Regulation of RcTNL Gene Expression
An analysis of the promoter region of the RcTNL gene revealed the presence of cis-elements related to plant hormones and the defense response (Figure 4, Table S5). Therefore, the transcriptome data pertaining to cut rose flowers treated with six hormones (2,4-D, ABA, GA, JA, NAA, and SA) were analyzed. Compared to the control, 15, 9, and 26 RcTNL genes were upregulated to varying degrees after GA, JA, and SA treatment, respectively, and responded to the hormones. As in plants such as cotton [17], cassava [35], and grape [68], we found that the TNL genes were significantly upregulated after the exogenous application of SA. In addition, the TNL genes involved in disease resistance in cassava [35] and grape [68] exhibited similar expression patterns during exogenous SA application and pathogen infection.
In the present study, the cis-elements related to defense and stress responsiveness on the RcTNL promoters were mainly TC-rich repeats (Table S5), which are also present in the promoter region of the VpTNL1 gene in Vitis pseudoreticulata. The promoter truncation experiment further suggested that this element may play an important role in the immune response to Erysiphe necator. In addition, TCA elements in the promoter region of this gene are associated with the response to SA [68]. In the present study, the SA response elements in the promoter region of the RcTNL genes were also mainly TCA elements (Table S5), and it is speculated that this element is also important for the SA regulation of RcTNL expression.
miRNA is involved in the expression regulation of genes as a negative regulator, and many studies have shown that it regulates the defense response of plants by targeting NLR genes [31]. The miR482 family, which reportedly targets the NLR gene family in various plants such as tomato [69], potato [70], and cotton [71], is involved in the defense response to pathogens. In the present study, we predicted possible miRNA binding sites on the RcTNL genes and found that the miR482 family targeted 69.79% of the RcTNL genes. Therefore, the miR482 family may be the main candidate for research into the regulation of RcTNL gene expression. The expression patterns of miRNAs and their targeted RcTNL genes in response to pathogens, combined with transgene, transcriptome, and degradation sequencing technologies, may provide further verification. It has been reported that miRNAs mainly target conserved regions of NLR genes, such as P-loop, Kinase-2, and MHDV motifs in the NB-ARC domain [31,72]. This may have been the reason for why a single miRNA was capable of targeting multiple RcTNL genes in the present study.
5. Conclusions
In the present study, 96 intact TNL family genes were identified in R. chinensis by bioinformatics. A phylogenetic analysis enabled us to divide the genes into six clades. It was predicted that RcTNL proteins would be mainly localized in the nucleus and would be unstable and hydrophilic. Eight of the RcTNL proteins contained IDs in addition to TIR, NBS, and LRR domains. Multiple conserved RcTNL protein motifs also existed in multiple species. Most of the RcTNL gene promoter regions contained cis-elements associated with hormones, stress, and defense, and there were miRNA target sites on the RcTNL genes. Segmental and tandem duplication events in the RcTNL genes may have contributed to the expansion of this gene family.
Compared to the control, 15, 9, and 26 RcTNL genes were upregulated and differentially expressed in cut rose petals after treatment with GA, JA, and SA, respectively. In addition, 12, 25, and 23 RcTNL genes responded to B. cinerea, M. rosae, and P. pannosa, respectively. Among these, RcTNL23, RcTNL24, and RcTNL90 all responded to the three pathogens, and RcTNL23 exhibited an upregulated expression. We isolated and identified the rose black spot pathogen as M. rosae, and inoculated rose leaves with it. The expression patterns of nine RcTNL genes in five stages of infection with M. rosae were obtained by qRT-PCR analysis, and the responses of the RcTNL genes to the pathogens were further verified. The results of the present study will lay the foundation for the functional study of TNL genes in roses, and may inform the mining of disease resistance genes and the selection and breeding of resistant rose varieties.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/biology12030426/s1, Figure S1: Distribution of cis-elements on the RcTNL gene promoters; Figure S2: Segmental duplicated RcTNL gene pairs in R. chinensis; Figure S3: Molecular identification of the black spot pathogen strain DBE24-1 in the rose; Figure S4: Black spot symptoms on ‘Old Blush’ leaves after inoculation with the DBE24-1 strain; Table S1: Information on TNL genes as resistance genes; Table S2: Collection and combination of TNL genes in Arabidopsis; Table S3: Characteristics of TNL genes in R. chinensis; Table S4: Conserved motifs information of TNL proteins in R. chinensis; Table S5: Cis-elements information of TNL gene promoters in R. chinensis; Table S6: Predicted binding sites on TNL genes targeted by miRNA in R. chinensis; Table S7: Information on TNL gene duplication events in R. chinensis; Table S8: Orthologous TNL gene pairs between R. chinensis and two other species; Table S9: TPM values of TNL genes in different tissues of R. chinensis; Table S10: TPM values of TNL genes differentially expressed in response to hormones in R. chinensis; Table S11: TPM values of TNL genes differentially expressed in response to Botrytis cinerea in R. chinensis; Table S12: TPM values of TNL genes differentially expressed in response to Marssonina rosae in R. chinensis; Table S13: TPM values of TNL genes differentially expressed in response to Podosphaera pannosa in R. chinensis; Table S14: Sequence information of primers. Table S15. qRT-PCR results of nine RcTNL genes after inoculation with Marssonina rosae.
Author Contributions
Conceptualization, F.X.; methodology, J.S.; software, J.S.; validation, B.L.; formal analysis, F.C.; investigation, J.Y., L.H. and J.G.; resources, B.L., C.G. and F.X.; data curation, J.S.; writing—original draft, J.S.; writing—review and editing, J.S.; visualization, J.S.; project administration, F.X. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the China Postdoctoral Science Foundation, grant number 2022M721077, the Hubei Agricultural Science and Technology Innovation Center Project, grant number 2021-620-000-001-008, and the Construction of a Modern Agricultural Industrial Technology System, grant number CRAS-23.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All data generated or analyzed during this study are included in this published article and its Supplementary Information Files.
Acknowledgments
We thank Guogui Ning from Huazhong Agricultural University for providing ‘Old Blush’; Chao He from Huazhong Agricultural University for his advice on genomic analysis; and Key Laboratory of Cotton Biology and Breeding in the Middle Reaches of the Yangtze River, Ministry of Agriculture, for providing CFX384TM Real-Time System.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Zipfel, C. Plant pattern-recognition receptors. Trends Immunol. 2014, 35, 345–351. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Feng, B.; He, P.; Shan, L. From chaos to harmony: Responses and signaling upon microbial pattern recognition. Annu. Rev. Phytopathol. 2017, 55, 109–137. [Google Scholar] [CrossRef] [PubMed]
- Toruño, T.Y.; Stergiopoulos, I.; Coaker, G. Plant-pathogen effectors: Cellular probes interfering with plant defenses in spatial and temporal manners. Annu. Rev. Phytopathol. 2016, 54, 419–441. [Google Scholar] [CrossRef]
- Monteiro, F.; Nishimura, M.T. Structural, functional, and genomic diversity of plant NLR proteins: An evolved resource for rational engineering of plant immunity. Annu. Rev. Phytopathol. 2018, 56, 243–267. [Google Scholar] [CrossRef] [PubMed]
- Lolle, S.; Stevens, D.; Coaker, G. Plant NLR-triggered immunity: From receptor activation to downstream signaling. Curr. Opin. Immunol. 2020, 62, 99–105. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Pruitt, R.N.; Nürnberger, T.; Wang, Y. Evasion of plant immunity by microbial pathogens. Nat. Rev. Microbiol. 2022, 20, 449–464. [Google Scholar] [CrossRef] [PubMed]
- Kourelis, J.; van der Hoorn, R.A.L. Defended to the nines: 25 years of resistance gene cloning identifies nine mechanisms for R protein function. Plant Cell 2018, 30, 285–299. [Google Scholar] [CrossRef]
- Dubey, N.; Singh, K. Role of NBS-LRR proteins in plant defense. In Molecular Aspects of Plant-Pathogen Interaction; Springer: Singapore, 2018; Volume 5, pp. 115–138. [Google Scholar]
- Shao, Z.-Q.; Xue, J.-Y.; Wu, P.; Zhang, Y.-M.; Wu, Y.; Hang, Y.-Y.; Wang, B.; Chen, J.-Q. Large-scale analyses of angiosperm nucleotide-binding site-leucine-rich repeat genes reveal three anciently diverged classes with distinct evolutionary patterns. Plant Physiol. 2016, 170, 2095–2109. [Google Scholar] [CrossRef]
- Meyers, B.C.; Dickerman, A.W.; Michelmore, R.W.; Sivaramakrishnan, S.; Sobral, B.W.; Young, N.D. Plant disease resistance genes encode members of an ancient and diverse protein family within the nucleotide-binding superfamily. Plant J. 1999, 20, 317–332. [Google Scholar] [CrossRef]
- Takken, F.L.; Albrecht, M.; Tameling, W.I. Resistance proteins: Molecular switches of plant defence. Curr. Opin. Plant Biol. 2006, 9, 383–390. [Google Scholar] [CrossRef]
- Dubey, N.; Chaudhary, A.; Singh, K. Genome-wide analysis of TIR-NBS-LRR gene family in potato identified StTNLC7G2 inducing reactive oxygen species in presence of Alternaria solani. Front. Genet. 2022, 12, 791055. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Yuan, Q.; Wu, Y.; Zhang, J.; Nie, J. Genome-wide identification and characterization of the CC-NBS-LRR gene family in Cucumber (Cucumis sativus L.). Int. J. Mol. Sci. 2022, 23, 5048. [Google Scholar] [CrossRef] [PubMed]
- Sarris, P.F.; Duxbury, Z.; Huh, S.U.; Ma, Y.; Segonzac, C.; Sklenar, J.; Derbyshire, P.; Cevik, V.; Rallapalli, G.; Saucet, S.B.; et al. A plant immune receptor detects pathogen effectors that target WRKY transcription factors. Cell 2015, 161, 1089–1100. [Google Scholar] [CrossRef]
- Kroj, T.; Chanclud, E.; Michel-Romiti, C.; Grand, X.; Morel, J.B. Integration of decoy domains derived from protein targets of pathogen effectors into plant immune receptors is widespread. New Phytol. 2016, 210, 618–626. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Ma, X.; Li, N.; Zhou, L.; Liu, Z.; Han, H.; Gui, Y.; Bao, Y.; Chen, J.; Dai, X. Genome-wide association study discovered candidate genes of verticillium wilt resistance in upland cotton (Gossypium hirsutum L.). Plant Biotechnol. J. 2017, 15, 1520–1532. [Google Scholar] [CrossRef] [PubMed]
- Li, T.G.; Wang, B.L.; Yin, C.M.; Zhang, D.D.; Wang, D.; Song, J.; Zhou, L.; Kong, Z.Q.; Klosterman, S.J.; Li, J.J.; et al. The Gossypium hirsutum TIR-NBS-LRR gene GhDSC1 mediates resistance against verticillium wilt. Mol. Plant Pathol. 2019, 20, 857–876. [Google Scholar] [CrossRef]
- Seo, Y.S.; Rojas, M.R.; Lee, J.Y.; Lee, S.W.; Jeon, J.S.; Ronald, P.; Lucas, W.J.; Gilbertson, R.L. A viral resistance gene from common bean functions across plant families and is up-regulated in a non-virus-specific manner. Proc. Natl. Acad. Sci. USA 2006, 103, 11856–11861. [Google Scholar] [CrossRef]
- Xun, H.; Yang, X.; He, H.; Wang, M.; Guo, P.; Wang, Y.; Pang, J.; Dong, Y.; Feng, X.; Wang, S.; et al. Over-expression of GmKR3, a TIR–NBS–LRR type R gene, confers resistance to multiple viruses in soybean. Plant Mol. Biol. 2018, 99, 95–111. [Google Scholar] [CrossRef]
- Paal, J.; Henselewski, H.; Muth, J.; Meksem, K.; Menéndez, C.M.; Salamini, F.; Ballvora, A.; Gebhardt, C. Molecular cloning of the potato Gro1-4 gene conferring resistance to pathotype Ro1 of the root cyst nematode Globodera rostochiensis, based on a candidate gene approach. Plant J. 2004, 38, 285–297. [Google Scholar] [CrossRef]
- Liu, Y.; Lan, J.; Li, Q.; Zhang, Y.; Wang, C.; Dai, H. Rapid location of Glomerella leaf spot resistance gene locus in apple by whole genome re-sequencing. Mol. Breed. 2017, 37, 96. [Google Scholar] [CrossRef]
- Lv, L.; Liu, Y.; Bai, S.; Turakulov, K.S.; Dong, C.; Zhang, Y. A TIR-NBS-LRR gene MdTNL1 regulates resistance to Glomerella leaf spot in apple. Int. J. Mol. Sci. 2022, 23, 6323. [Google Scholar] [CrossRef] [PubMed]
- Hibrand Saint-Oyant, L.; Ruttink, T.; Hamama, L.; Kirov, I.; Lakhwani, D.; Zhou, N.N.; Bourke, P.M.; Daccord, N.; Leus, L.; Schulz, D.; et al. A high-quality genome sequence of Rosa chinensis to elucidate ornamental traits. Nat. Plants 2018, 4, 473–484. [Google Scholar] [CrossRef]
- Debener, T.; Byrne, D.H. Disease resistance breeding in rose: Current status and potential of biotechnological tools. Plant Sci. 2014, 228, 107–117. [Google Scholar] [CrossRef] [PubMed]
- Dean, R.; Van Kan, J.A.L.; Pretorius, Z.A.; Hammond-Kosack, K.E.; Di Pietro, A.; Spanu, P.D.; Rudd, J.J.; Dickman, M.; Kahmann, R.; Ellis, J.; et al. The Top 10 fungal pathogens in molecular plant pathology. Mol. Plant Pathol. 2012, 13, 414–430. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Li, X.; Liu, X.; Zhang, Z. Comprehensive analysis of bZIP gene family and function of RcbZIP17 on Botrytis resistance in rose (Rosa chinensis). Gene 2023, 849, 146867. [Google Scholar] [CrossRef]
- Liu, X.; Li, D.; Zhang, S.; Xu, Y.; Zhang, Z. Genome-wide characterization of the rose (Rosa chinensis) WRKY family and role of RcWRKY41 in gray mold resistance. BMC Plant Biol. 2019, 19, 522. [Google Scholar] [CrossRef]
- Li, D.; Liu, X.; Shu, L.; Zhang, H.; Zhang, S.; Song, Y.; Zhang, Z. Global analysis of the AP2/ERF gene family in rose (Rosa chinensis) genome unveils the role of RcERF099 in Botrytis resistance. BMC Plant Biol. 2020, 20, 533. [Google Scholar] [CrossRef]
- Yang, S.; Zhang, X.; Yue, J.-X.; Tian, D.; Chen, J.-Q. Recent duplications dominate NBS-encoding gene expansion in two woody species. Mol. Genet. Genom. 2008, 280, 187–198. [Google Scholar] [CrossRef]
- Arya, P.; Kumar, G.; Acharya, V.; Singh, A.K. Genome-wide identification and expression analysis of NBS-encoding genes in Malus x domestica and expansion of NBS genes family in Rosaceae. PLoS ONE 2014, 9, e107987. [Google Scholar] [CrossRef]
- Zhang, Y.; Xia, R.; Kuang, H.; Meyers, B.C. The diversification of plant NBS-LRR defense genes directs the evolution of microRNAs that target them. Mol. Biol. Evol. 2016, 33, 2692–2705. [Google Scholar] [CrossRef]
- Wang, T.; Jia, Z.-H.; Zhang, J.-Y.; Liu, M.; Guo, Z.-R.; Wang, G. Identification and analysis of NBS-LRR genes in Actinidia chinensis genome. Plants 2020, 9, 1350. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.; You, C.; Zhang, H.; Wang, Y.; Zhang, R. Genome-wide analysis of NBS-LRR genes in Rosaceae species reveals distinct evolutionary patterns. Front. Genet. 2022, 13, 1052191. [Google Scholar] [CrossRef] [PubMed]
- Qian, L.-H.; Wu, J.-Y.; Wang, Y.; Zou, X.; Zhou, G.-C.; Sun, X.-Q. Genome-wide analysis of NBS-LRR genes from an early-diverging angiosperm Euryale ferox. Front. Genet. 2022, 13, 880071. [Google Scholar] [CrossRef]
- Zhang, H.; Ye, Z.; Liu, Z.; Sun, Y.; Li, X.; Wu, J.; Zhou, G.; Wan, Y. The cassava NBS-LRR genes confer resistance to cassava bacterial blight. Front. Plant Sci. 2022, 13, 790140. [Google Scholar] [CrossRef]
- Raymond, O.; Gouzy, J.; Just, J.; Badouin, H.; Verdenaud, M.; Lemainque, A.; Vergne, P.; Moja, S.; Choisne, N.; Pont, C.; et al. The Rosa genome provides new insights into the domestication of modern roses. Nat. Genet. 2018, 50, 772–777. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Chao, J.; Li, Z.; Sun, Y.; Aluko, O.O.; Wu, X.; Wang, Q.; Liu, G. MG2C: A user-friendly online tool for drawing genetic maps. Mol. Hortic. 2021, 1, 16. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Bailey, T.L.; Johnson, J.; Grant, C.E.; Noble, W.S. The MEME Suite. Nucleic Acids Res. 2015, 43, W39–W49. [Google Scholar] [CrossRef] [PubMed]
- Dai, X.; Zhuang, Z.; Zhao, P.X. psRNATarget: A plant small RNA target analysis server (2017 release). Nucleic Acids Res. 2018, 46, W49–W54. [Google Scholar] [CrossRef]
- Wang, Y.; Tang, H.; DeBarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.-h.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef] [PubMed]
- Jung, S.; Lee, T.; Cheng, C.H.; Buble, K.; Zheng, P.; Yu, J.; Humann, J.; Ficklin, S.P.; Gasic, K.; Scott, K.; et al. 15 years of GDR: New data and functionality in the Genome Database for Rosaceae. Nucleic Acids Res. 2019, 47, D1137–D1145. [Google Scholar] [CrossRef]
- Liu, X.; Cao, X.; Shi, S.; Zhao, N.; Li, D.; Fang, P.; Chen, X.; Qi, W.; Zhang, Z. Comparative RNA-Seq analysis reveals a critical role for brassinosteroids in rose (Rosa hybrida) petal defense against Botrytis cinerea infection. BMC Genet. 2018, 19, 62. [Google Scholar] [CrossRef] [PubMed]
- Neu, E.; Domes, H.S.; Menz, I.; Kaufmann, H.; Linde, M.; Debener, T. Interaction of roses with a biotrophic and a hemibiotrophic leaf pathogen leads to differences in defense transcriptome activation. Plant Mol. Biol. 2019, 99, 299–316. [Google Scholar] [CrossRef] [PubMed]
- Bray, N.L.; Pimentel, H.; Melsted, P.; Pachter, L. Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 2016, 34, 525–527. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Zurn, J.D.; Zlesak, D.C.; Holen, M.; Bradeen, J.M.; Hokanson, S.C.; Bassil, N.V. Mapping a novel black spot resistance locus in the climbing rose Brite Eyes™ (‘RADbrite’). Front. Plant Sci. 2018, 9, 1730. [Google Scholar] [CrossRef]
- Song, J.; Li, B.; Cui, Y.; Zhuo, C.; Gu, Y.; Hu, K.; Wen, J.; Yi, B.; Shen, J.; Ma, C.; et al. QTL mapping and diurnal transcriptome analysis identify candidate genes regulating Brassica napus flowering time. Int. J. Mol. Sci. 2021, 22, 7559. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Sarris, P.F.; Cevik, V.; Dagdas, G.; Jones, J.D.G.; Krasileva, K.V. Comparative analysis of plant immune receptor architectures uncovers host proteins likely targeted by pathogens. BMC Biol. 2016, 14, 8. [Google Scholar] [CrossRef] [PubMed]
- López-Márquez, D.; Del-Espino, Á.; López-Pagán, N.; Rodríguez-Negrete, E.A.; Rubio-Somoza, I.; Ruiz-Albert, J.; Bejarano, E.R.; Beuzón, C.R.; Murray, J. miR825-5p targets the TIR-NBS-LRR gene MIST1 and down-regulates basal immunity against Pseudomonas syringae in Arabidopsis. J. Exp. Bot. 2021, 72, 7316–7334. [Google Scholar] [CrossRef] [PubMed]
- Cui, X.; Yan, Q.; Gan, S.; Xue, D.; Dou, D.; Guo, N.; Xing, H. Overexpression of gma-miR1510a/b suppresses the expression of a NB-LRR domain gene and reduces resistance to Phytophthora sojae. Gene 2017, 621, 32–39. [Google Scholar] [CrossRef]
- Dey, S.; Sarkar, A.; Chowdhury, S.; Singh, R.; Mukherjee, A.; Ghosh, Z.; Kundu, P. Heightened miR6024-NLR interactions facilitate necrotrophic pathogenesis in tomato. Plant Mol. Biol. 2022, 109, 717–739. [Google Scholar] [CrossRef]
- Asad, M.; Chen, J.; Liao, J.; Liu, D.; Yu, J.; Yang, G. Genome-wide identification, expression profiling, and characterization of cyclin-like genes reveal their role in the fertility of the diamondback moth. Biology 2022, 11, 1493. [Google Scholar] [CrossRef]
- Liu, X.; Wu, J.; Ji, F.; Cao, X.; Zhao, Q.; Cheng, C.; Ma, N.; Zhou, X.; Zhang, Z. Transcriptomic profiling of rose flower under treatment of various phytohormones and plant growth regulators. Sci. Data 2022, 9, 669. [Google Scholar] [CrossRef]
- Xu, T.; Wu, Y.; Yi, X.; Tan, J.; Zhao, H.; Yu, C.; Luo, L.; Cheng, T.; Wang, J.; Pan, H.; et al. Reinforcement of resistance of modern rose to black spot disease via hybridization with Rosa rugosa. Euphytica 2018, 214, 175. [Google Scholar] [CrossRef]
- Whitaker, V.M.; Zuzek, K.; Hokanson, S.C. Resistance of 12 rose genotypes to 14 isolates of Diplocarpon rosae Wolf (rose blackspot) collected from eastern North America. Plant Breed. 2007, 126, 83–88. [Google Scholar] [CrossRef]
- Gachomo, E.W.; Dehne, H.W.; Steiner, U. Microscopic evidence for the hemibiotrophic nature of Diplocarpon rosae, cause of black spot disease of rose. Physiol. Mol. Plant Pathol. 2006, 69, 86–92. [Google Scholar] [CrossRef]
- Lau, J.; Young, E.L.; Collins, S.; Windham, M.T.; Klein, P.E.; Byrne, D.H.; Riera-Lizarazu, O. Rose rosette disease resistance loci detected in two interconnected tetraploid garden rose populations. Front. Plant Sci. 2022, 13, 916231. [Google Scholar] [CrossRef] [PubMed]
- Williams, S.J.; Sohn, K.H.; Wan, L.; Bernoux, M.; Sarris, P.F.; Segonzac, C.; Ve, T.; Ma, Y.; Saucet, S.B.; Ericsson, D.J.; et al. Structural basis for assembly and function of a heterodimeric plant immune receptor. Science 2014, 344, 299–303. [Google Scholar] [CrossRef]
- Tameling, W.I.L.; Vossen, J.H.; Albrecht, M.; Lengauer, T.; Berden, J.A.; Haring, M.A.; Cornelissen, B.J.C.; Takken, F.L.W. Mutations in the NB-ARC domain of I-2 that impair ATP hydrolysis cause autoactivation. Plant Physiol. 2006, 140, 1233–1245. [Google Scholar] [CrossRef] [PubMed]
- Riedl, S.J.; Li, W.; Chao, Y.; Schwarzenbacher, R.; Shi, Y. Structure of the apoptotic protease-activating factor 1 bound to ADP. Nature 2005, 434, 926–933. [Google Scholar] [CrossRef] [PubMed]
- Ellis, J.G.; Lawrence, G.J.; Luck, J.E.; Dodds, P.N. Identification of regions in alleles of the flax rust resistance gene L that determine differences in gene-for-gene specificity. Plant Cell 1999, 11, 495–506. [Google Scholar] [CrossRef] [PubMed]
- Krasileva, K.V.; Dahlbeck, D.; Staskawicz, B.J. Activation of an Arabidopsis resistance protein is specified by the in planta association of its leucine-rich repeat domain with the cognate oomycete effector. Plant Cell 2010, 22, 2444–2458. [Google Scholar] [CrossRef]
- Wen, Z.; Yao, L.; Singer, S.D.; Muhammad, H.; Li, Z.; Wang, X. Constitutive heterologous overexpression of a TIR-NB-ARC-LRR gene encoding a putative disease resistance protein from wild Chinese Vitis pseudoreticulata in Arabidopsis and tobacco enhances resistance to phytopathogenic fungi and bacteria. Plant Physiol. Biochem. 2017, 112, 346–361. [Google Scholar] [CrossRef]
- Canto-Pastor, A.; Santos, B.A.M.C.; Valli, A.A.; Summers, W.; Schornack, S.; Baulcombe, D.C. Enhanced resistance to bacterial and oomycete pathogens by short tandem target mimic RNAs in tomato. Proc. Natl. Acad. Sci. USA 2019, 116, 2755–2760. [Google Scholar] [CrossRef]
- Yang, L.; Mu, X.; Liu, C.; Cai, J.; Shi, K.; Zhu, W.; Yang, Q. Overexpression of potato miR482e enhanced plant sensitivity to Verticillium dahliae infection. J. Integr. Plant Biol. 2015, 57, 1078–1088. [Google Scholar] [CrossRef]
- Zhu, Q.H.; Jin, S.; Yuan, Y.; Liu, Q.; Zhang, X.; Wilson, I. CRISPR/Cas9-mediated saturated mutagenesis of the cotton MIR482 family for dissecting the functionality of individual members in disease response. Plant Direct 2022, 6, e410. [Google Scholar] [CrossRef]
- Shivaprasad, P.V.; Chen, H.M.; Patel, K.; Bond, D.M.; Santos, B.A.; Baulcombe, D.C. A microRNA superfamily regulates nucleotide binding site-leucine-rich repeats and other mRNAs. Plant Cell 2012, 24, 859–874. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).