A TaqMan® Assay Allows an Accurate Detection and Quantification of Fusarium spp., the Causal Agents of Tomato Wilt and Rot Diseases
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Fungal Isolates
2.2. Plant Sampling
2.3. gDNA Extraction from Fungal Isolates and Tomato Samples
2.4. Design of a Fusarium spp.-Specific qPCR Assay
2.5. Specificity, Sensitivity, and Reliability of the qPCR Assay
2.6. gDNA Calibrator Plasmid
3. Results
3.1. Specificity and Sensitivity of the Fusarium spp.-Specific qPCR TaqMan® Assay
3.2. Calibration Curves for Quantification of Fusarium spp. gDNA
3.3. Applicability of the Fusarium spp.-Specific qPCR TaqMan® Assay in Tomato Plants
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Panno, S.; Davino, S.; Caruso, A.G.; Bertacca, S.; Crnogorac, A.; Mandi, A. A Review of the Most Common and Economically Important Diseases That Undermine the Cultivation of Tomato Crop in the Mediterranean Basin. Agronomy 2021, 11, 2188. [Google Scholar] [CrossRef]
- Jones, J.B.; Zitter, T.A.; Momol, T.M.; Miller, S.A. Compendium of Tomato Diseases and Pests, Second Edition; The American Phytopathological Society: St. Paul, MN, USA, 2016; ISBN 978-0-89054-434-1. [Google Scholar]
- Srinivas, C.; Nirmala Devi, D.; Narasimha Murthy, K.; Mohan, C.D.; Lakshmeesha, T.R.; Singh, B.P.; Kalagatur, N.K.; Niranjana, S.R.; Hashem, A.; Alqarawi, A.A.; et al. Fusarium oxysporum f. sp. lycopersici causal agent of vascular wilt disease of tomato: Biology to diversity—A review. Saudi J. Biol. Sci. 2019, 26, 1315–1324. [Google Scholar] [CrossRef] [PubMed]
- Campos, M.D.; Félix, M.R.; Patanita, M.; Materatski, P.; Varanda, C. High throughput sequencing unravels tomato-pathogen interactions towards a sustainable plant breeding. Hortic. Res. 2021, 8, 171. [Google Scholar] [CrossRef] [PubMed]
- Bodah, E.T. Root Rot Diseases in Plants: A Review of Common Causal Agents and Management Strategies. Agric. Res. Technol. Open Access J. 2017, 5, 555661. [Google Scholar] [CrossRef]
- Yadeta, K.A.; Thomma, B.P.H.J. The xylem as battleground for plant hosts and vascular wilt pathogens. Front. Plant Sci. 2013, 4, 97. [Google Scholar] [CrossRef] [PubMed]
- De la Lastra, E.; Basallote-Ureba, M.J.; De los Santos, B.; Miranda, L.; Vela-Delgado, M.D.; Capote, N. A TaqMan real-time polymerase chain reaction assay for accurate detection and quantification of Fusarium solani in strawberry plants and soil. Sci. Hortic. 2018, 237, 128–134. [Google Scholar] [CrossRef]
- Ji, X.; Deng, T.; Xiao, Y.; Jin, C.; Lyu, W.; Wu, Z.; Wang, W.; Wang, X.; He, Q.; Yang, H. Emerging Alternaria and Fusarium mycotoxins in tomatoes and derived tomato products from the China market: Occurrence, methods of determination, and risk evaluation. Food Control 2023, 145, 109464. [Google Scholar] [CrossRef]
- Maschietto, V.; Colombi, C.; Pirona, R.; Pea, G.; Strozzi, F.; Marocco, A.; Rossini, L.; Lanubile, A. QTL mapping and candidate genes for resistance to Fusarium ear rot and fumonisin contamination in maize. BMC Plant Biol. 2017, 17, 20. [Google Scholar] [CrossRef]
- Debbi, A.; Boureghda, H.; Monte, E.; Hermosa, R. Distribution and genetic variability of Fusarium oxysporum associated with tomato diseases in Algeria and a biocontrol strategy with indigenous Trichoderma spp. Front. Microbiol. 2018, 9, 282. [Google Scholar] [CrossRef]
- McGovern, R.J. Management of tomato diseases caused by Fusarium oxysporum. Crop Prot. 2015, 73, 78–92. [Google Scholar] [CrossRef]
- Rozlianal, F.S.; Sariah, M. Characterization of Malaysian Isolates of Fusarium from Tomato and Pathogenicity Testing. Res. J. Microbiol. 2006, 1, 266–272. [Google Scholar]
- Murad, N.B.A.; Kusai, N.A.; Zainudin, N.A.I.M. Identification and diversity of Fusarium species isolated from tomato fruits. J. Plant Prot. Res. 2016, 56, 231–236. [Google Scholar] [CrossRef]
- Akbar, A.; Hussain, S.; Ali, G.S. Germplasm Evaluation of Tomato for Resistance to the Emerging Wilt Pathogen Fusarium equiseti. J. Agric. Stud. 2018, 5, 174. [Google Scholar] [CrossRef]
- Akbar, A.; Hussain, S.; Ullah, K.; Fahim, M.; Ali, G.S. Detection, virulence and genetic diversity of Fusarium species infecting tomato in Northern Pakistan. PLoS ONE 2018, 13, e0203613. [Google Scholar] [CrossRef]
- Ribeiro, J.A.; Albuquerque, A.; Materatski, P.; Patanita, M.; Varanda, C.M.R.; Félix, M.R.; Campos, M.D. Tomato Response to Fusarium spp. Infection under Field Conditions: Study of Potential Genes Involved. Horticulturae 2022, 8, 433. [Google Scholar] [CrossRef]
- Patanita, M.; Campos, M.D.; Félix, M.D.R.; Carvalho, M.; Brito, I. Effect of tillage system and cover crop on maize mycorrhization and presence of Magnaporthiopsis maydis. Biology 2020, 9, 46. [Google Scholar] [CrossRef]
- Le, K.D.; Kim, J.; Yu, N.H.; Kim, B.; Lee, C.W.; Kim, J.C. Biological Control of Tomato Bacterial Wilt, Kimchi Cabbage Soft Rot, and Red Pepper Bacterial Leaf Spot Using Paenibacillus elgii JCK-5075. Front. Plant Sci. 2020, 11, 775. [Google Scholar] [CrossRef]
- Malik, M.S.; Haider, S.; Rehman, A.; Rehman, S.U.; Jamil, M.; Naz, I.; Anees, M. Biological control of fungal pathogens of tomato (Lycopersicon esculentum) by chitinolytic bacterial strains. J. Basic Microbiol. 2022, 62, 48–62. [Google Scholar] [CrossRef]
- de Almeida, A.B.; Concas, J.; Campos, M.D.; Materatski, P.; Varanda, C.; Patanita, M.; Murolo, S.; Romanazzi, G.; Félix, M.R. Endophytic fungi as potential biological control agents against grapevine trunk diseases in alentejo region. Biology 2020, 9, 420. [Google Scholar] [CrossRef]
- Heo, A.Y.; Koo, Y.M.; Choi, H.W. Biological Control Activity of Plant Growth Promoting Rhizobacteria Burkholderia contaminans AY001 against Tomato Fusarium Wilt and Bacterial Speck Diseases. Biology 2022, 11, 619. [Google Scholar] [CrossRef]
- Campos, M.D.; Zellama, M.S.; Varanda, C.; Materatski, P.; Peixe, A.; Chaouachi, M.; Félix, M.R. Establishment of a sensitive qPCR methodology for detection of the olive-infecting viruses in portuguese and tunisian orchards. Front. Plant Sci. 2019, 10, 694. [Google Scholar] [CrossRef] [PubMed]
- Azevedo-Nogueira, F.; Gomes, S.; Lino, A.; Carvalho, T.; Martins-Lopes, P. Real-time PCR assay for Colletotrichum acutatum sensu stricto quantification in olive fruit samples. Food Chem. 2021, 339, 127858. [Google Scholar] [CrossRef] [PubMed]
- Lukianova, A.A.; Evseev, P.V.; Stakheev, A.A.; Kotova, I.B.; Zavriev, S.K.; Ignatov, A.N.; Miroshnikov, K.A. Development of qpcr detection assay for potato pathogen Pectobacterium atrosepticum based on a unique target sequence. Plants 2021, 10, 355. [Google Scholar] [CrossRef] [PubMed]
- Thomas, W.J.; Borland, T.G.; Bergl, D.D.; Claassen, B.J.; Flodquist, T.A.; Montgomery, A.S.; Rivedal, H.M.; Woodhall, J.; Ocamb, C.M.; Gent, D.H. A Quantitative PCR Assay for Detection and Quantification of Fusarium sambucinum. Plant Dis. 2022, 106, 2601–2606. [Google Scholar] [CrossRef]
- Campos, M.D.; Patanita, M.; Campos, C.; Materatski, P.; Varanda, C.M.R.; Brito, I.; Félix, M.R. Detection and quantification of Fusarium spp. (F. oxysporum, F. verticillioides, F. graminearum) and Magnaporthiopsis maydis in maize using real-time PCR targeting the ITS region. Agronomy 2019, 9, 45. [Google Scholar] [CrossRef]
- Varanda, C.M.R.; Materatski, P.; Landum, M.; Campos, M.D.; Félix, M.R. Fungal communities associated with peacock and cercospora leaf spots in olive. Plants 2019, 8, 169. [Google Scholar] [CrossRef]
- Doyle, J.J.; Doyle, J.L. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull. 1987, 19, 11–15. [Google Scholar]
- Varanda, C.M.R.; Oliveira, M.; Materatski, P.; Landum, M.; Clara, M.I.E.; Félix, M.R. Fungal endophytic communities associated to the phyllosphere of grapevine cultivars under different types of management. Fungal Biol. 2016, 120, 1525–1536. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: A multiple sequence alignment method with reduced time and space complexity. BMC Bioinform. 2004, 5, 113. [Google Scholar] [CrossRef]
- White, T.J.; Bruns, S.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phyologenetics. PCR Protoc. A Guide Methods Appl. 1990, 18, 315–322. [Google Scholar]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Campos, M.D.; Valadas, V.; Campos, C.; Morello, L.; Braglia, L.; Breviario, D.; Cardoso, H.G. A TaqMan real-time PCR method based on alternative oxidase genes for detection of plant species in animal feed samples. PLoS ONE 2018, 13, e0190668. [Google Scholar] [CrossRef]
- Materatski, P.; Varanda, C.; Carvalho, T.; Dias, A.B.; Campos, M.D.; Gomes, L.; Nobre, T.; Rei, F.; Félix, M.R. Effect of long-term fungicide applications on virulence and diversity of Colletotrichum spp. Associated to olive anthracnose. Plants 2019, 8, 311. [Google Scholar] [CrossRef]
- Ruijter, J.M.; Ramakers, C.; Hoogaars, W.M.H.; Karlen, Y.; Bakker, O.; van den hoff, M.J.B.; Moorman, A.F.M. Amplification efficiency: Linking baseline and bias in the analysis of quantitative PCR data. Nucleic Acids Res. 2009, 37, e45. [Google Scholar] [CrossRef]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef]
- Kralik, P.; Ricchi, M. A basic guide to real time PCR in microbial diagnostics: Definitions, parameters, and everything. Front. Microbiol. 2017, 8, 108. [Google Scholar] [CrossRef]
- Hariharan, G.; Prasannath, K. Recent Advances in Molecular Diagnostics of Fungal Plant Pathogens: A Mini Review. Front. Cell. Infect. Microbiol. 2021, 10, 600234. [Google Scholar] [CrossRef]
- Validov, S.Z.; Kamilova, F.D.; Lugtenberg, B.J.J. Monitoring of pathogenic and non-pathogenic Fusarium oxysporum strains during tomato plant infection. Microb. Biotechnol. 2011, 4, 82–88. [Google Scholar] [CrossRef]
- Johnson, G.; Nolan, T.; Bustin, S.A. Real-time quantitative PCR, pathogen detection and MIQE. Methods Mol. Biol. 2013, 943, 1–16. [Google Scholar] [CrossRef]
- Coats, K.; Debauw, A.; Lakshman, D.K.; Roberts, D.P.; Ismaiel, A.; Chastagner, G. Detection and Molecular Phylogenetic-Morphometric Characterization of Rhizoctonia tuliparum, Causal Agent of Gray Bulb Rot of Tulips and Bulbous Iris. J. Fungi 2022, 8, 163. [Google Scholar] [CrossRef]
- Pavón, M.A.; González, I.; Martín, R.; García Lacarra, T. ITS-based detection and quantification of Alternaria spp. in raw and processed vegetables by real-time quantitative PCR. Food Microbiol. 2012, 32, 165–171. [Google Scholar] [CrossRef]
- Lofgren, L.A.; Uehling, J.K.; Branco, S.; Bruns, T.D.; Martin, F.; Kennedy, P.G. Genome-based estimates of fungal rDNA copy number variation across phylogenetic scales and ecological lifestyles. Mol. Ecol. 2019, 28, 721–730. [Google Scholar] [CrossRef]
- Lavrinienko, A.; Jernfors, T.; Koskimäki, J.J.; Pirttilä, A.M.; Watts, P.C. Does Intraspecific Variation in rDNA Copy Number Affect Analysis of Microbial Communities? Trends Microbiol. 2021, 29, 19–27. [Google Scholar] [CrossRef]
- Kulik, T.; Jestoi, M.; Okorski, A. Development of TaqMan assays for the quantitative detection of Fusarium avenaceum/Fusarium tricinctum and Fusarium poae esyn1 genotypes from cereal grain. FEMS Microbiol. Lett. 2011, 314, 49–56. [Google Scholar] [CrossRef]
- Bhagat, N.; Magotra, S.; Gupta, R.; Sharma, S.; Verma, S. Invasion and Colonization of Pathogenic Fusarium oxysporum R1 in Crocus sativus L. during Corm Rot Disease Progression. J. Fungy 2022, 8, 1246. [Google Scholar] [CrossRef]
- Broeders, S.; Huber, I.; Grohmann, L.; Berben, G.; Taverniers, I.; Mazzara, M.; Roosens, N.; Morisset, D. Guidelines for validation of qualitative real-time PCR methods. Trends Food Sci. Technol. 2014, 37, 115–126. [Google Scholar] [CrossRef]
- Perea-Domínguez, X.P.; Hernández-Gastelum, L.Z.; Olivas-Olguin, H.R.; Espinosa-Alonso, L.G.; Valdez-Morales, M.; Medina-Godoy, S. Phenolic composition of tomato varieties and an industrial tomato by-product: Free, conjugated and bound phenolics and antioxidant activity. J. Food Sci. Technol. 2018, 55, 3453–3461. [Google Scholar] [CrossRef]
- Schrader, C.; Schielke, A.; Ellerbroek, L.; Johne, R. PCR inhibitors—occurrence, properties and removal. J. Appl. Microbiol. 2012, 113, 1014–1026. [Google Scholar] [CrossRef]
- Azevedo-Nogueira, F.; Rego, C.; Gonçalves, H.M.R.; Fortes, A.M.; Gramaje, D.; Martins-Lopes, P. The road to molecular identification and detection of fungal grapevine trunk diseases. Front. Plant Sci. 2022, 13, 960289. [Google Scholar] [CrossRef]

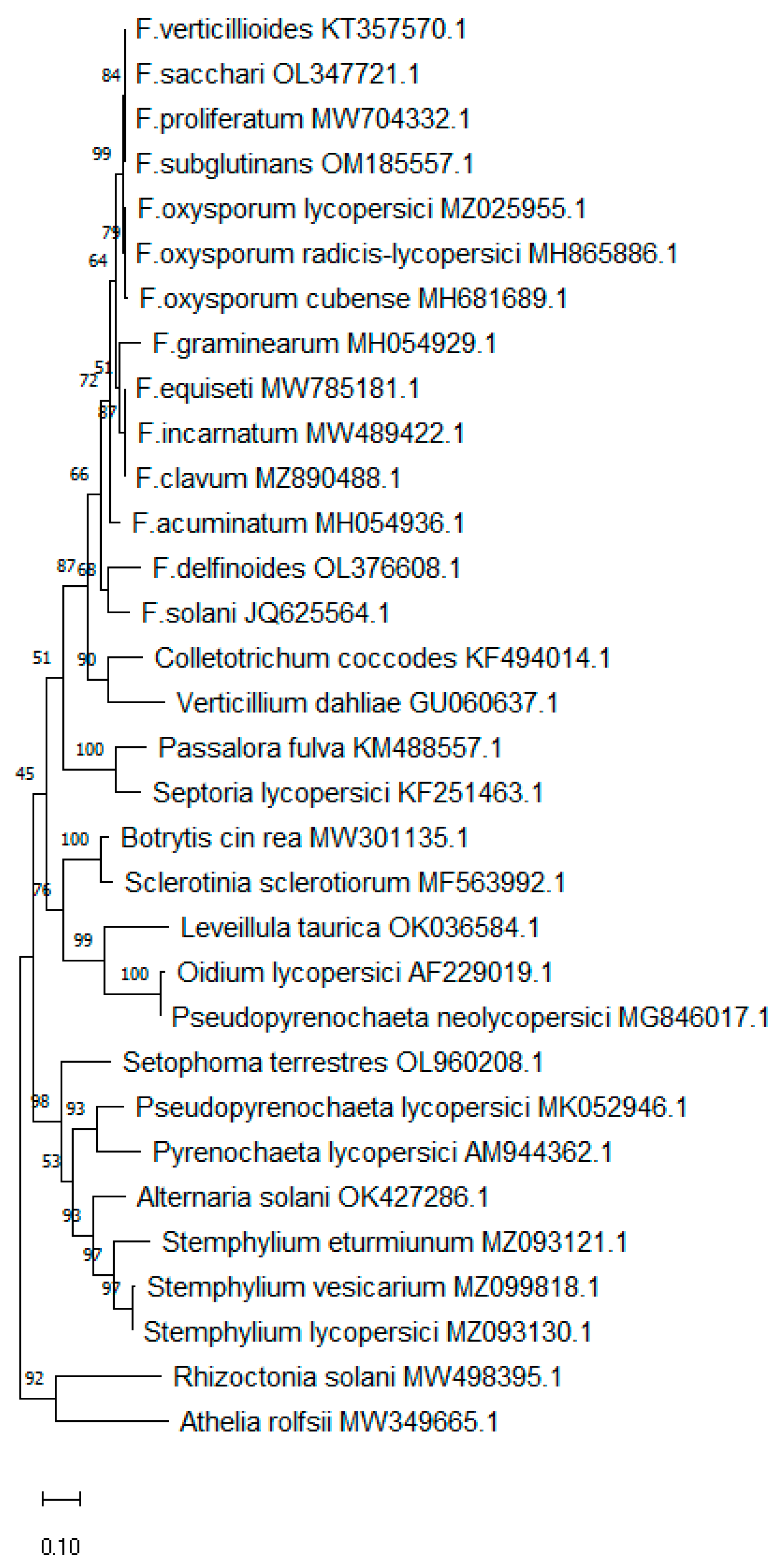

| Species | Isolate Ref. | Cq Value |
|---|---|---|
| F. incarnatum | M_4F | 22.90 |
| F. oxysporum | M_45-2 | 22.86 |
| F. oxysporum | M_67 | 20.48 |
| F. oxysporum | M_84 | 21.53 |
| F. oxysporum | M_90 | 25.05 |
| F. oxysporum | M_91 | 20.31 |
| F. oxysporum | M_92 | 20.26 |
| F. oxysporum | M_100 | 19.29 |
| F. oxysporum | A_56 | 25.08 |
| F. oxysporum | A_57 | 22.76 |
| F. oxysporum | A_122 | 23.57 |
| F. oxysporum | A_127a | 26.42 |
| F. oxysporum | A_127b | 30.32 |
| F. oxysporum | A_137 | 25.65 |
| F. oxysporum | C_4 | 18.42 |
| F. oxysporum f.sp. radicis-lycopersici | C_3 | 19.20 |
| F. solani | M_215c | 21.51 |
| F. verticillioides | A_118 | 29.29 |
| Fusarium nelsonii | A_82c | 20.17 |
| Fusarium nelsonii | A_82e | 20.17 |
| Fusarium sp. | M_62 | 20.01 |
| Fusarium sp. | M_207 | 20.16 |
| Fusarium sp. | M_208 | 22.19 |
| Fusarium sp. | M_209 | 20.28 |
| Fusarium sp. | P_Q7 | 19.95 |
| Fusarium sp. | A_123 | 24.63 |
| Fusarium sp. | A_126 | 25.91 |
| Fusarium sp. | C_1 | 21.48 |
| Fusarium sp. | C_2 | 21.06 |
| Alternaria alternata | A_59 | N.D. |
| Alternaria alternata | A_72 | N.D. |
| Alternaria alternata | C_6 | N.D. |
| Alternaria tenuissima | P_75B | N.D. |
| Botrytis cinerea | A_43 | N.D. |
| Botrytis cinerea | A_64 | N.D. |
| Botrytis cinerea | A_115 | N.D. |
| Botrytis cinerea | A_119 | N.D. |
| Botrytis cinerea | A_132 | N.D. |
| Botrytis cinerea | C_9 | N.D. |
| Cladosporium cladosporioides | P_70D | N.D. |
| Colletotrichum sp. | A_71 | N.D. |
| Epicoccum nigrum | P_R46 | N.D. |
| Phytium sp. | C_10 | N.D. |
| Phytophthora sp. | C_5 | N.D. |
| Verticillium dahliae | C_8 | N.D. |
| Dilution | gDNA in PCR (ng) | Cq Value (±SD) | TCN |
|---|---|---|---|
| P.C. | 100.00 | 14.67 (±0.22) | 39,606,399.4 |
| 2−1 | 50.00 | 15.53 (±0.27) | 22,779,380.0 |
| 2−2 | 25.00 | 16.72 (±0.27) | 10,595,944.3 |
| 2−3 | 12.50 | 17.52 (±0.29) | 6,333,969.6 |
| 2−4 | 6.25 | 18.57 (±0.27) | 3,223,887.6 |
| 2−5 | 3.13 | 19.61 (±0.17) | 1,651,494.4 |
| 2−6 | 1.56 | 20.50 (±0.08) | 931,695.0 |
| 2−7 | 7.81 × 10−1 | 21.73 (±0.26) | 422,375.1 |
| 2−8 | 3.91 × 10−1 | 23.09 (±0.13) | 176,120.6 |
| 2−9 | 1.95 × 10−1 | 24.52 (±0.25) | 70,205.1 |
| 2−10 | 9.77 × 10−2 | 25.82 (±0.19) | 30,425.7 |
| 2−11 | 4.88 × 10−2 | 26.74 (±0.07) | 16,836.7 |
| 2−12 | 2.44 × 10−2 | 28.38 (±0.16) | 5863.5 |
| 2−13 | 1.22 × 10−2 | 30.27 (±0.34) | 1738.7 |
| 2−14 | 6.10 × 10−3 | 32.52 (±0.59) | 409.0 |
| 2−15 | 3.05 × 10−3 | 33.42 (±0.29) | 229.3 |
| 2−16 | 1.53 × 10−3 | N.D. | N.D. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Campos, M.D.; Varanda, C.; Patanita, M.; Amaro Ribeiro, J.; Campos, C.; Materatski, P.; Albuquerque, A.; Félix, M.d.R. A TaqMan® Assay Allows an Accurate Detection and Quantification of Fusarium spp., the Causal Agents of Tomato Wilt and Rot Diseases. Biology 2023, 12, 268. https://doi.org/10.3390/biology12020268
Campos MD, Varanda C, Patanita M, Amaro Ribeiro J, Campos C, Materatski P, Albuquerque A, Félix MdR. A TaqMan® Assay Allows an Accurate Detection and Quantification of Fusarium spp., the Causal Agents of Tomato Wilt and Rot Diseases. Biology. 2023; 12(2):268. https://doi.org/10.3390/biology12020268
Chicago/Turabian StyleCampos, Maria Doroteia, Carla Varanda, Mariana Patanita, Joana Amaro Ribeiro, Catarina Campos, Patrick Materatski, André Albuquerque, and Maria do Rosário Félix. 2023. "A TaqMan® Assay Allows an Accurate Detection and Quantification of Fusarium spp., the Causal Agents of Tomato Wilt and Rot Diseases" Biology 12, no. 2: 268. https://doi.org/10.3390/biology12020268
APA StyleCampos, M. D., Varanda, C., Patanita, M., Amaro Ribeiro, J., Campos, C., Materatski, P., Albuquerque, A., & Félix, M. d. R. (2023). A TaqMan® Assay Allows an Accurate Detection and Quantification of Fusarium spp., the Causal Agents of Tomato Wilt and Rot Diseases. Biology, 12(2), 268. https://doi.org/10.3390/biology12020268










