Short-Term UVB Irradiation Leads to Persistent DNA Damage in Limbal Epithelial Stem Cells, Partially Reversed by DNA Repairing Enzymes
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Primary Human Limbal Epithelial (HLE) Cell Culture
2.2. UVB Irradiation and T4 Endonuclease Treatment
2.3. Harvesting and Irradiating Mouse Eyes
2.4. Immunohistochemistry: CPD Staining in Mouse Cornea and Human Epithelial Cells
2.5. Proteomics
2.5.1. Labelling for SP3
2.5.2. Quantitation
2.5.3. Data Processing
2.6. Image Analysis and Statistical Analysis
3. Results
3.1. T4N5 Repairs CPDs in Human Limbal Epithelial Cells

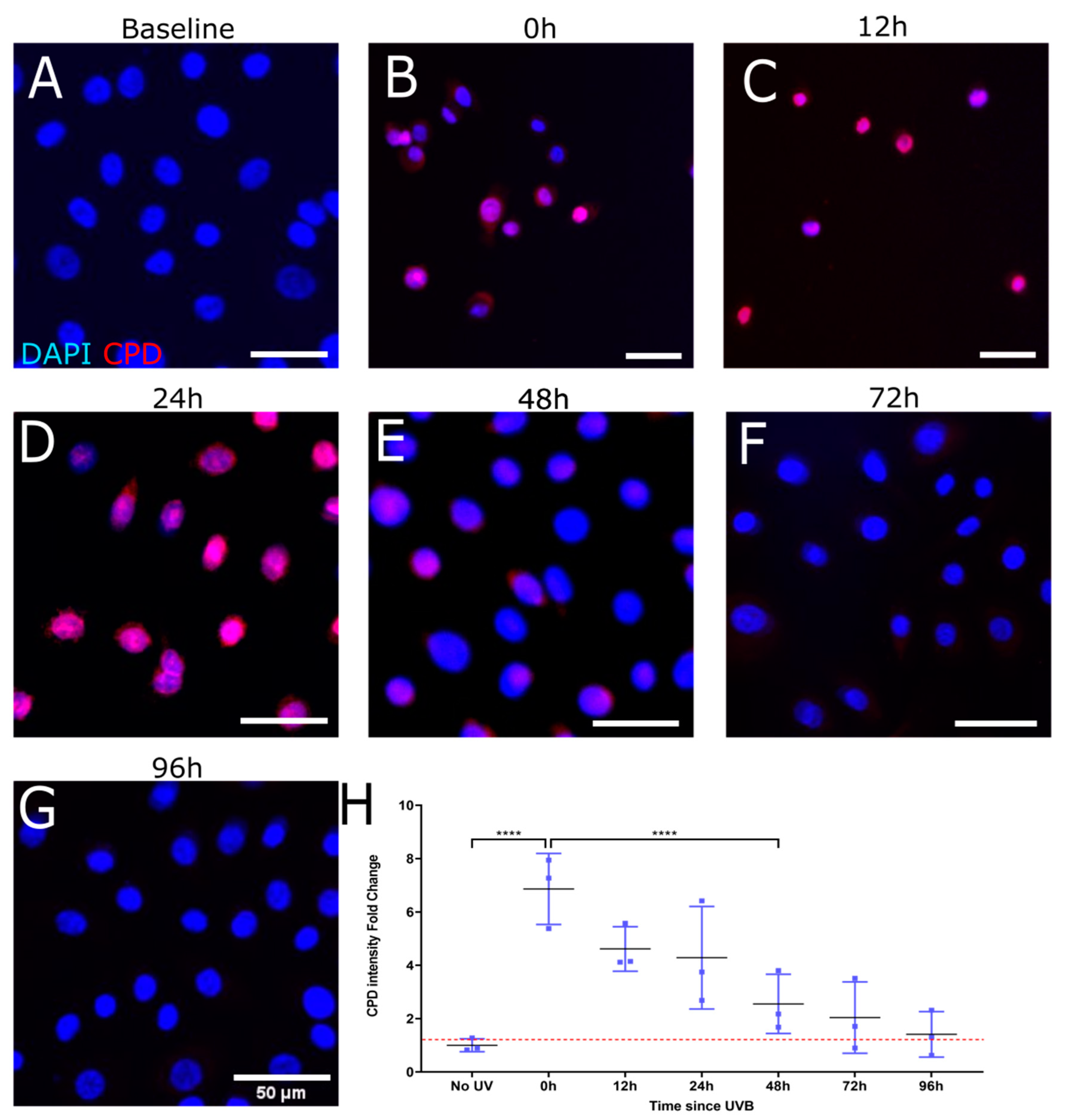
3.2. T4N5 Repair Time Course in Human Limbal Epithelial Cells

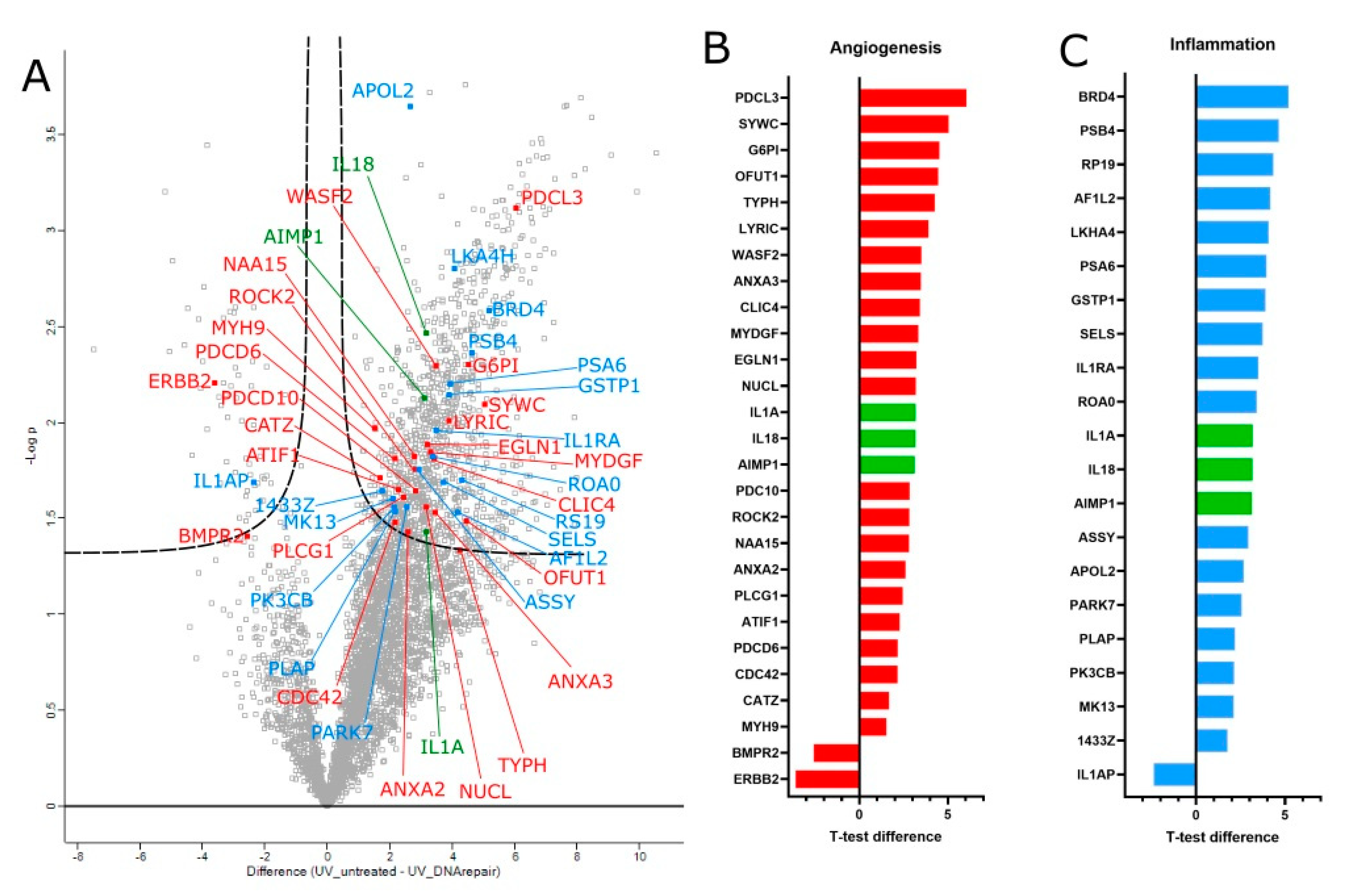
3.3. Proteomic Analysis of T4 Endonuclease Treatment
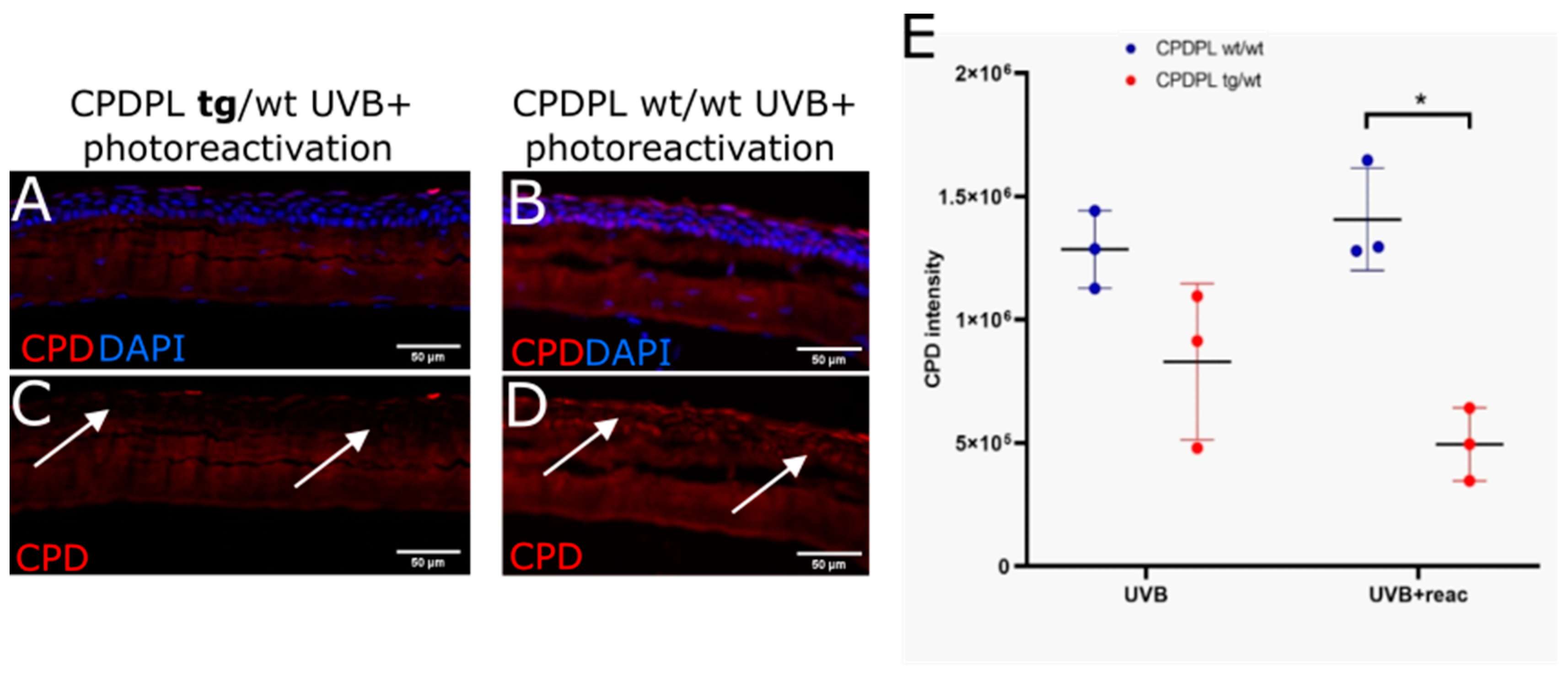
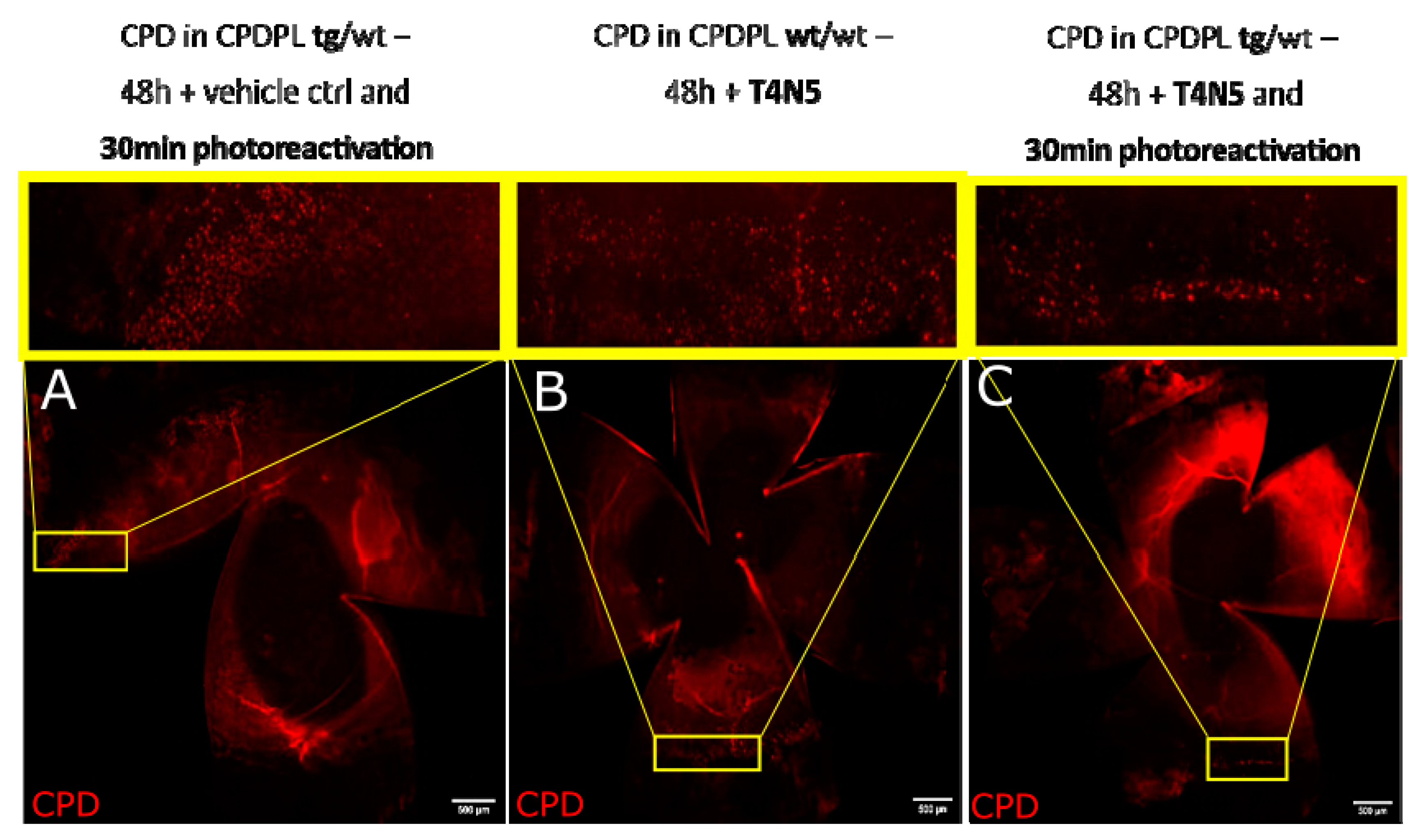
3.4. CPD Recovery in Sections of Whole Mouse Cornea
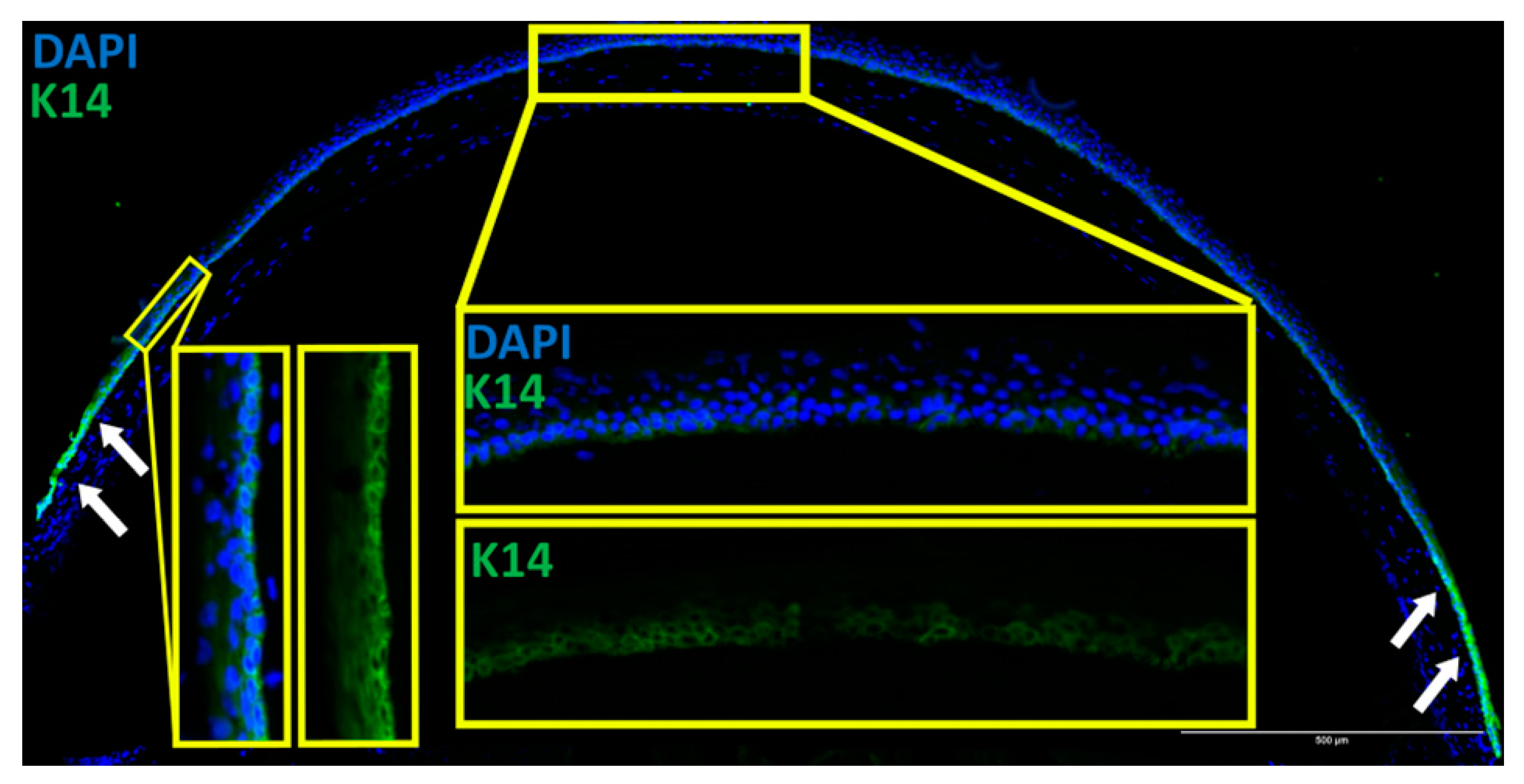
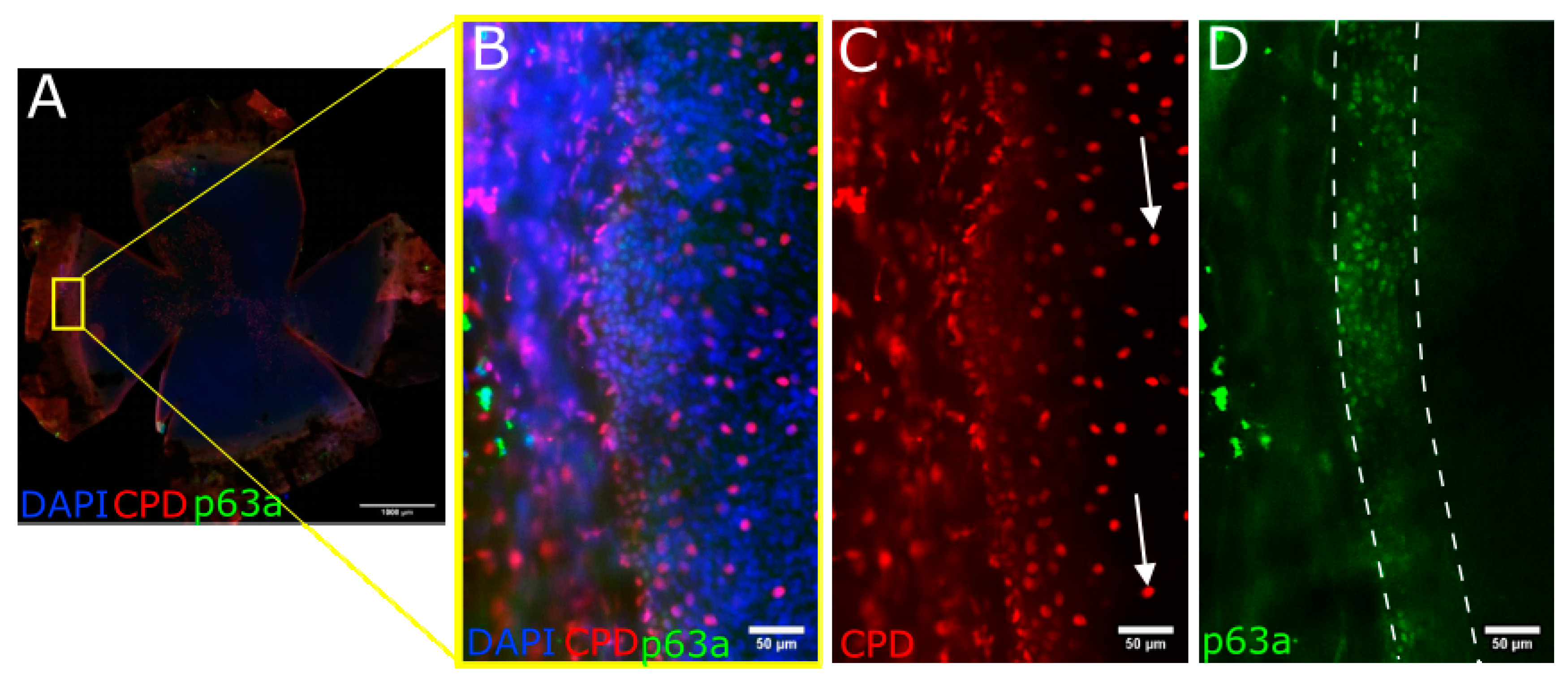
3.5. CPD Recovery in the Limbus of Whole Mouse Cornea
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Tong, L.; Corrales, R.M.; Chen, Z.; Villarreal, A.L.; De Paiva, C.S.; Beuerman, R.; Li, D.Q.; Pflugfelder, S.C. Expression and regulation of cornified envelope proteins in human corneal epithelium. Invest. Ophthalmol. Vis. Sci. 2006, 47, 1938–1946. [Google Scholar] [CrossRef] [PubMed]
- Wilson, S.E.; Mohan, R.R.; Mohan, R.R.; Ambrosio, R., Jr.; Hong, J.; Lee, J. The corneal wound healing response: Cytokine-mediated interaction of the epithelium, stroma, and inflammatory cells. Prog. Retin. Eye Res. 2001, 20, 625–637. [Google Scholar] [CrossRef]
- Huang, M.; Wang, B.; Wan, P.; Liang, X.; Wang, X.; Liu, Y.; Zhou, Q.; Wang, Z. Roles of limbal microvascular net and limbal stroma in regulating maintenance of limbal epithelial stem cells. Cell Tissue Res. 2015, 359, 547–563. [Google Scholar] [CrossRef] [PubMed]
- Papas, E.B. The limbal vasculature. Contact Lens Anterior Eye 2003, 26, 71–76. [Google Scholar] [CrossRef]
- Kruse, F.E. Stem cells and corneal epithelial regeneration. Eye 1994, 8, 170–183. [Google Scholar] [CrossRef]
- Zieske, J.D. Perpetuation of stem cells in the eye. Eye 1994, 8, 163–169. [Google Scholar] [CrossRef] [PubMed]
- Yoon, J.J.; Ismail, S.; Sherwin, T. Limbal stem cells: Central concepts of corneal epithelial homeostasis. World J. Stem Cells 2014, 6, 391–403. [Google Scholar] [CrossRef] [PubMed]
- Notara, M.; Behboudifard, S.; Kluth, M.A.; Masslo, C.; Ganss, C.; Frank, M.H.; Schumacher, B.; Cursiefen, C. UV light-blocking contact lenses protect against short-term UVB-induced limbal stem cell niche damage and inflammation. Sci. Rep. 2018, 8, 12564. [Google Scholar] [CrossRef]
- Notara, M.; Refaian, N.; Brown, G.; Steven, P.; Bock, F.; Cursiefen, C. Effects of UVB irradiation on limbal stem cell niche and its role in cornea lymphangiogenesis. Investig. Ophthalmol. Vis. Sci. 2015, 56, 5622. [Google Scholar]
- Notara, M.; Refaian, N.; Braun, G.; Steven, P.; Bock, F.; Cursiefen, C. Short-Term Ultraviolet A Irradiation Leads to Dysfunction of the Limbal Niche Cells and an Antilymphangiogenic and Anti-inflammatory Micromilieu. Investig. Ophthalmol. Vis. Sci. 2016, 57, 928–939. [Google Scholar] [CrossRef]
- Notara, M.; Braun, G.; Dreisow, M.L.; Bock, F.; Cursiefen, C. Avastin effects on human limbal epithelial cell function and phenotype in vitro. Investig. Ophthalmol. Vis. Sci. 2016, 57, 4343. [Google Scholar]
- Gao, X.; Guo, K.; Santosa, S.M.; Montana, M.; Yamakawa, M.; Hallak, J.A.; Han, K.Y.; Doh, S.J.; Rosenblatt, M.I.; Chang, J.H.; et al. Application of corneal injury models in dual fluorescent reporter transgenic mice to understand the roles of the cornea and limbus in angiogenic and lymphangiogenic privilege. Sci. Rep. 2019, 9, 12331. [Google Scholar] [CrossRef]
- Azar, D.T. Corneal angiogenic privilege: Angiogenic and antiangiogenic factors in corneal avascularity, vasculogenesis, and wound healing (an American Ophthalmological Society thesis). Trans. Am. Ophthalmol. Soc. 2006, 104, 264–302. [Google Scholar] [PubMed]
- Klyce, S.D. 12. Endothelial pump and barrier function. Exp. Eye Res. 2020, 198, 108068. [Google Scholar] [CrossRef] [PubMed]
- Ljubimov, A.V.; Saghizadeh, M. Progress in corneal wound healing. Prog. Retin. Eye Res. 2015, 49, 17–45. [Google Scholar] [CrossRef] [PubMed]
- Sage, E. Distribution and repair of photolesions in DNA: Genetic consequences and the role of sequence context. Photochem. Photobiol. 1993, 57, 163–174. [Google Scholar] [CrossRef] [PubMed]
- Rochette, P.J.; Bastien, N.; Todo, T.; Drouin, R. Pyrimidine (6-4) pyrimidone photoproduct mapping after sublethal UVC doses: Nucleotide resolution using terminal transferase-dependent PCR. Photochem. Photobiol. 2006, 82, 1370–1376. [Google Scholar] [CrossRef]
- Liu, Z.; Tan, C.; Guo, X.; Kao, Y.T.; Li, J.; Wang, L.; Sancar, A.; Zhong, D. Dynamics and mechanism of cyclobutane pyrimidine dimer repair by DNA photolyase. Proc. Natl. Acad. Sci. USA 2011, 108, 14831–14836. [Google Scholar] [CrossRef]
- Torizawa, T.; Ueda, T.; Kuramitsu, S.; Hitomi, K.; Todo, T.; Iwai, S.; Morikawa, K.; Shimada, I. Investigation of the cyclobutane pyrimidine dimer (CPD) photolyase DNA recognition mechanism by NMR analyses. J. Biol. Chem. 2004, 279, 32950–32956. [Google Scholar] [CrossRef]
- Hutchinson, F. Induction of tandem-base change mutations. Mutat. Res. 1994, 309, 11–15. [Google Scholar] [CrossRef]
- Yamada, D.; Dokainish, H.M.; Iwata, T.; Yamamoto, J.; Ishikawa, T.; Todo, T.; Iwai, S.; Getzoff, E.D.; Kitao, A.; Kandori, H. Functional Conversion of CPD and (6-4) Photolyases by Mutation. Biochemistry 2016, 55, 4173–4183. [Google Scholar] [CrossRef]
- Mees, A.; Klar, T.; Gnau, P.; Hennecke, U.; Eker, A.P.; Carell, T.; Essen, L.O. Crystal structure of a photolyase bound to a CPD-like DNA lesion after in situ repair. Science 2004, 306, 1789–1793. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, D.L.; Nairn, R.S. The biology of the (6-4) photoproduct. Photochem. Photobiol. 1989, 49, 805–819. [Google Scholar] [CrossRef] [PubMed]
- Boros, G.; Miko, E.; Muramatsu, H.; Weissman, D.; Emri, E.; van der Horst, G.T.; Szegedi, A.; Horkay, I.; Emri, G.; Kariko, K.; et al. Identification of Cyclobutane Pyrimidine Dimer-Responsive Genes Using UVB-Irradiated Human Keratinocytes Transfected with In Vitro-Synthesized Photolyase mRNA. PLoS ONE 2015, 10, e0131141. [Google Scholar] [CrossRef] [PubMed]
- Boros, G.; Kariko, K.; Muramatsu, H.; Miko, E.; Emri, E.; Hegedus, C.; Emri, G.; Remenyik, E. Transfection of Human Keratinocytes with Nucleoside-Modified mRNA Encoding CPD-Photolyase to Repair DNA Damage. Methods Mol. Biol. 2016, 1428, 219–228. [Google Scholar] [PubMed]
- Todo, T.; Tsuji, H.; Otoshi, E.; Hitomi, K.; Kim, S.T.; Ikenaga, M. Characterization of a human homolog of (6-4) photolyase. Mutat. Res. 1997, 384, 195–204. [Google Scholar] [CrossRef]
- Li, Y.F.; Kim, S.T.; Sancar, A. Evidence for lack of DNA photoreactivating enzyme in humans. Proc. Natl. Acad. Sci. USA 1993, 90, 4389–4393. [Google Scholar] [CrossRef] [PubMed]
- Van der Spek, P.J.; Kobayashi, K.; Bootsma, D.; Takao, M.; Eker, A.P.; Yasui, A. Cloning, tissue expression, and mapping of a human photolyase homolog with similarity to plant blue-light receptors. Genomics 1996, 37, 177–182. [Google Scholar] [CrossRef] [PubMed]
- Munoz, M.J.; Nieto Moreno, N.; Giono, L.E.; Cambindo Botto, A.E.; Dujardin, G.; Bastianello, G.; Lavore, S.; Torres-Mendez, A.; Menck, C.F.M.; Blencowe, B.J.; et al. Major Roles for Pyrimidine Dimers, Nucleotide Excision Repair, and ATR in the Alternative Splicing Response to UV Irradiation. Cell Rep. 2017, 18, 2868–2879. [Google Scholar] [CrossRef]
- Hsu, P.H.; Hanawalt, P.C.; Nouspikel, T. Nucleotide excision repair phenotype of human acute myeloid leukemia cell lines at various stages of differentiation. Mutat. Res. Mol. Mech. Mutagen. 2007, 614, 3–15. [Google Scholar] [CrossRef]
- Marteijn, J.A.; Lans, H.; Vermeulen, W.; Hoeijmakers, J.H. Understanding nucleotide excision repair and its roles in cancer and ageing. Nat. Rev. Mol. Cell Biol. 2014, 15, 465–481. [Google Scholar] [CrossRef]
- Cohen, V.M.L.; O’Day, R.F. Management Issues in Conjunctival Tumours: Ocular Surface Squamous Neoplasia. Ophthalmol. Ther. 2020, 9, 181–190. [Google Scholar] [CrossRef] [PubMed]
- Darwich, R.; Ghazawi, F.M.; Le, M.; Rahme, E.; Alghazawi, N.; Zubarev, A.; Moreau, L.; Sasseville, D.; Burnier, M.N., Jr.; Litvinov, I.V. Epidemiology of invasive ocular surface squamous neoplasia in Canada during 1992–2010. Br. J. Ophthalmol. 2020, 104, 1368–1372. [Google Scholar] [CrossRef]
- Yadav, S.; Gupta, N.; Singh, R.; Patil, M.; Meel, R.; Vanathi, M.; Kashyap, S.; Tandon, R. Role of Conjunctival Ultraviolet Autofluorescence in Ocular Surface Squamous Neoplasia. Ocul. Oncol. Pathol. 2020, 6, 422–429. [Google Scholar] [CrossRef] [PubMed]
- Vempuluru, V.S.; Pattnaik, M.; Ghose, N.; Kaliki, S. Bilateral ocular surface squamous neoplasia: A study of 25 patients and review of literature. Eur. J. Ophthalmol. 2021, 32, 1620–1627. [Google Scholar] [CrossRef] [PubMed]
- Walsh, J.E.; Bergmanson, J.P.; Wallace, D.; Saldana, G.; Dempsey, H.; McEvoy, H.; Collum, L.M. Quantification of the ultraviolet radiation (UVR) field in the human eye in vivo using novel instrumentation and the potential benefits of UVR blocking hydrogel contact lens. Br. J. Ophthalmol. 2001, 85, 1080–1085. [Google Scholar] [CrossRef] [PubMed]
- Kao, A.A.; Galor, A.; Karp, C.L.; Abdelaziz, A.; Feuer, W.J.; Dubovy, S.R. Clinicopathologic correlation of ocular surface squamous neoplasms at Bascom Palmer Eye Institute: 2001 to 2010. Ophthalmology 2012, 119, 1773–1776. [Google Scholar] [CrossRef] [PubMed]
- Cardenas-Cantu, E.; Zavala, J.; Valenzuela, J.; Valdez-Garcia, J.E. Molecular Basis of Pterygium Development. Semin. Ophthalmol. 2016, 31, 567–583. [Google Scholar] [CrossRef] [PubMed]
- Yasuda, S.; Sekiguchi, M. T4 endonuclease involved in repair of DNA. Proc. Natl. Acad. Sci. USA 1970, 67, 1839–1845. [Google Scholar] [CrossRef]
- Yarosh, D.B. Enhanced DNA repair of cyclobutane pyrimidine dimers changes the biological response to UV-B radiation. Mutat. Res. 2002, 509, 221–226. [Google Scholar] [CrossRef]
- Yarosh, D.; Bucana, C.; Cox, P.; Alas, L.; Kibitel, J.; Kripke, M. Localization of liposomes containing a DNA repair enzyme in murine skin. J. Investig. Dermatol. 1994, 103, 461–468. [Google Scholar] [CrossRef] [PubMed]
- Yarosh, D.; Klein, J.; O’Connor, A.; Hawk, J.; Rafal, E.; Wolf, P. Effect of topically applied T4 endonuclease V in liposomes on skin cancer in xeroderma pigmentosum: A randomised study. Xeroderma Pigmentosum Study Group. Lancet 2001, 357, 926–929. [Google Scholar] [CrossRef] [PubMed]
- Cafardi, J.A.; Elmets, C.A. T4 endonuclease V: Review and application to dermatology. Expert. Opin. Biol. Ther. 2008, 8, 829–838. [Google Scholar] [CrossRef] [PubMed]
- Zahid, S.; Brownell, I. Repairing DNA damage in xeroderma pigmentosum: T4N5 lotion and gene therapy. J. Drugs Dermatol. 2008, 7, 405–408. [Google Scholar]
- Piquero-Casals, J.; Morgado-Carrasco, D.; Gilaberte, Y.; Del Rio, R.; Macaya-Pascual, A.; Granger, C.; Lopez-Estebaranz, J.L. Management Pearls on the Treatment of Actinic Keratoses and Field Cancerization. Dermatol. Ther. 2020, 10, 903–915. [Google Scholar] [CrossRef]
- Gomes, C.; Silva, A.C.; Marques, A.C.; Lobo, J.S.; Amaral, M.H. Biotechnology Applied to Cosmetics and Aesthetic Medicines. Cosmetics 2020, 7, 33. [Google Scholar] [CrossRef]
- Moscarella, E.; Argenziano, G.; Longo, C.; Aladren, S. Management of cancerization field with a medical device containing photolyase: A randomized, double-blind, parallel-group pilot study. J. Eur. Acad. Dermatol. 2017, 31, E410–E413. [Google Scholar] [CrossRef]
- Puig, S.; Granger, C.; Garre, A.; Trullas, C.; Sanmartin, O.; Argenziano, G. Review of Clinical Evidence over 10 Years on Prevention and Treatment of a Film-Forming Medical Device Containing Photolyase in the Management of Field Cancerization in Actinic Keratosis. Dermatol. Ther. 2019, 9, 259–270. [Google Scholar] [CrossRef]
- Eibenschutz, L.; Silipo, V.; De Simone, P.; Buccini, P.L.; Ferrari, A.; Carbone, A.; Catricala, C. A 9-month, randomized, assessor-blinded, parallel-group study to evaluate clinical effects of film-forming medical devices containing photolyase and sun filters in the treatment of field cancerization compared with sunscreen in patients after successful photodynamic therapy for actinic keratosis. Br. J. Dermatol. 2016, 175, 1391–1393. [Google Scholar]
- Stege, H. Effect of xenogenic repair enzymes on photoimmunology and photocarcinogenesis. J. Photochem. Photobiol. B 2001, 65, 105–108. [Google Scholar] [CrossRef] [PubMed]
- Stege, H.; Roza, L.; Vink, A.A.; Grewe, M.; Ruzicka, T.; Grether-Beck, S.; Krutmann, J. Enzyme plus light therapy to repair DNA damage in ultraviolet-B-irradiated human skin. Proc. Natl. Acad. Sci. USA 2000, 97, 1790–1795. [Google Scholar] [CrossRef] [PubMed]
- Yarosh, D.; Alas, L.G.; Yee, V.; Oberyszyn, A.; Kibitel, J.T.; Mitchell, D.; Rosenstein, R.; Spinowitz, A.; Citron, M. Pyrimidine dimer removal enhanced by DNA repair liposomes reduces the incidence of UV skin cancer in mice. Cancer Res. 1992, 52, 4227–4231. [Google Scholar] [PubMed]
- Yarosh, D.; Klein, J.; Kibitel, J.; Alas, L.; O’Connor, A.; Cummings, B.; Grob, D.; Gerstein, D.; Gilchrest, B.A.; Ichihashi, M.; et al. Enzyme therapy of xeroderma pigmentosum: Safety and efficacy testing of T4N5 liposome lotion containing a prokaryotic DNA repair enzyme. Photodermatol. Photoimmunol. Photomed. 1996, 12, 122–130. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, K.; Hayakawa, H.; Sekiguchi, M.; Okada, Y. Specific action of T4 endonuclease V on damaged DNA in xeroderma pigmentosum cells in vivo. Proc. Natl. Acad. Sci. USA 1977, 74, 2958–2962. [Google Scholar] [CrossRef]
- Srinivasan, S.; Meyer, R.D.; Lugo, R.; Rahimi, N. Identification of PDCL3 as a Novel Chaperone Protein Involved in the Generation of Functional VEGF Receptor 2. J. Biol. Chem. 2013, 288, 23171–23181. [Google Scholar] [CrossRef]
- Derynck, R.; Budi, E.H. Specificity, versatility, and control of TGF-beta family signaling. Sci. Signal. 2019, 12, eaav5183. [Google Scholar] [CrossRef]
- Kamil, S.; Mohan, R.R. Corneal stromal wound healing: Major regulators and therapeutic targets. Ocul. Surf. 2021, 19, 290–306. [Google Scholar] [CrossRef]
- Rosenzweig, B.L.; Imamura, T.; Okadome, T.; Cox, G.N.; Yamashita, H.; Tendijke, P.; Heldin, C.H.; Miyazono, K. Cloning and Characterization of a Human Type-Ii Receptor for Bone Morphogenetic Proteins. Proc. Natl. Acad. Sci. USA 1995, 92, 7632–7636. [Google Scholar] [CrossRef]
- Mohan, R.R.; Kim, W.J.; Mohan, R.R.; Chen, L.; Wilson, S.E. Bone morphogenic proteins 2 and 4 and their receptors in the adult human cornea. Investig. Ophthalmol. Vis. Sci. 1998, 39, 2626–2636. [Google Scholar]
- Zhao, X.; Das, A.V.; Thoreson, W.B.; James, J.; Wattnem, T.E.; Rodriguez-Sierra, J.; Ahmad, I. Adult corneal limbal epithelium: A model for studying neural potential of non-neural stem cells/progenitors. Dev. Biol. 2002, 250, 317–331. [Google Scholar] [CrossRef]
- Sircoulomb, F.; Bekhouche, I.; Finetti, P.; Adelaide, J.; Ben Hamida, A.; Bonansea, J.; Raynaud, S.; Innocenti, C.; Charafe-Jauffret, E.; Tarpin, C.; et al. Genome profiling of ERBB2-amplified breast cancers. BMC Cancer 2010, 10, 539. [Google Scholar] [CrossRef] [PubMed]
- Xu, K.P.; Riggs, A.; Ding, Y.; Yu, F.S.X. Role of ErbB2 in corneal epithelial wound healing. Investig. Ophthalmol. Vis. Sci. 2004, 45, 4277–4283. [Google Scholar] [CrossRef] [PubMed]
- Bachu, M.; Dey, A.; Ozato, K. Chromatin Landscape of the IRF Genes and Role of the Epigenetic Reader BRD4. J. Interferon Cytokine Res. 2016, 36, 470–475. [Google Scholar] [CrossRef]
- Devaiah, B.N.; Singer, D.S. Two faces of brd4: Mitotic bookmark and transcriptional lynchpin. Transcription 2013, 4, 13–17. [Google Scholar] [CrossRef]
- Zuber, J.; Shi, J.W.; Wang, E.; Rappaport, A.R.; Herrmann, H.; Sison, E.A.; Magoon, D.; Qi, J.; Blatt, K.; Wunderlich, M.; et al. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature 2011, 478, 524–528. [Google Scholar] [CrossRef]
- Lee, J.E.; Park, Y.K.; Park, S.; Jang, Y.; Waring, N.; Dey, A.; Ozato, K.; Lai, B.; Peng, W.Q.; Ge, K. Brd4 binds to active enhancers to control cell identity gene induction in adipogenesis and myogenesis. Nat. Commun. 2017, 8, 2217. [Google Scholar] [CrossRef] [PubMed]
- Najafova, Z.; Tirado-Magallanes, R.; Subramaniam, M.; Hossan, T.; Schmidt, G.; Nagarajan, S.; Baumgart, S.J.; Mishra, V.K.; Bedi, U.; Hesse, E.; et al. BRD4 localization to lineage-specific enhancers is associated with a distinct transcription factor repertoire. Nucleic Acids Res. 2017, 45, 127–141. [Google Scholar] [CrossRef]
- Donati, B.; Lorenzini, E.; Ciarrocchi, A. BRD4 and Cancer: Going beyond transcriptional regulation. Mol. Cancer 2018, 17, 164. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Xue, Y.H.; Li, W.; Liu, Z.G. Transcription Factor Brd4, a Potential Marker of Corneal Epithelial Stem Cells. Investig. Ophthalmol. Vis. Sci. 2014, 55, 503. [Google Scholar]
- Qu, M.L.; Zhang, X.P.; Hu, X.L.; Dong, M.C.; Pan, X.J.; Bian, J.; Zhou, Q.J. BRD4 inhibitor JQ1 inhibits and reverses mechanical injury-induced corneal scarring. Cell Death Discov. 2018, 4, 64. [Google Scholar] [CrossRef]
- Zhang, Y.; Chen, X.Y.; Wang, H.M.; Gordon-Mitchell, S.; Sahu, S.; Bhagat, T.D.; Choudhary, G.; Aluri, S.; Pradhan, K.; Sahu, P.; et al. Innate immune mediator, Interleukin-1 receptor accessory protein (IL1RAP), is expressed and pro-tumorigenic in pancreatic cancer. J. Hematol. Oncol. 2022, 15, 70. [Google Scholar] [CrossRef]
- Daull, P.; Barabino, S.; Feraille, L.; Kessal, K.; Docquier, M.; Parsadaniantz, S.M.; Baudouin, C.; Garrigue, J.S. Modulation of Inflammation-Related Genes in the Cornea of a Mouse Model of Dry Eye upon Treatment with Cyclosporine Eye Drops. Curr. Eye Res. 2019, 44, 476–485. [Google Scholar] [CrossRef] [PubMed]
- Dana, M.R.; Zhu, S.N.; Yamada, J. Topical modulation of interleukin-1 activity in corneal neovascularization. Cornea 1998, 17, 403–409. [Google Scholar] [CrossRef]
- Lu, P.R.; Li, L.B.A.; Liu, G.Q.; Zhang, X.G.; Mukaida, N. Enhanced Experimental Corneal Neovascularization along with Aberrant Angiogenic Factor Expression in the Absence of IL-1 Receptor Antagonist. Investig. Ophthalmol. Vis. Sci. 2009, 50, 4761–4768. [Google Scholar] [CrossRef] [PubMed]
- Yamada, J.; Dana, M.R.; Sotozono, C.; Kinoshita, S. Local suppression of IL-1 by receptor antagonist in the rat model of corneal alkali injury. Exp. Eye Res. 2003, 76, 161–167. [Google Scholar] [CrossRef]
- Yoon, H.; Choi, S.I.; Kim, E.K. Uptake of cell debris and enhanced expression of inflammatory factors in response to dead cells in corneal fibroblast cells. Exp. Eye Res. 2020, 194, 108017. [Google Scholar] [CrossRef]
- Abd Ghafar, N.; Jalil, N.A.A.; Kamarudin, T.A. Wound healing of the corneal epithelium: A review. Asian Biomed. 2021, 15, 199–212. [Google Scholar] [CrossRef]
- Di Zazzo, A.; Lee, S.M.; Sung, J.; Niutta, M.; Coassin, M.; Mashaghi, A.; Inomata, T. Variable Responses to Corneal Grafts: Insights from Immunology and Systems Biology. J. Clin. Med. 2020, 9, 586. [Google Scholar] [CrossRef]
- Dinarello, C.A. Biologic basis for interleukin-1 in disease. Blood 1996, 87, 2095–2147. [Google Scholar] [CrossRef] [PubMed]
- Clements, J.L.; Dana, R. Inflammatory Corneal Neovascularization: Etiopathogenesis. Semin. Ophthalmol. 2011, 26, 235–245. [Google Scholar] [CrossRef]
- Korhonen, E.; Bisevac, J.; Hyttinen, J.M.T.; Piippo, N.; Hytti, M.; Kaarniranta, K.; Petrovski, G.; Kauppinen, A. UV-B-Induced Inflammasome Activation Can Be Prevented by Cis-Urocanic Acid in Human Corneal Epithelial Cells. Investig. Ophthalmol. Vis. Sci. 2020, 61, 7. [Google Scholar] [CrossRef] [PubMed]
- Korhonen, E.; Piippo, N.; Hytti, M.; Hyttinen, J.M.T.; Kaarniranta, K.; Kauppinen, A. Only IL-1 beta release is inflammasome-dependent upon ultraviolet B irradiation although IL-18 is also secreted. Faseb J. 2020, 34, 6437–6448. [Google Scholar] [CrossRef] [PubMed]
- Park, S.G.; Kang, Y.S.; Ahn, Y.H.; Lee, S.H.; Kim, K.R.; Kim, K.W.; Koh, G.Y.; Ko, Y.G.; Kim, S. Dose-dependent biphasic activity of tRNA synthetase-associating factor, p43, in angiogenesis. J. Biol. Chem. 2002, 277, 45243–45248. [Google Scholar] [CrossRef]
- Park, S.G.; Shin, H.; Shin, Y.K.; Lee, Y.; Choi, E.C.; Park, B.J.; Kim, S. The novel cytokine p43 stimulates dermal fibroblast proliferation and wound repair. Am. J. Pathol. 2005, 166, 387–398. [Google Scholar] [CrossRef] [PubMed]
- Proteomic Data. Available online: https://www.ebi.ac.uk/pride (accessed on 11 January 2023).


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Volatier, T.; Schumacher, B.; Meshko, B.; Hadrian, K.; Cursiefen, C.; Notara, M. Short-Term UVB Irradiation Leads to Persistent DNA Damage in Limbal Epithelial Stem Cells, Partially Reversed by DNA Repairing Enzymes. Biology 2023, 12, 265. https://doi.org/10.3390/biology12020265
Volatier T, Schumacher B, Meshko B, Hadrian K, Cursiefen C, Notara M. Short-Term UVB Irradiation Leads to Persistent DNA Damage in Limbal Epithelial Stem Cells, Partially Reversed by DNA Repairing Enzymes. Biology. 2023; 12(2):265. https://doi.org/10.3390/biology12020265
Chicago/Turabian StyleVolatier, Thomas, Björn Schumacher, Berbang Meshko, Karina Hadrian, Claus Cursiefen, and Maria Notara. 2023. "Short-Term UVB Irradiation Leads to Persistent DNA Damage in Limbal Epithelial Stem Cells, Partially Reversed by DNA Repairing Enzymes" Biology 12, no. 2: 265. https://doi.org/10.3390/biology12020265
APA StyleVolatier, T., Schumacher, B., Meshko, B., Hadrian, K., Cursiefen, C., & Notara, M. (2023). Short-Term UVB Irradiation Leads to Persistent DNA Damage in Limbal Epithelial Stem Cells, Partially Reversed by DNA Repairing Enzymes. Biology, 12(2), 265. https://doi.org/10.3390/biology12020265






